Molecular determinants of the Ska-Ndc80 interaction and their influence on microtubule tracking and force-coupling
Figures
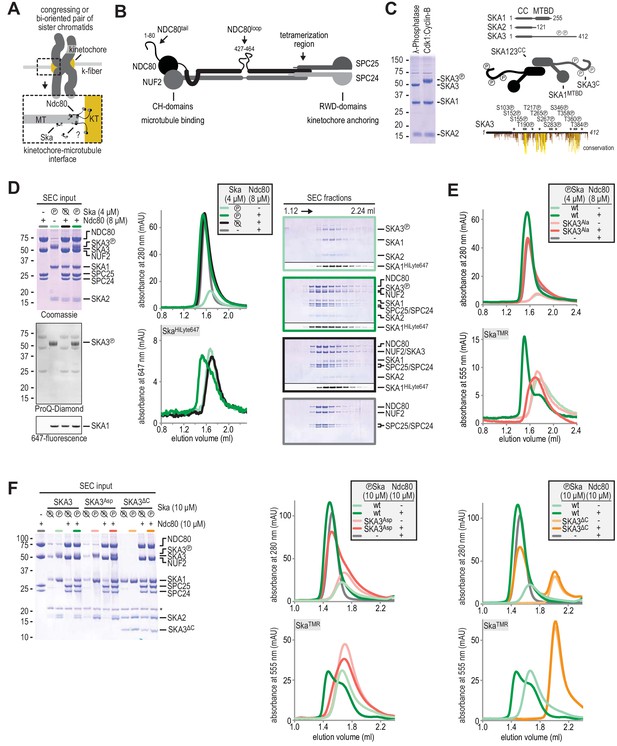
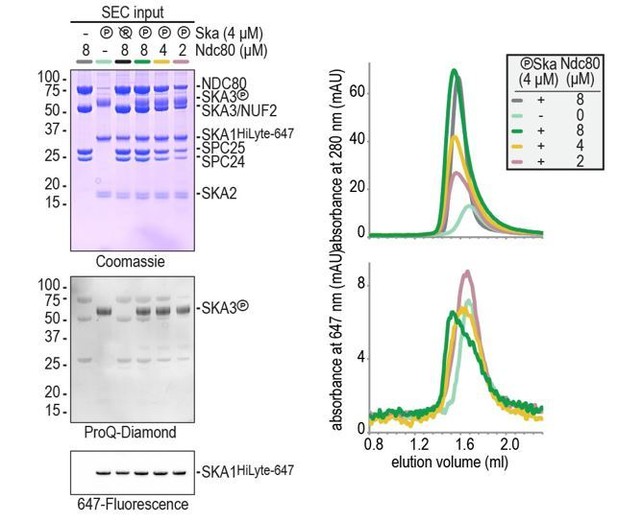
Formation of a Ska:Ndc80 complex upon SKA3 phosphorylation by CDK1.
(A) Schematic representation of Ndc80 and Ska at the kinetochore-microtubule interface. (B) Overview of important regions in the Ndc80 complex. (C) The Ska complex. SKA1, SKA2, and SKA3 contain an N-terminal coiled coil (CC) region that mediates complex formation and dimerization. SKA1 contains a microtubule binding domain (MTBD). The largely unstructured C-terminal region of SKA3 is phosphorylated during mitosis. Multisite in vitro phosphorylation of purified Ska by CDK1:Cyclin-B altered the migration of SKA3 on SDS-PAGE. Identified phosphorylation sites and the conservation of SKA3 are shown. (D) Analysis of a Ska:Ndc80 mixture by size-exclusion chromatography (SEC) using a superose 6 increase 5/150 column shows that a stable complex is formed between Ska that is phosphorylated by CDK1:Cyclin-B and Ndc80. Elution of Ska from the column can be followed specifically through the fluorescently labelled SKA1. In-gel fluorescence of SKA1 in the SEC fractions analyzed by SDS-PAGE is also shown. (E) Phosphorylated Ska with SKA3T358A/T360A does not bind to Ndc80. Analysis of fractions is shown in Figure 1—figure supplement 3. (F) Ska without SKA3104-412 as well as SKA3T358D/T360D does not interaction with Ndc80. A comparison with phosphatase-treated Ska on the input gel indicates the effective phosphorylation of the mutated SKA3. These chromatograms originate from one experiment and wild-type Ska (green) and Ndc80 (gray) are shown in both panels for comparison.
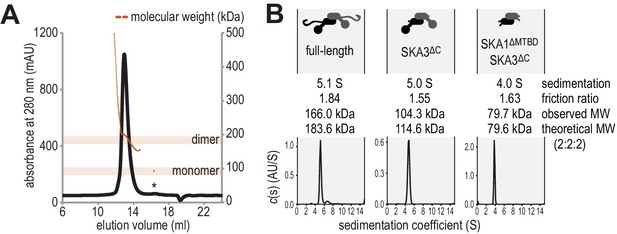
Hydrodynamic analysis of the SKA complex.
(A) SEC-MALS analysis reveals that Ska is predominantly a dimer of a heterotrimer at a concentration of 1 mg/ml. Small amounts of monomeric Ska were determined as monomeric (eluting ~16.5 ml) or multimeric (eltuting before the dimeric Ska). (B) Sedimentation velocity AUC of Ska highlighted sample purity and the dimeric and elongated nature of the complex. Truncation of the C-terminal region of SKA3 and the MTBD of SKA1 show that the N-terminal coiled-coils of SKA1, SKA2, and SKA3 are sufficient for dimerization.
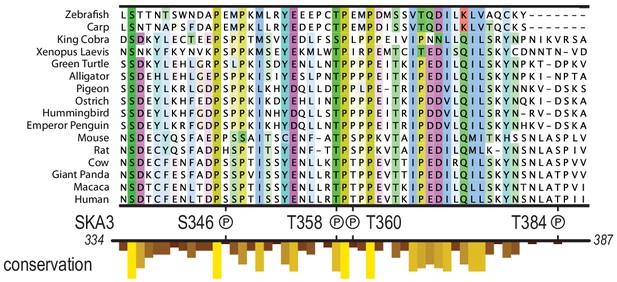
Multiple species alignment of the region in SKA3 that has been implicated in the binding to Ndc80.
The P symbols mark residues that were phosphorylated by CDK1:Cyclin-B in vitro. The conservation analysis is part of the track shown in Figure 1C.
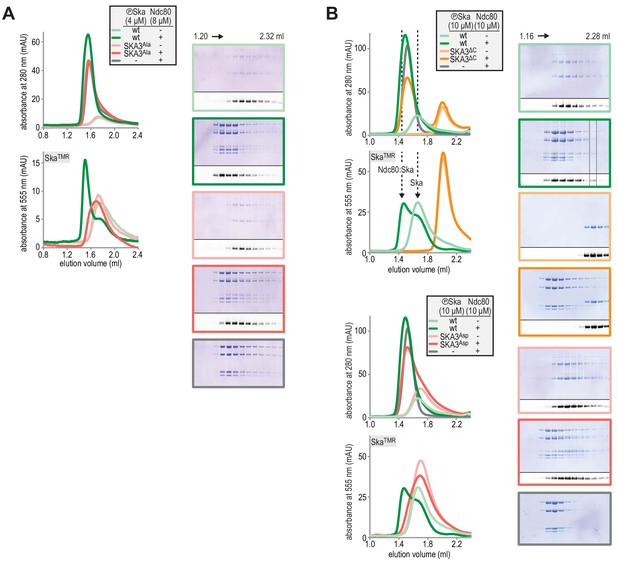
Fractions of SEC experiments shown in Figure 1E and Figure 1F were analysed by Coomassie staining and in-gel fluorescence following SDS-PAGE.
The chromatograms are shown again for clarity.
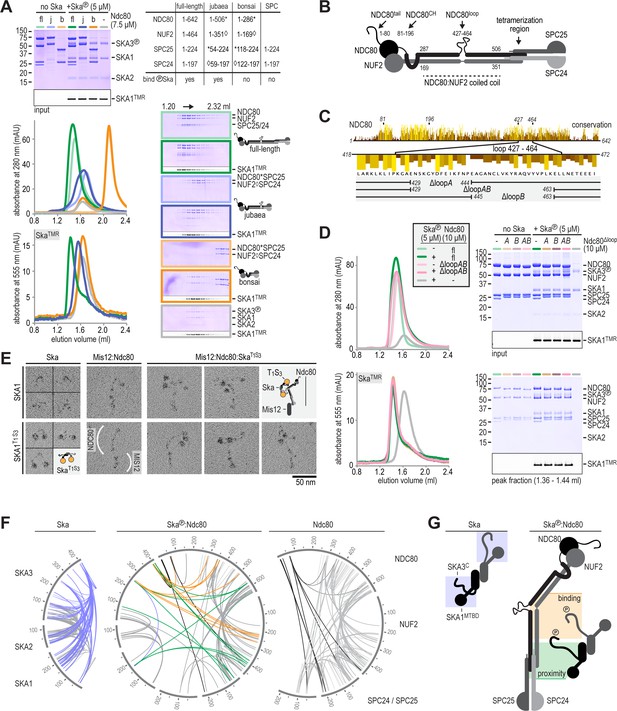
Ska binds the NDC80:NUF2 coiled-coil and the Ndc80-loop is dispensable.
(A) Full-length (fl), jubaea (j), and bonsai (b) Ndc80 complexes were tested for their ability to bind phosphorylated Ska. Symbols (* and ◊) indicate fusion proteins in jubaea and bonsai Ndc80. (B) Overview of Ndc80 as in Figure 1B. NDC80287-504 and NUF2169-351, the Ska binding region that is present in Ndc80Csequoia and absent in Ndc80Cbonsai, is indicated. (C) Overview of the three tested constructs that lack different parts of the NDC80-loop. The conservation of Ndc80 and its loop-region are shown. (D) Ndc80 lacking the Ndc80-loop still binds phosphorylated Ska. (E). Ska and Mis12:Ndc80:Ska were visualized by electron microscopy after glycerol-spraying and low-angle metal shadowing. SKA1MTBD-biotin (30 kDa) was conjugated with the biotin-binding globular T1S3 (88 kDa) to facilitate the recognition of Ska in micrographs. The presence of Mis12 (20 nm) marks the SPC24:SPC25 side of Ndc80 (62 nm). See Figure 2—figure supplement 2 for detailed sample preparation information. (F) Intra- and intermolecular crosslinks for Ska, Ska:Ndc80, and Ndc80. Contacts between SKA3C and the rest of Ska are highlighted in blue. Contacts between SKA3C and the NDC80:NUF2 coiled coil and the Ndc80 tetramerization domainare shown in orange and green, respectively. The SKA1MTBD is also proximal to the tetramerization domain. (G) A schematic representation of proximities.
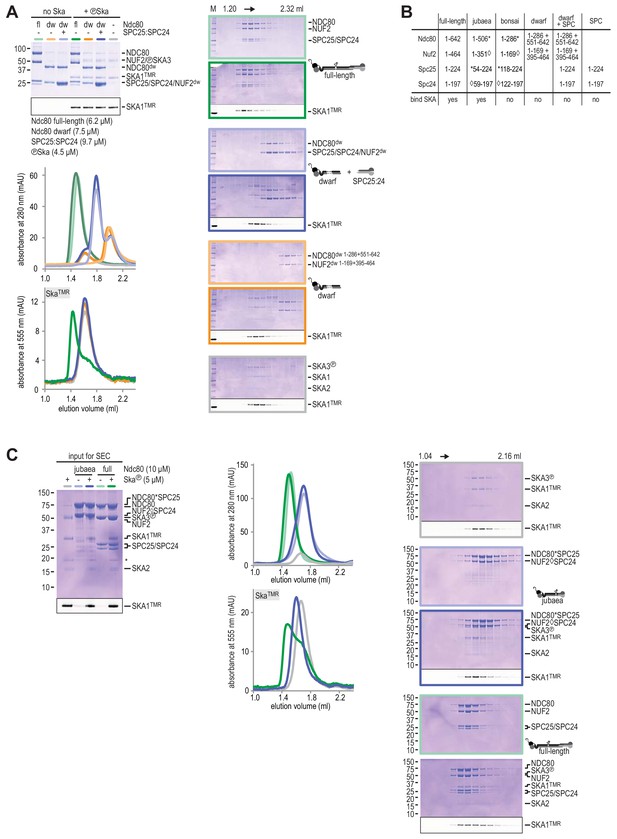
Testing Ska binding interactions with various Ndc80 constructs.
(A) Ndc80, Ndc80-dwarf, and Ndc80-dwarf in the presence of an excess of SPC24:SPC25 were tested for their ability to bind phosphorylated Ska. Ndc80-dwarf binds SPC24:SPC25 stoichiometrically. Phosphorylated Ska only binds to full-length Ndc80. (B) An overview of the various Ndc80 constructs that were used. (C) During the course of our studies, we realized that the Ndc80jubaea construct used for the experiments in Figure 2A contained a V15M mutation in Nuf2. After correcting the mutation in the Ndc80jubaea construct, we repeated the Ska-binding assays, obtaining essentially identical results. Thus, the presence of the V15M mutation does not modify our conclusions on the ability of Ndc80jubaea to bind Ska.
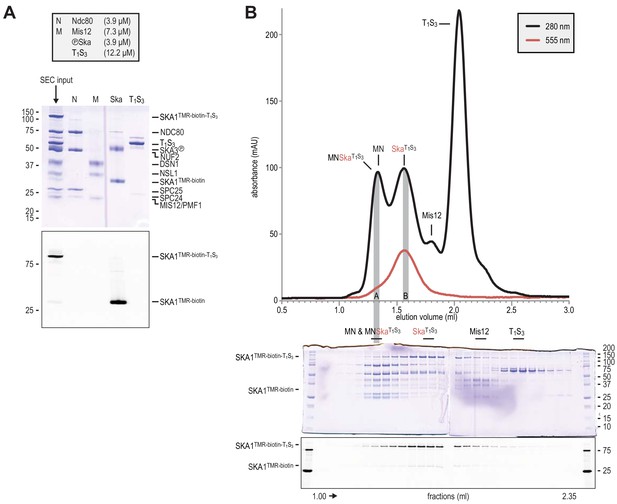
Preparation of Traptavidin-Streptavidin-labelled Ska for rotary shadowing electron microscopy.
(A) The input for the preparative size-exclusion was analysed by SDS-PAGE followed by Coomassie staining and in-gel fluorescence of SKA1. The samples were not boiled prior to electrophoresis, and T1S3 remained tetrameric. The shift of SKA1TMR-biotin-T1S3 compared to SKA1TMR-biotin indicates successful and near complete labeling of SKA1 with T1S3. Ndc80 (N), Mis12 (M), Ska, and T1S3 are shown for comparison. (B) Size-exclusion chromatography was used to separate the input sample shown in panel A. Fractions were analyzed as indicated and labeled. Fractions indicated with A and B were analysed by electron microscopy (see Figure 2E).
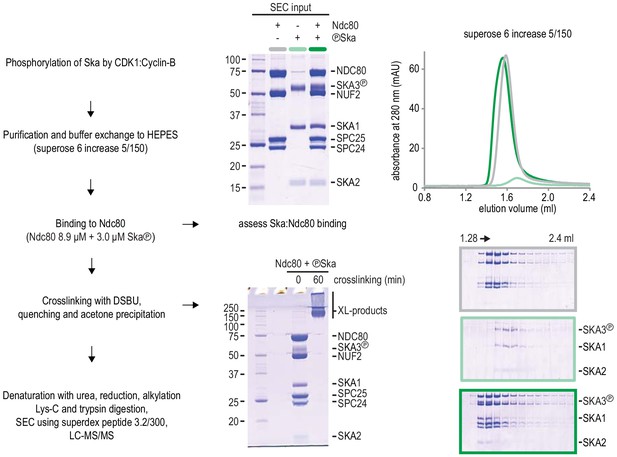
Protocol of Ska:Ndc80 complex preparation for chemical cross-linking.
Complex formation was assessed with a fraction of the material prior to the DSBU treatment. The near complete shift of Ska (light green) into a Ska:Ndc80 complex (dark green) is important to interpret the proximity map resulting from the cross-linking mass spectrometry analysis.
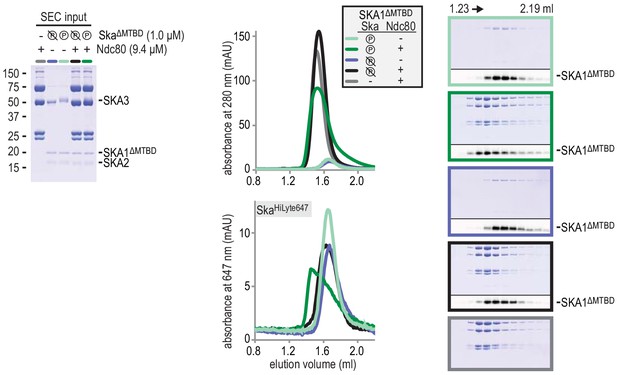
Deletion of the MTBD of SKA1 does not interfere with the phosphorylayion-dependent binding of Ska to Ndc80.
All Skasubunit are visible in the input gel, but only the larger SKA3 is visible by Coomassie staining in the analysed fractions after size-exclusion chromatography. SKA1 was labeled with HiLyte-647 and could be followed specifically during size-exclusion chromatography and through in-gel fluorescence.
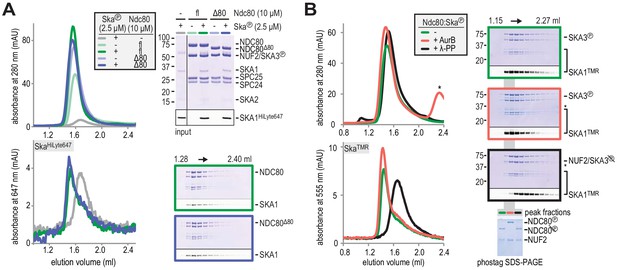
Aurora B kinase activity does not disrupt Ska:Ndc80 binding.
(A) Full-length (fl) and tailless (Δ80) Ndc80 both bind phosphorylated Ska, as analyzed by SEC and SDS-PAGE. (B) A Ska:Ndc80 complex was exposed to Aurora B kinase or lambda-phosphatase. Low amounts of Ska:Ndc80 impair the detection of Ska by Coomassie but fluorescent SKA1 was detected. Dephosphorylation disrupted the Ska:Ndc80 complex but Aurora B phosphorylation did not. Kinase activity is indicated by the shift in elution volume of Aurora B treated Ndc80:Ska and by the altered migration of phosphorylated NDC80 after gel filtration on the phostag SDS-PAGE. The asterisk marks Aurora B kinase.
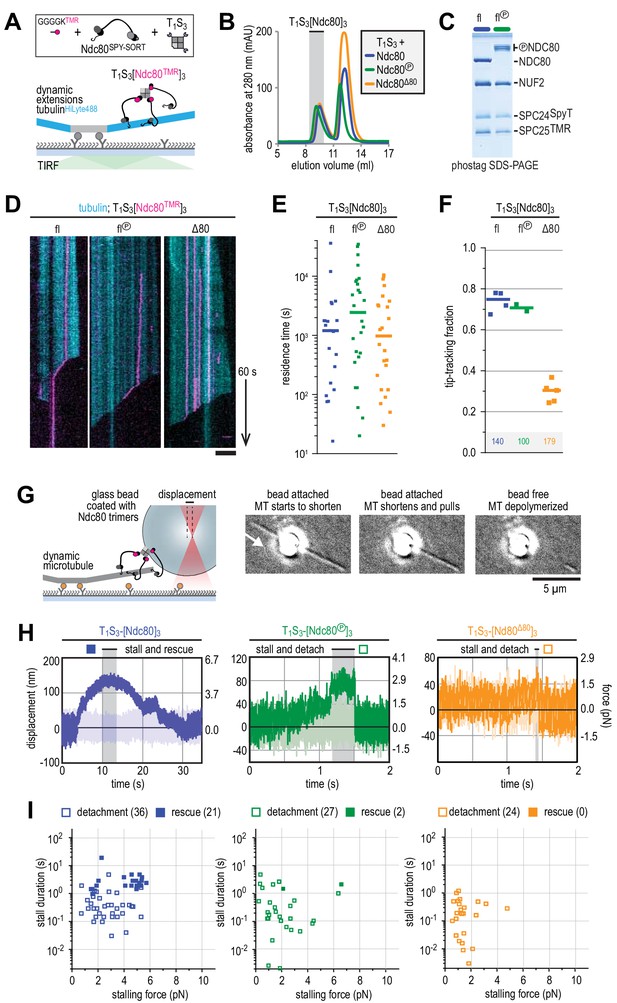
Ndc80-tail is required for interaction with a microtubule end.
(A) Experimental setup featuring trimeric T1S3[Ndc80]3 modules visualized on dynamic microtubules using TIRF microscopy. (B) Size-exclusion chromatography traces showing separation of monomeric Ndc80 from trivalent T1S3-Ndc80 modules using full-length Ndc80 (fl, blue), Aurora B phosphorylated full-length Ndc80 (fl-phosphorylated, green), and Δ80-Ndc80 (orange). (C) Phostag gel demonstrating successful phosphorylation of the NDC80 subunit by Aurora B. (D) Kymographs showing Ndc80 trimers (magenta) on dynamic microtubules (cyan). Scale bars: vertical (60 s), horizontal (5 μm). (E) Residence times of Ndc80 trimers on taxol-stabilized microtubules. Horizontal bars: median. T-tests indicate the following two-tailed p-values: T1S3[Ndc80]3 vs T1S3[Ndc80℗]3: p=0.26; T1S3[Ndc80]3 vs T1S3[Ndc80Δ80]3: p=0.07; T1S3[Ndc80℗]3 vs T1S3[Ndc80Δ80]3: p=0.63. (F) Fraction of Ndc80 trimers that initiate movement in the direction of microtubule shortening upon encounter with a depolymerizing end. Squares: fractions in an individual experiment, horizontal bars: median. T-tests indicate the following two-tailed p-values: T1S3[Ndc80]3 vs T1S3[Ndc80℗]3: p=0.48; T1S3[Ndc80]3 vs T1S3[Ndc80Δ80]3: p<10−5; T1S3[Ndc80℗]3 vs T1S3[Ndc80Δ80]3: p<10−3. (G) Left: setup of an optical trap experiment. Right: DIC images showing a microtubule depolymerizing past an optically trapped bead. Scale bar: 5 μm. (H) Representative traces of a microtubule pulling on a bead coated with Ndc80 trimers (blue), Aurora B phosphorylated wild type Ndc80 trimers (green) or trimers containing Ndc80Δ80 (orange). (I) Correlation between the stalling force and the duration of the stall for each individual stall event resulting in a detachment (open symbols) or rescue (filled symbols) for Ndc80 trimers (blue), Aurora B phosphorylated Ndc80 trimers (green) or Ndc80 trimers containing NDC80Δ80 (orange). Two-sided Fisher exact testing indicates different detachment-rescue distributions between untreated Ndc80 (36-21) and Aurora B phosphorylated (27–2, p=0.004) or Ndc80Δ80(24–0, p=0.0002). The distribution does not differ significantly between Aurora B phosphorylated and Ndc80Δ80 (p=0.49).
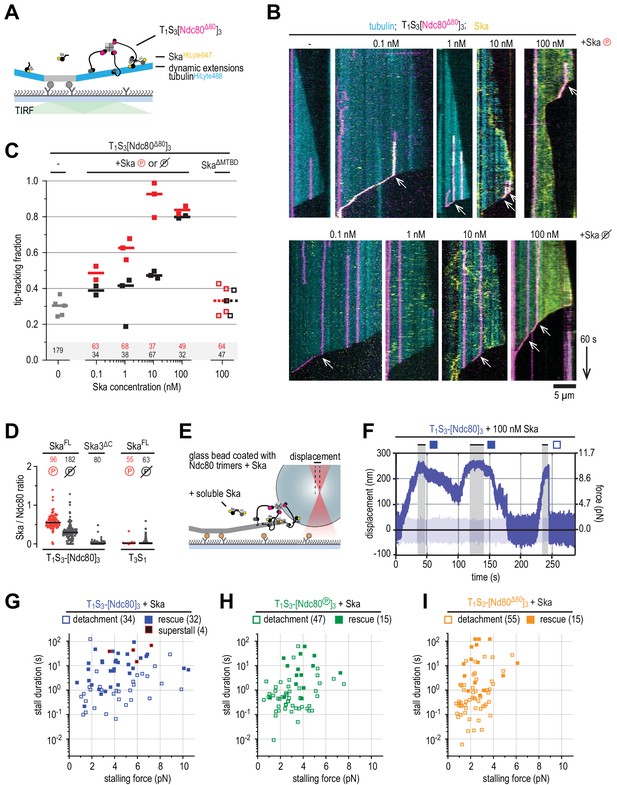
Ska bound to Ndc80 presents an additional microtubule end-binding site that stabilizes the stalled microtubule ends.
(A) Ska and trimeric Ndc80Δ80 modules were imaged simultaneously on dynamic microtubules using TIRF microscopy. (B) Trimeric Ndc80Δ80 modules (magenta) on dynamic microtubules (cyan) in the presence of increasing concentrations of phosphorylated (top row) or dephophosphorylated (bottom row) Ska (yellow). Scale bars: vertical (60 s), horizontal (5 μm). Arrows indicate successful end-tracking events. (C) Fraction of trimeric Ndc80Δ80 modules that initiate movement in the direction of microtubule shortening upon encounter with a depolymerizing end in the presence of Ska. Squares: fractions in an individual experiment, horizontal bars: median. (D) Ratio of Ska to Ndc80 after incubation of the beads coated with Ndc80 trimers and then Ska (400 nM; see also Figure 5—figure supplement 1). Horizontal lines: median. A t-test indicates significant different between the addition of phosphorylated or dephosphorylated Ska: T1S3[Ndc80]3 + Ska vs Ska℗: p<10−17. Other two-tailed p-values are T1S3[Ndc80]3 + Ska vs Ska3ΔC: p<10−22; T1S3[Ndc80]3 + Ska℗ vs Ska3ΔC: p<10−34; T3S1 + Ska vs Ska℗: p=0.012; T3S1 + Ska vs Ska3ΔC: p=0.99; T3S1 + Ska℗ vs Ska3ΔC: p=0.012; Ska℗ + T1S3[Ndc80]3 vs T3S1: p<10−28; Ska + T1S3[Ndc80]3 vs T3S1: p<10−30. (E) Optically trapped bead coated with Ndc80 trimers and Ska in a chamber with dynamic microtubules and additional soluble Ska. (F) An example force trace obtained in the presence of dephosphorylated Ska and a bead coated with non-phosphorylated Ndc80 trimers. (G–I) Correlation between the stalling force and the duration of the stall for each individual stall event in the presence of 10–100 nM Ska resulting in a detachment (open symbols), rescue (filled symbols) or superstall (black symbols) for the beads coated with non-phosphorylated Ndc80 trimers (G, blue symbols), Aurora B phosphorylated Ndc80 trimers (H), green symbols) or trimers containing Ndc80Δ80 (i), orange symbols). Two-sided Fisher exact testing indicates different detachment-rescue distributions between untreated Ndc80 (34-32) and Aurora B phosphorylated (47–15, p=0.006) or Ndc80Δ80(55–15, p=0.001). The addition of Ska (in comparison to Figure 4I) did change the detachment-rescue distribution for Ndc80Δ80 (p=0.010), but not significantly for untreated and Aurora B phosphorylated Ndc80 (p=0.207 and 0.081, respectively). See Figure 5—figure supplements 2 and 3 for data separated per Ska concentration and phosphorylation state.
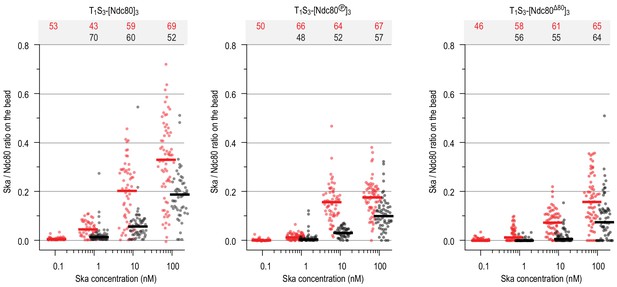
Ratio of Ska copy number to Ndc80 copy number after incubation of beads coated with untreated, Aurora-B-phosphorylated or tail-less Ndc80 trimers, and then Ska in indicated concentration.
Horizontal lines: median.
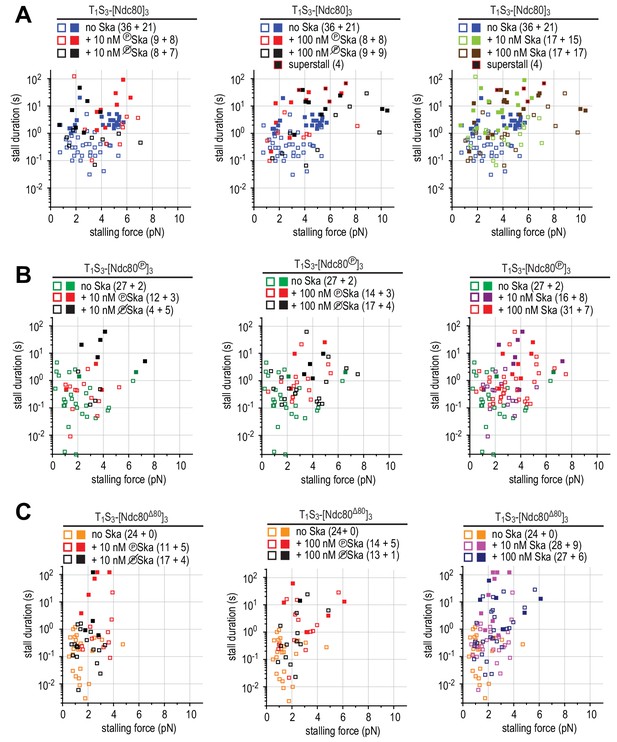
Correlations between the stalling force and the duration of the stall for each individual stall event in the presence of 10 or 100 nM Ska resulting in a detachment (open symbols), rescue (filled symbols) or superstall (black symbols) for the beads coated with non-phosphorylated Ndc80 trimers (A), Aurora B-phosphorylated Ndc80 trimers (B), or tail-less Ndc80 trimers (C).
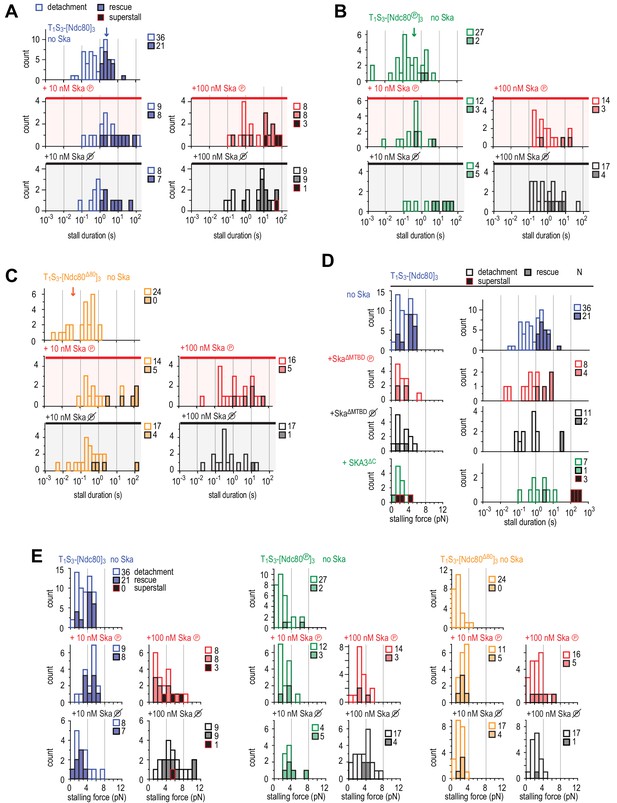
Distributions of durations and forces of stalls with Ndc80 and plus or minus Ska.
(A–C) Distributions of the duration of Ndc80-mediated microtubule stall in the absence or presence of 10 or 100 nM Ska (either dephosphorylated or phosphorylated) performed with beads coated with untreated (A), Aurora-B-phosphorylated (B) or tail-less (C) Ndc80 trimers. The force traces that are shown in Figure 4H come from events indicated with an arrow. Light bars indicate detachment events, filled bars rescue events, and black bars indicate ‘superstalls’. (D) Distributions of stalling forces (left) and stall durations (right) obtained with beads coated with untreated Ndc80 in the absence of Ska (blue), in the presence of phosphorylated (red) or dephosphorylated (black) Ska lacking SKA1MTBD, or in the presence of Ska lacking SKA3C (green). (E) Distributions of stalling forces in the absence or presence of 10 or 100 nM Ska (either dephosphorylated or phosphorylated) performed with beads coated with untreated (blue), Aurora-B-phosphorylated (green) or tail-less (orange) Ndc80 trimers.

Molecular determinants of the Ska-Ndc80 interaction and their influence on microtubule tracking and force-coupling.
(A) Schematic representation of force-coupling before, during, and after a force-induced stall of microtubule depolymerization that was followed by microtubule re-growth. (B) A density plot of stall durations and forces resulting in detachment or rescue in the absence of Ska (left) or in the presence of 10–100 nM Ska (right). Data are pooled for all three types of Ndc80 trimers. (C) The fraction of rescues was plotted against stalling force or stall duration after binning of data from the different Ndc80 complexes in the presence or absence of 10–100 nM Ska (as shown in Figures 4 and 5). (D) The fraction of rescues was plotted against stall duration after pooling and binning of all data. Detachment is more likely in the absence of Ska and when the Ndc80-tail is phosphorylated. An unphosphorylated Ndc80-tail and the presence of Ska increase the attachment survival rate.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | pBIG1 Ska | This study | Following the biGBac system (Weissmann et al., 2016): SKA1SORT-HIS (CasI), SKA2 (CasII), SKA3 (CasII) | |
| Recombinant DNA reagent | pBIG1 SkaΔMTBD | This study | SKA11-108 SORT-HIS (CasI), SKA2 (CasII), SKA3 (CasII) | |
| Recombinant DNA reagent | pBIG1 Ska3ΔC | This study | SKA1SORT-HIS (CasI), SKA2 (CasII), SKA31-101 (CasII) | |
| Recombinant DNA reagent | pBIG1 Ska3ΔCΔMTBD | This study | SKA11-108 SORT-HIS (CasI), SKA2 (CasII), SKA31-101 (CasII) | |
| Recombinant DNA reagent | pBIG1 Ska3ΔC | This study | SKA1SORT-HIS (CasI), SKA2 (CasII), SKA31-101 (CasII) | |
| Recombinant DNA reagent | pBIG1 SkaT358AT360A | This study | SKA1SORT-HIS (CasI), SKA2 (CasII), SKA3T358AT360A (CasII) | |
| Recombinant DNA reagent | pBIG1 SkaT358DT360D | This study | SKA1SORT-HIS (CasI), SKA2 (CasII), SKA3T358DT360D (CasII) | |
| Recombinant DNA reagent | pBIG1 Ndc80 | Musacchio laboratory, Huis in 't Veld et al., 2016 | NDC80 (CasI), NUF2 (CasII), SPC25HIS (CasIII), SPC24 (CasIV) | |
| Recombinant DNA reagent | pBIG1 Ndc80 | Musacchio laboratory, Volkov et al., 2018 | NDC80 (CasI), NUF2 (CasII), SPC25SORT-HIS(CasIII), SPC24SPY (CasIV) | |
| Recombinant DNA reagent | pBIG1 Ndc80 | Musacchio laboratory, Volkov et al., 2018 | NDC80 (CasI), NUF2 (CasII), SPC25SORT-HIS(CasIII), SPC24SPY (CasIV) | |
| Recombinant DNA reagent | pBIG1 Ndc80Δ80 | this study | NDC80Δ1-80 (CasI), NUF2 (CasII), SPC25SORT-HIS (CasIII), SPC24SPY (CasIV) | |
| Recombinant DNA reagent | pBIG1 Ndc80ΔloopA | this study | NDC80Δ429-444 (CasI), NUF2 (CasII), SPC25SORT-HIS (CasIII), SPC24SPY (CasIV) | |
| Recombinant DNA reagent | pBIG1 Ndc80ΔloopB | this study | NDC80Δ445-463 (CasI), NUF2 (CasII), SPC25SORT-HIS (CasIII), SPC24SPY (CasIV) | |
| Recombinant DNA reagent | pBIG1 Ndc80ΔloopAB | this study | NDC80Δ429-463 (CasI), NUF2 (CasII), SPC25SORT-HIS (CasIII), SPC24SPY (CasIV) | |
| Recombinant DNA reagent | pGEX 2-rbs Ndc80bonsai | Musacchio laboratory, Ciferri et al., 2008 | GST-PreScissionNUF21-169:SPC24122-197(CasI), NDC801-286:SPC25118-224 (CasII) | |
| Recombinant DNA reagent | pGEX 2-rbs Ndc80jubaea | this study | GST-PreScissionNUF21-351:SPC2459-197(CasI), NDC801-506:SPC2554-224 (CasII) | |
| Recombinant DNA reagent | pGEX 2-rbs Ndc80dwarf | this study | GST-PreScissionNUF21-169 & 395-464 (CasI), NDC801-286 & 551-642 (CasII) | |
| Recombinant DNA reagent | pGEX 2-rbs SPC25:SPC24 | Musacchio laboratory, Ciferri et al., 2005 | GST-PreScissionSPC25 (CasI), SPC24 (CasII) | |
| Recombinant DNA reagent | pET30b-7M-SrtA | Hidde Ploegh laboratory | addgene 51141 | Sortase (Ca2+ independent) |
| Recombinant DNA reagent | pET21a-Traptavidin | Mark Howarth laboratory | addgene 26054 | Core Traptavidin (T) |
| Recombinant DNA reagent | pET21a-DCatch | Mark Howarth laboratory | addgene 59547 | Biotin-binding dead streptavidin-SpyCatcher (S) |
| Peptide, recombinant protein | Cdk1:Cyclin-B kinase | Musacchio laboratory, Huis in 't Veld et al., 2016 | ||
| Peptide, recombinant protein | Aurora B kinase | Musacchio laboratory, Girdler et al., 2008 | Aurora B45-344:INCENP835-903 | |
| Peptide, recombinant protein | Lambda-phosphatase | Generated in-house | ||
| Peptide, recombinant protein | GGGG[Lys-HiLyte647] | Life Technologies | ||
| Peptide, recombinant protein | GGGG[Lys-TMR] | GenScript | ||
| Peptide, recombinant protein | GGGG[Lys-PEG2-Biotin][Lys-TMR] | GenScript |
| no Ska | + 100 nM Ska | |
|---|---|---|
| T1S3-[Ndc80]3 | 0.35 (p = 0.009) | 0.42 (p = 0.014) |
| T1S3-[Ndc80-P]3 | -0.23 (p = 0.23) | 0.27 (p = 0.03) |
| T1S3-[Ndc80∆80]3 | -0.16 (p = 0.45) | 0.47 (p = 0.003) |
Additional files
-
Supplementary file 1
(a,b) In vitro phosphorylation of the Ska complex by CDK1:Cyclin-B (1a, concise report; 1b, full report). Phosphopeptides were detected in fractions after size-exclusion chromatography for dephosphorylated Ska + Ndc80 (Figure 1D; black; 1.60–1.76 ml; SDS-PAGE lanes 7 and 8), phosphorylated Ska (Figure 1D; light green; 1.60–1.76 ml; SDS-PAGE lanes 7 and 8), and phosphorylated Ska + Ndc80 (Figure 1D; dark green; 1.44–1.60 ml; SDS-PAGE lanes 5 and 6). Sites with a localization probability >0.5 and an Andromeda search engine score >100 are shown. Additional information is available in a separate table. (c,d) Identified crosslinks for Ska, Ndc80, and Ska:Ndc80 (1 c, concise report; 1d, full report) The unique crosslinks obtained for Ska (90 inter; 97 intra), Ndc80:Ska (86 inter; 71 intra), and Ndc80 (as in Pan et al., 2018) inter; 72 intra;) are reported. (e,f) In vitro phosphorylation of Ndc80 by Aurora B (1e, concise report; 1 f, full report) Ndc80 was Aurora B treated (or untreated) and assembled into T1S3-[Ndc80]3 modules (see Figure 4B–C). Ndc80 phosphopeptides with a localization probability >0.5 are shown. (g) Table of experiments on optical tweezers reporting force measurements, duration, and outcome of stall.
- https://cdn.elifesciences.org/articles/49539/elife-49539-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/49539/elife-49539-transrepform-v2.docx