Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles
Figures
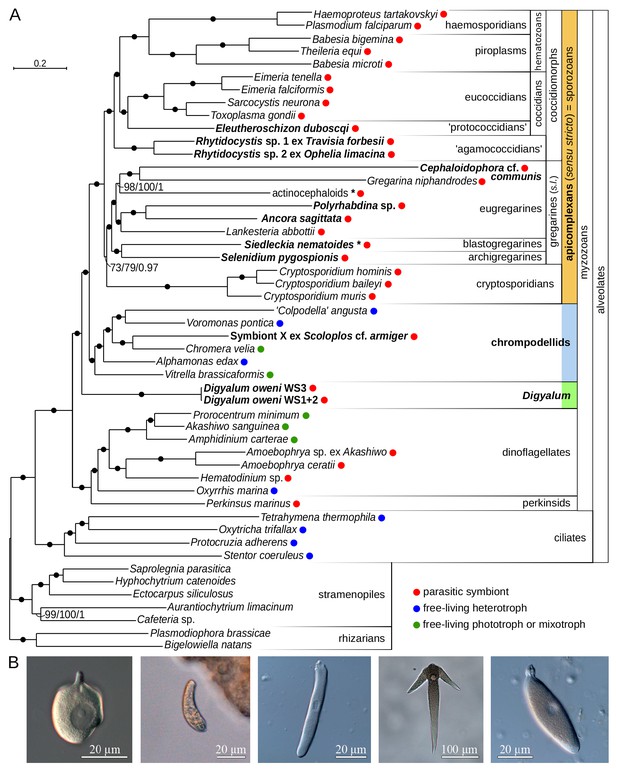
Multiprotein phylogeny of apicomplexans and related taxa.
(A) Maximum likelihood tree (IQ-TREE) of apicomplexans and their relatives based on 296 concatenated protein markers. Species newly sequenced in this study are in bold. Values at branches correspond to UFBoot2 supports (1000 replicates, LG+G4+F+C60+PMSF model), non-parametric bootstraps (100 replicates, LG+G4+F+C60+PMSF model), and Bayesian posterior probabilities (PhyloBayes, consensus of 10 independent runs, CAT+GTR+G4 model). Black dots indicate 100/100/1 support. Actinocephaloids and Siedleckia nematoides are hybrid taxa (* symbol) composed from sequences of three parasites and two distant sequence variants, respectively (see Figure 1—figure supplement 1). Values in parentheses behind species names show % of missing data in the phylogenetic matrix. Sequence sources and the phylogenetic matrix are found in Supplementary file 2 and Figure 1—source data 1, respectively. Single quotation marks indicate potentially problematic taxonomic assignments (formal group names are in Figure 1—figure supplement 1). (B) Light micrographs of some species studied, left to right: Digyalum oweni, Symbiont X, Selenidium pygospionis, Ancora sagittata, Polyrhabdina sp., with the anterior end facing up.
-
Figure 1—source data 1
Phylogenetic matrix used in Figure 1A analysis in FASTA format.
- https://doi.org/10.7554/eLife.49662.005
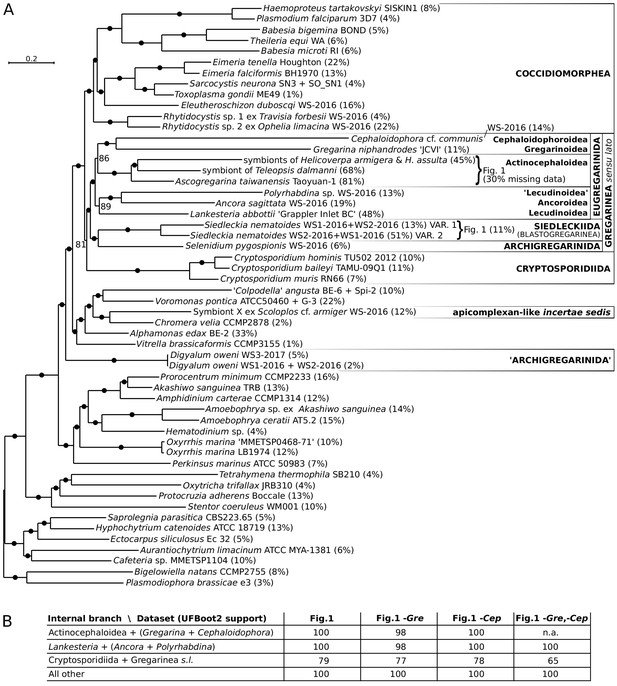
Multiprotein phylogenies of apicomplexans and related taxa.
(A) Maximum likelihood tree (IQ-TREE) of apicomplexans and their relatives (54 species) based on 296 concatenated protein markers (99948 sites, 14.4% missing data). Values at branches correspond to UFBoot2 supports, black dots indicate 100 support (1000 replicates, LG+G4+F+C60+PMSF model). Sequences of three actinocephaloids and of the two distinct sequence variants in Siedleckia nematoides were merged in the Figure 1A dataset, as shown. Values in parentheses behind species names show % of missing data in the final phylogenetic matrix. Single quotation marks indicate potentially problematic taxonomic assignments. Sequence sources are in Supplementary file 2. (B) Maximum likelihood phylogenies on datasets that excluded the two fastest evolving taxa, Gregarina and Cephaloidophora, either individually or together (IQ-TREE). UFBoot2 supports are shown (LG+G4+F+C60+PMSF model); note that all trees were topologically congruent.
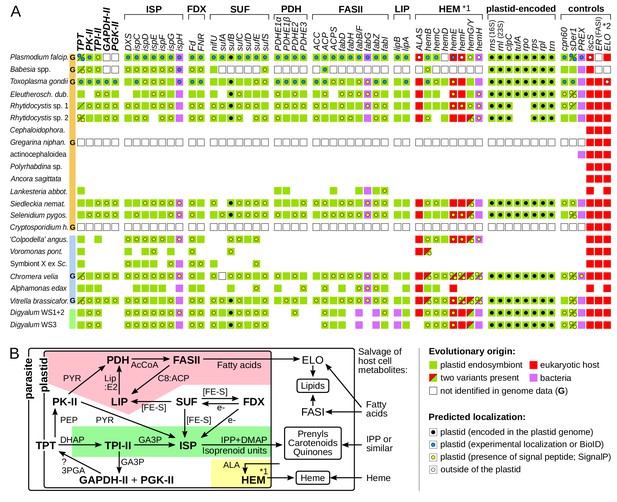
Core plastid metabolism in apicomplexans and their relatives.
(A) Presence of genes and pathway modules (top; abbreviations in Supplementary file 4) in representative genomes (G) and transcriptomes (left. Each gene (box) is color-coded as to its evolutionary origin, as determined by a maximum likelihood phylogeny (plastid-encoded rps, rpl, rpo, trn genes were not analyzed). Empty boxes indicate gene absence in completed genomes and blank spaces indicate absence in transcriptomes. Intracellular localization of corresponding proteins is shown by a circle inside the box and summarizes known experimental data (Supplementary file 4) or de novo prediction in silico by SignalP v4.1 (Supplementary file 5); it is missing in proteins with incomplete N-termini. Note that only some enzymes of the heme pathway (HEM) are localized in the plastid (*1) and that signal peptides in FAS:ER and ELO were not predicted (*2). (B) Dependence network of plastid protein modules for the biosynthesis of key metabolites – isoprenoid precursors IPP and DMAP, fatty acids and heme – which underlie dependency on the plastid organelle in Apicomplexa. Colored regions contain modules specific to one pathway: fatty acid (pink), isoprenoid precursor (pale green) and heme biosynthesis (yellow). Interactions are reconstructed from the literature and substrates are shown near arrows (PYR = pyruvate; AcCoA = acetyl coenzyme A, Lip:E2 = lipoylation on PDHE2; C8:ACP = octanoyl:acyl carrier protein; [FE-S]=iron sulphur cluster; PEP = phosphoenolpyruvate; e-=electron reductive power; GA3p=glyceraldehyde-3-phosphate; 3PGA = 3 phosphoglycerate; ALA = δ-aminolevulinic acid; ?=uncertainty).
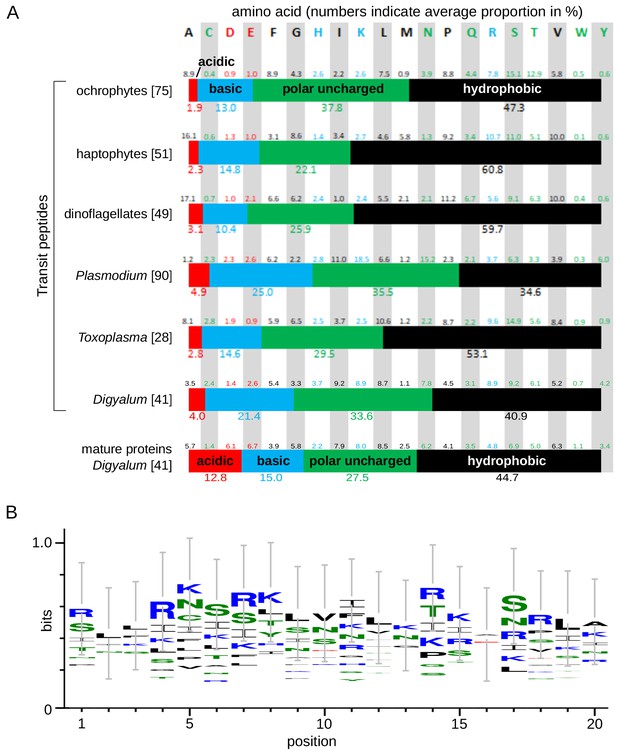
Transit peptides in Digyalum plastid proteins have positive charge but lack a conserved phenylalanine in the first position.
(A) Amino acid composition of plastid transit peptides in Digyalum in comparison with downstream sequences of mature plastid proteins and transit peptides in other complex plastids as reported by Patron and Waller (2007) (only the first 14 residues were analyzed to enable consistent comparison) (Patron and Waller, 2007). Average proportions of individual amino acids and their classes based on charge (colored bars) are shown: acidic (D + E) in red, basic (H + K + R) in blue, polar uncharged (C + N + Q + S + T + W + Y) in green and hydrophobic (A + F + G + I + L + M + P + V) in black. Number of proteins analyzed are indicated in square brackets behind species/group names. (B) WebLogo3 diagram of the relative composition of the first 20 amino acids in 41 target peptides of Digyalum oweni. Letter size indicates the probability of an amino acid occurring at a specific position, weighted to account for the relative abundance of each amino acids natural occurrence and unrepresentative results arising from a small sample size.
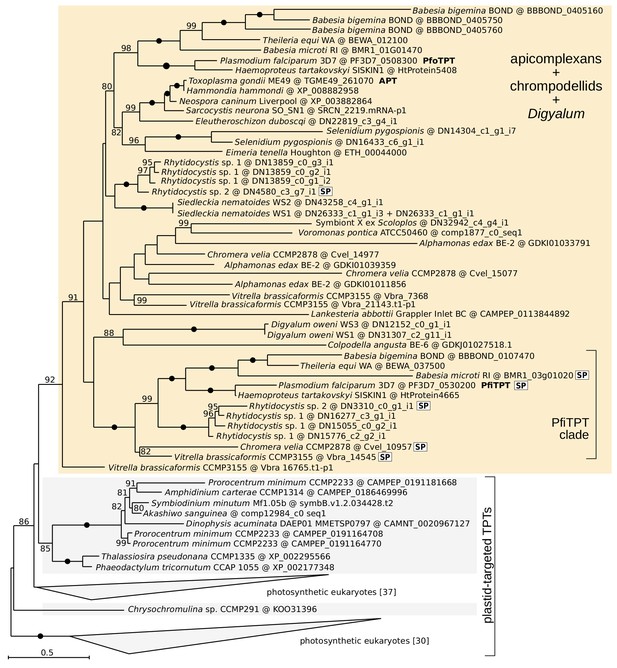
Maximum likelihood phylogeny of the triose phosphate translocator, TPT (LG+F+R7 model in IQ-TREE with 10000 UFBoot2 replicates;≥80 are shown; black dots indicate full support).
Apicomplexans, chrompodellids and Digyalum are shown on orange background and other eukaryotes on gray background. Selected characterized proteins are highlighted in bold.
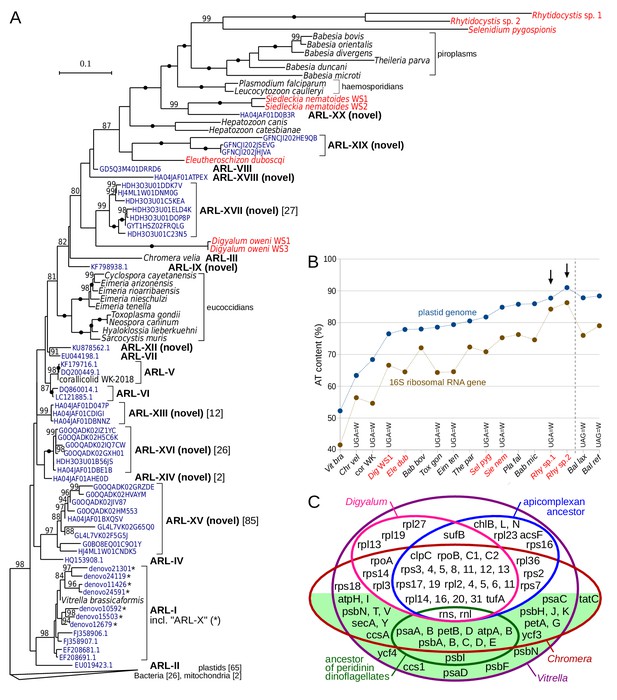
Plastid genomes in apicomplexans and their relatives are widespread and highly divergent.
(A) Maximum likelihood phylogeny of plastid-encoded 16S ribosomal RNA genes (rDNA) reveals 10 novel apicomplexan-related lineages (ARL-IX and ARL-XII to ARL-XX; Supplementary file 6). Tree was computed with the best-fit TVMe+R5 model in IQ-TREE with UFBoot2 supports at branches (10000 replicates;≥80 are shown; black dots indicate 100 support). Environmental sequences (dark blue) are derived from GenBank or VAMPS (97% identity cluster centroids; numbers of reads are shown in square brackets where > 1). Plastid 16S rDNA transcripts of newly sequenced species are shown in red. Note that sequences in the tree vary greatly in their AT content and substitution rates, which can induce a misleading topology - deep relationships in the tree should therefore be interpreted with caution. The fast-evolving sequences of peridinin dinoflagellates were not included. (B) Extremely high AT content in rhytidocystid plastid genomes (arrows). AT content of representative species from part A and Balanophora laxiflora and B. reflexa parasitic plants (Su et al., 2019), all abbreviated to first three letters, is shown for 16S rDNA and plastid genomes. Plastid genomes in the newly sequenced species are only partially reconstructed from transcripts (red color; see Supplementary file 7). Altered genetic codes are indicated. (C) Euler diagram of plastid genome contents in apicomplexans, dinoflagellates (ancestral gene sets for each), Digyalum, Chromera, and Vitrella. Genes on the green background are associated solely with photosynthesis. Small RNA genes are not shown.
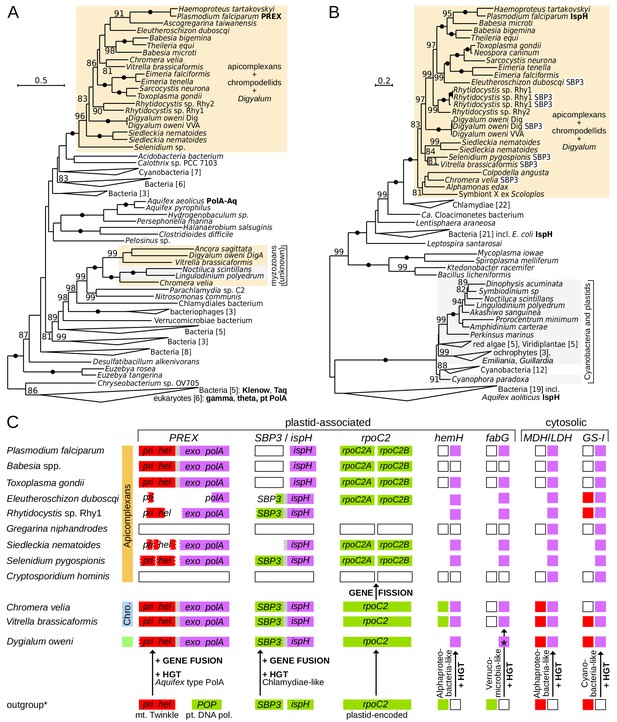
Innovations in plastid and cytosolic genes define early evolution of apicomplexans and their relatives.
(A) Maximum likelihood phylogeny of the exonuclease/polymerase subunit of the plastid replication and repair complex, PREX (LG+R6 model). (B) Maximum likelihood phylogeny of 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IspH (LG+I+G4 model). Trees were derived from protein sequences in IQ-TREE and have UFBoot2 supports at branches (10000 replicates;≥80 are shown; black dots indicate 100 support). Apicomplexans, chrompodellids and Digyalum are highlighted in orange, and other eukaryotes in gray. Characterized enzymes are highlighted in bold. Fusion ispH genes with SBP3 at the N-terminus are shown in white boxes. (C) Predicted gain, loss, fusion and fission events in the evolution of five plastid-associated and two cytosolic genes. Genes are shown by boxes (jagged edges indicate truncated genes) and are color-coded by origin as in Figure 2A (plastid endosymbiont = green, eukaryotic host = red, bacteria = purple, absent in genome data = white, absence of evidence = blank area). Outgroup (*) shows the state in the closest relevant comparator: dinoflagellates (IspH, HemH, MDH/LDH, GS-I) or other algae (PREX, SBP3, RpoC2, FabG). Note that the ferrochelatase (HemH) is mitochondrial in Plasmodium but probably plastidial in Chromera and some apicomplexans. Abbreviations: POP = plant organellar DNA polymerase, Twinkle = mitochondrial primase/helicase, SBP3 = sedoheptulose-1,7-bisphosphatase form 3, MDH/LDH = malate/lactate dehydrogenase (other abbreviations in Supplementary file 3).

The region of the split in the apicomplexan plastid-encoded rpoC2.
Plastid genome sequences and their three-frame amino acid translations are shown for representative apicomplexan species (snapshots from Artemis 17.0.1). Coding sequences are highlighted by horizontal rectangles. STOP codons are shown by # (=TAA), + (=TAG), and * (=TGA) symbols. In some taxa, TGA encodes for tryptophan (W). Note that the split occurs in a non-conserved part of rpoC2 so the corresponding region cannot be shown for a non-apicomplexan outgroup - rpoC2 genes in chrompodellids, Digyalum and all other plastids nevertheless lack the split.
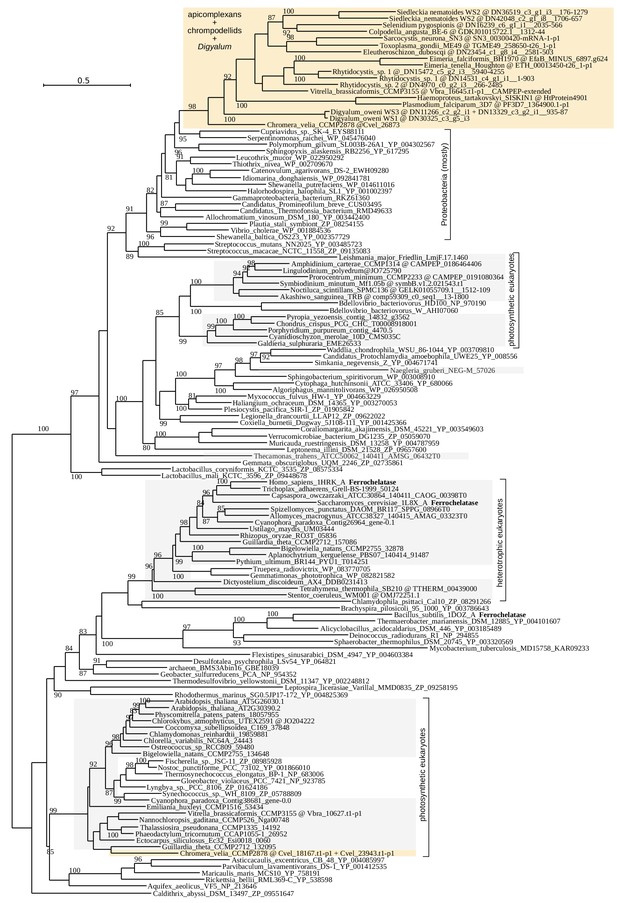
Maximum likelihood phylogeny of the ferrochelatase, HemH (LG+R8 model in IQ-TREE with 10000 UFBoot2 replicates;≥80 are shown).
Apicomplexans, chrompodellids and Digyalum are shown on orange background and other eukaryotes on gray background. Selected characterized enzymes are highlighted in bold.
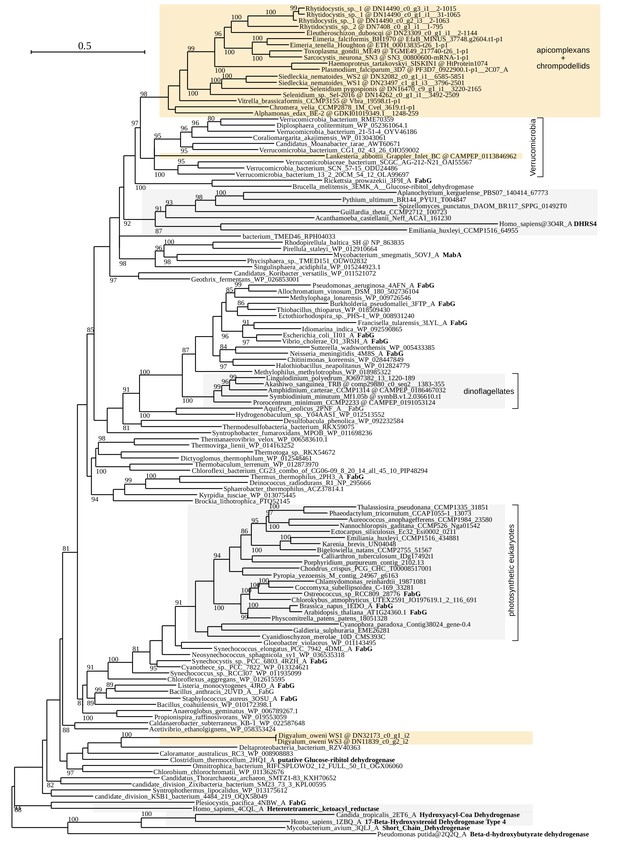
Maximum likelihood phylogeny of the beta-ketoacyl-acyl carrier protein reductase, FabG (LG+R6 model in IQ-TREE with 10000 UFBoot2 replicates;≥80 are shown).
Apicomplexans, chrompodellids and Digyalum are shown on orange background and other eukaryotes on gray background. Selected characterized enzymes are highlighted in bold.
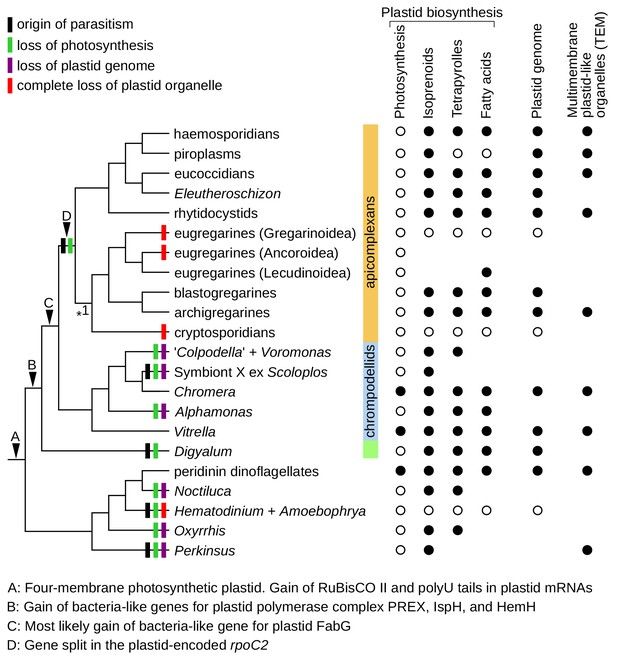
Plastid evolution in apicomplexans and their relatives.
Plastid-related characteristics (A–D), origin of parasitism, and predicted losses of photosynthesis, plastid genomes, and plastid organelles were mapped on the updated phylogeny. The phylogeny is fully resolved except one branch where more support is needed (*1). Predicted core plastid anabolic capabilities, presence of plastid genomes, and transmission electron microscopy evidence (TEM) for multimembrane organelles corresponding to plastids by their size, appearance, and position within cells are shown on the right.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Commercial assay or kit | RNAqueous-Micro Total RNA Isolation Kit | ThermoFisher | cat no. AM1931 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit | Takara | cat no. 634888 | |
| Software, algorithm | MAFFT v7.402 | Katoh and Standley, 2013 | RRID:SCR_011811 | |
| Software, algorithm | IQ-TREE v1.6.5 | Nguyen et al., 2015 | RRID:SCR_017254 | |
| Software, algorithm | PhyloBayes v4.1c (and MPI v1.7b) | Lartillot et al., 2009 | RRID:SCR_006402 |
Additional files
-
Supplementary file 1
Information about parasites collected (single quotation marks indicate problematic affiliations; asterisk denotes a species complex).
- https://doi.org/10.7554/eLife.49662.015
-
Supplementary file 2
Sources of sequence datasets used in this study (single quotation marks indicate arbitrarily assigned strain name).
- https://doi.org/10.7554/eLife.49662.016
-
Supplementary file 3
Results of seven tree topology tests as computed in IQ-TREE (of 105 tested topologies those not rejected at p=0.001 in at least one test are listed; those not rejected at p=0.01 are highlighted in bold).
- https://doi.org/10.7554/eLife.49662.017
-
Supplementary file 4
Names, abbreviations, accessions and localizations of plastidial proteins and modules (plastid-localized = green; mitochondrion-localized = blue; endoplasmic reticulum = red).
- https://doi.org/10.7554/eLife.49662.018
-
Supplementary file 5
Prediction statistics for signal (SignalP v4.1: noTM, Dmaxcut = 0.45) and transit peptides (ChloroP v1.1).
- https://doi.org/10.7554/eLife.49662.019
-
Supplementary file 6
Reference 16S rDNA sequences for apicomplexan related lineages (ARLs), Digyalum, and newly sequenced apicomplexans.
- https://doi.org/10.7554/eLife.49662.020
-
Supplementary file 7
Plastid transcriptomic contigs in species newly sequenced in this study.
- https://doi.org/10.7554/eLife.49662.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49662.022





