Mouse retinal cell behaviour in space and time using light sheet fluorescence microscopy
Figures
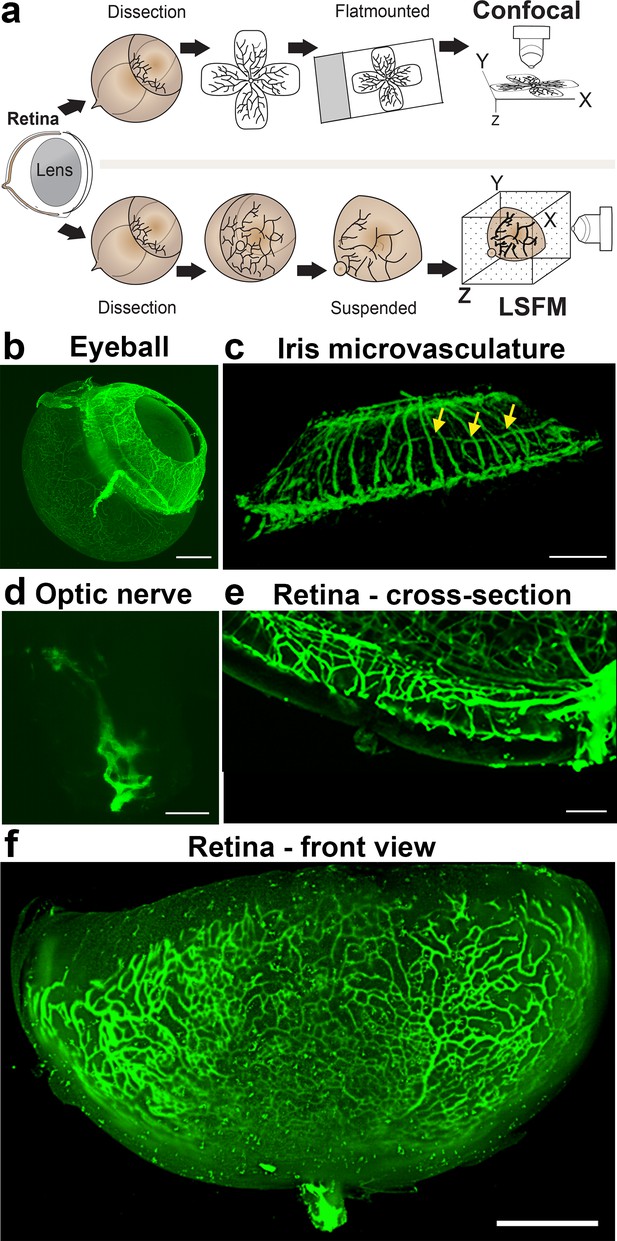
Imaging of the whole eye using light sheet microscopy.
(a) Schematic of retina preparation for imaging. For conventional confocal microscopy, four incisions are made to enable flat-mounting of the retina onto a cover slip. For LSFM, pieces of the retina are suspended and imaged from a right angle. (b) Maximum intensity projection (MIP) of a P15 mouse eyeball (z = 274 slices). Vessels were visualised with IsoB4 staining. Scale bar, 500 µm. (c) 3D-rendered image of the adult iris microvasculature (z = 263 slices). Vessels were visualised with IsoB4 (yellow arrows). Scale bar, 250 µm. (d) MIP of the optic nerve. Vessels were visualised with IsoB4 staining (z = 176 slices). Scale bar, 50 µm. (e) MIP of a cross section of a P10 mouse retina. Vessels were visualised with IsoB4 staining. Scale bar, 100 µm. (f) MIP of a whole P10 mouse retina suspended and imaged intact (z = 176 slices). Vessels were visualised with IsoB4 staining. Scale bar, 500 µm. See Figure 1—figure supplement 1 for additional images.
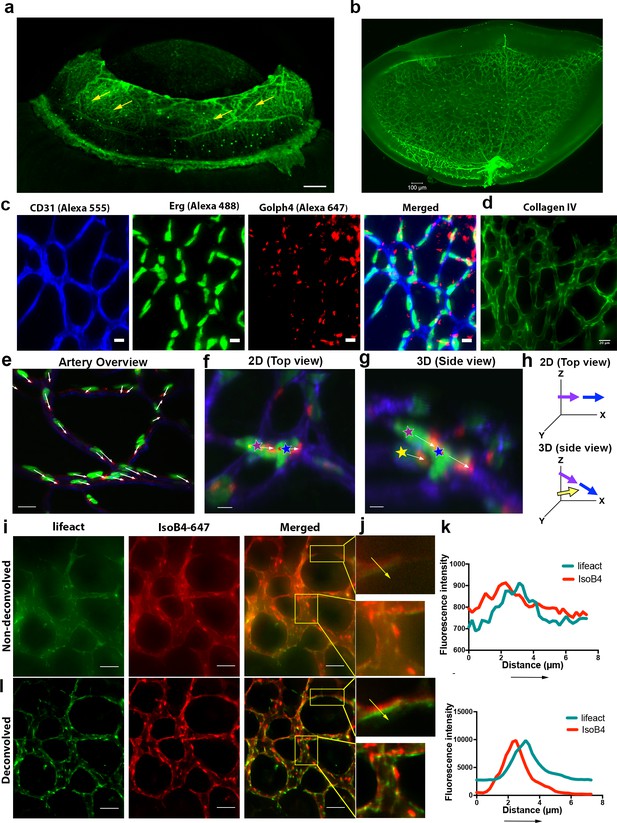
Iris microvasculature and Golgi-polarity as revealed by LSFM.
(a) Maximum intensity projection of the iris microvasculature of two-week old mouse, LSFM. Vessels were visualised with IsoB4 (green). Scale bar, 200 µm; (b) the full unprocessed retinal image used for the cropped figure in Figure 1e. (c) P12 retina stained using anti-CD31(Alexa 555), Anti-Erg (alexa488) and anti-Golgi (Golph4, Alexa 647) imaged using LSFM, Scale bar is 10 µm. (d) P12 Retina stained for CollagenIV followed by LSFM imaging. Scale bar is 20 µm. (e) representative image showing aligned cell polarisation in an artery of P12 GNrep mouse retina, Scale bar = 15 µm (f,g) zoom in region from a different P12 GNrep mouse retina highlighting the benefit of 3D rotational viewing in LSFM to standard 2D analysis of max projection (as viewed from the top) as in f only two cells are discernible (purple and blue stars) whereas in g when the image stack is rotated to the side view a hidden cell (yellow star) in a vessel crossing underneath the one viewable from above is revealed, Scale bar in f = 7 μm, scale bar in g = 5 μm h, schematic to illustrate the additional 3D information that LSFM gives to Golgi-nuclei axis analysis (arrow colour corresponds to start colour of each cell analysed in g,h). (i) lifeAct (green) mouse retinas stained for vessels (IsoB4 - red), merged images (j) shows the overlap between these two channels together with the plot profile of the selected area and the yellow arrow (k, upper). (h) the same channels were deconvolved using Huygens (l) and the same area was plotted to compare the image resolution after deconvolution. Values were normalised to the maximum fluorescence intensity across both images at each pixel.
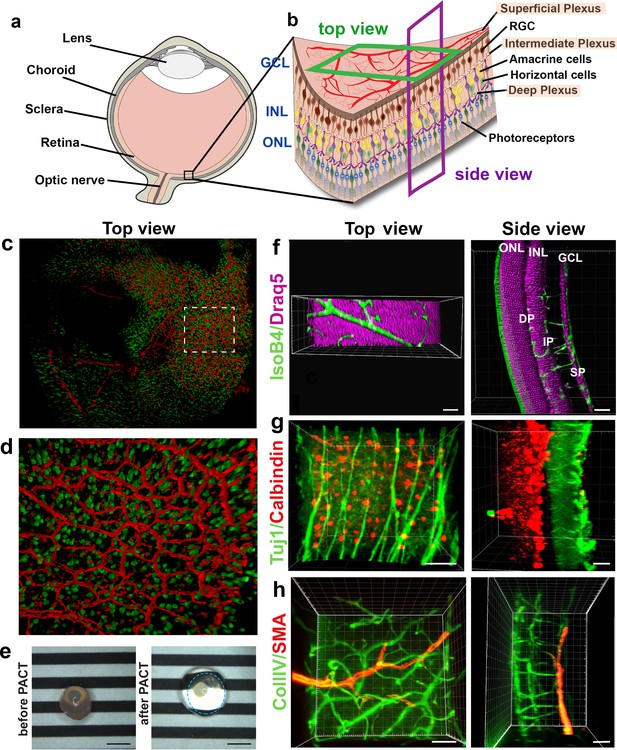
3D reconstruction of nerves and vessels in one image.
(a) Schematic of an eyeball. (b) Schematic of the retina and its cell types. (c) Retinal eye cups expressing yellow fluorescent protein, YFP (green) were harvest from Thy1-YFP mice and stained with Isolectin IB4 (red). The retinal eye cups were mounted and imaged with LSFM. (d) Enlarged region of c. (e) Representative image of an eyeball before clearing (left panel), and an eyeball after PACT clearing (right panel). The circle around the cleared eyeball depicts the outline of the eyeball. Scale bar, 2 mm. (f) Draq5 staining (magenta) visualises the inner nuclear layer (INL) and outer nuclear layer (ONL) of the adult mouse PACT cleared retina. Vessels were visualised by IsoB4 staining (green). (g) Tuj1 (green) and calbindin (red) visualise the ganglion and horizontal cells in the mouse PACT cleared retina. (h) Smooth muscle actin (SMA, red) and Collagen IV staining (Coll.IV, green) visualise the three vascular layers and smooth muscle cells in the mouse PACT cleared retina. Scale bars, 50 µm.
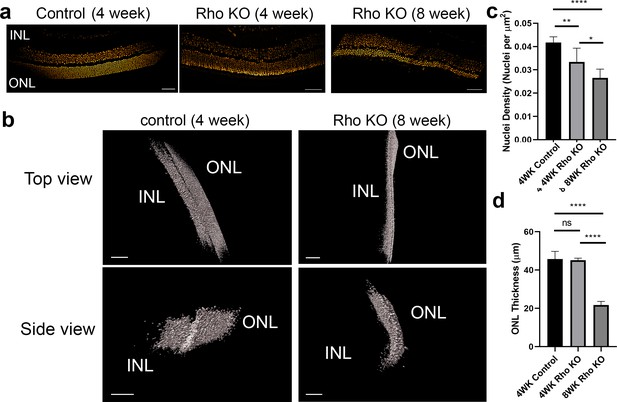
Changes in neuronal density in Rho KO degeneration model.
(a) Slices of control mouse retinas at four weeks, and retinas from Rho KO mutants at four and eight weeks, revealing ruffling of the outer nuclear layer and inner nuclear layer, potentially due to nuclear drop-out. (b) 3D renders of control and Rho KO (8 weeks) retinas. Scale bars = 70 μm (top left), 50 μm (bottom left), 50 μm (top right), 40 μm (bottom right) (c) Plot of nuclei density of control retinas at 4 weeks and Rho KO at 4 and 8 weeks, showing a decrease in nuclear density upon removal of Rho over time (p=0.0188) and compared to the control (vs. 4WK Rho KO - p=0.0061, vs. 8WK Rho KO – p<0.0001). p-values were calculated using unpaired t-test (one retina per condition). This was performed at multiple points (n = approx. 8 ROIs, incremented by 50 slices in z per condition, one retina per condition). (d) Plot of ONL thickness of control retinas at 4 weeks and Rho KO at 4 and 8 weeks, demonstrating a reduction in ONL thickness in Rho mutants as they developed (p<0.0001) and compared to the control (p<0.0001). No difference could be discerned when mutants were 4 weeks old compared to control. p-values were calculated using unpaired t-test. (n = approx. 8 ROIs, incremented by 50 slices in z per condition, one retina per condition).
-
Figure 2—figure supplement 1—source data 1
Excel file containing source data pertaining to Figure 2—figure supplement 1c and d.
- https://cdn.elifesciences.org/articles/49779/elife-49779-fig2-figsupp1-data1-v3.xlsx
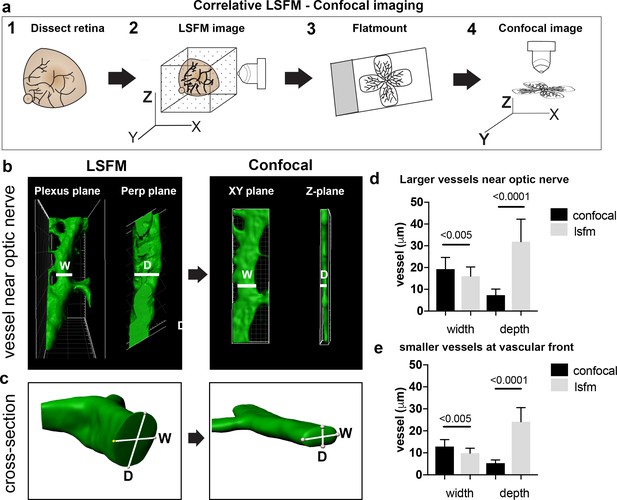
Vessel depth distortion in confocal due to flatmounting.
(a) Schematic showing the correlative LSFM-Confocal imaging approach used to quantify vessel distortion incurred by flatmounting. (b) The same large vessel segment imaged first with LSFM then confocal (surface rendered in Imaris). By orienting with the surrounding vessel connections to determine the plexus plane (equivalent to the XY plane in confocal) and the plane perpendicular to it (‘perp plane’), which is equivalent to the Z plane in confocal, comparative width (W) and depth (D) measurements can be made of the same vessel segment. (c) Cross sectional views of another representative large vessel near the optic nerve shows how the aspect ratio of W and D is shifted to an ellipse in confocal. Near Optic: n = 60 vessels from six retinas (seven images). Vascular Front n = 28 vessels in from four retinas (four images) for each confocal and LSFM.
-
Figure 3—source data 1
Excel file containing source data pertaining to Figure 3d and e.
- https://cdn.elifesciences.org/articles/49779/elife-49779-fig3-data1-v3.xlsx
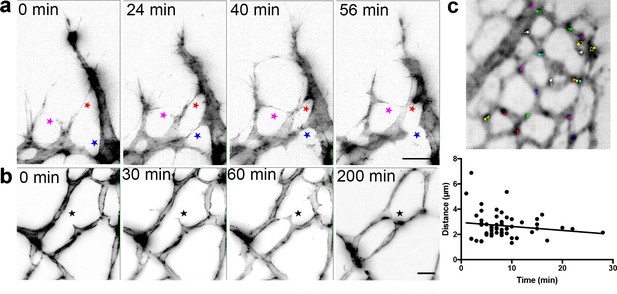
Live-imaging of the retinal vasculature.
(a) Single maximum intensity projections (MIP) of an hour time lapse Video show long, slender filopodia, and rapid fusion and disconnection of tip cells at the vascular front of mT/mG x Cdh5 (PAC) CreERT2 mice (stars). (b) MIPs of a time lapse Video reveal the connection between two branches in the capillary plexus (star). (c) lifeAct-EGFP mouse retina at P4/5 were live imaged for 40 min with an interval of one minute per frame. Actin-rich bundles were tracked manually using ImageJ/Fiji. Each color represents one bundle trajectory tracked over time, scale bar is 10 µm. Plot (below) shows each actin bundle’s distance travelled over time, average speed was 2.56 µm/min, n = 6 retinas (all uncleared).
-
Figure 4—source data 1
Excel file containing source data pertaining to Figure 4c.
- https://cdn.elifesciences.org/articles/49779/elife-49779-fig4-data1-v3.xlsx
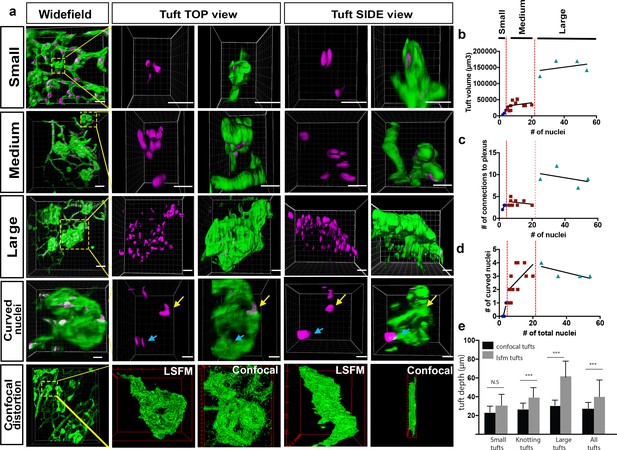
Analysis of three subclasses of OIR vascular tuft.
(a) Representative 3D-rendered LSFM images of small (1st row), medium (2nd row), and large (3rd row) tufts showing the vasculature (IsoB4, green) and endothelial nuclei (ERG, magenta), scale bar = 10 µm. For all widefield images, scale bar = 40 µm, yellow box indicates tuft in situ; 4th row - representative 3D-rendered images showing curved nuclei in a medium tuft, yellow arrows indicate curved nuclei, blue arrows indicate flat nuclei parallel to each other. Scale bar, 10 um. 5th row: correlative LSFM-confocal microscopy of the same tuft reveals the tuft depth distortion (side view) incurred with confocal flatmounting versus LSFM. (b) The volume of the tufts versus the number of nuclei per tuft. (c) The number of vessel connections between the tuft and the underlying vascular plexus versus the number of nuclei. (d) Quantification of the number of curved nuclei per tuft versus the total number of total nuclei per tuft. (e) Quantifications of tuft depths per subclass size in LSFM vs confocal images, significant difference shown using unpaired t-test, *** means p<0.0001.
-
Figure 5—source data 1
Excel file containing source data pertaining to Figure 5b–e.
- https://cdn.elifesciences.org/articles/49779/elife-49779-fig5-data1-v3.xlsx
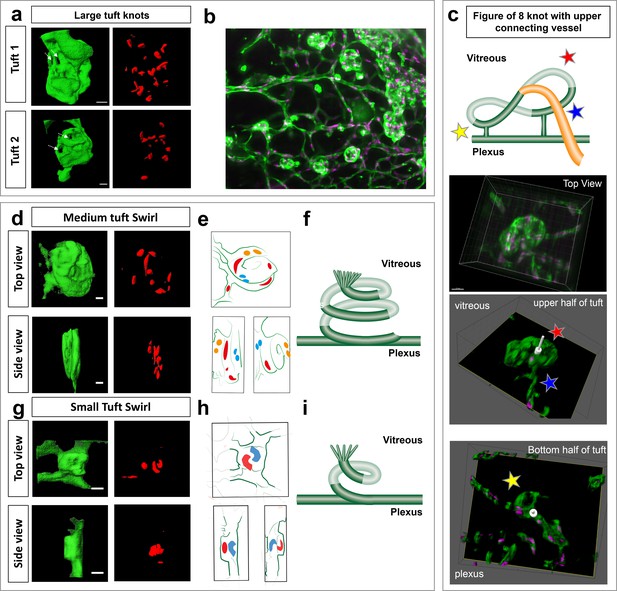
Knotted morphology of neovascular tufts revealed with LSFM.
(a,d,g) Representative 3D-rendered images (generated using IMARIS software) of large (upper panel), medium and small tufts showing the vasculature (IsoB4, green) and endothelial nuclei (ERG, red) from rotational Videos 14 and 15 See Figure 6—figure supplement 1a for further views from different angles of the large tufts. White dashed arrows indicates a hole through the tuft. Scale bar, 30 µm. (b) Widefield LSFM of OIR retina demonstrates that the knotted morphology is hard to discern from afar. (c) Detailed 3D clipping plane and 3D rotational drawings of an individual knot reveal a figure of eight structure with two clear holes through the tuft as well a vessel connecting from the upper, vitreous facing surface of the tuft to the plexus below (blue star). Stars mark corresponding regions from the illustration to the images - lower tuft loop nearer plexus (yellow star), upper tuft loop nearer vitreous (red star). See also Figure 6—figure supplement 1b for detailed 3D drawings made from each rotational view of this tuft with clipping planes, and Video 13. (e,h) 3D sketches made from rotational Videos 16 and 17 to better elucidate nuclei: blue nuclei - bottom of tuft (near plexus), red nuclei – middle of tuft (in e), top of tuft (facing vitreous) in (h) yellow nuclei - top of tuft (facing vitreous) in e, (f,i,) schematic illustrating the swirling tuft morphology observed in (d-h) with three layers for the medium tuft (f) and two for the small one (i).
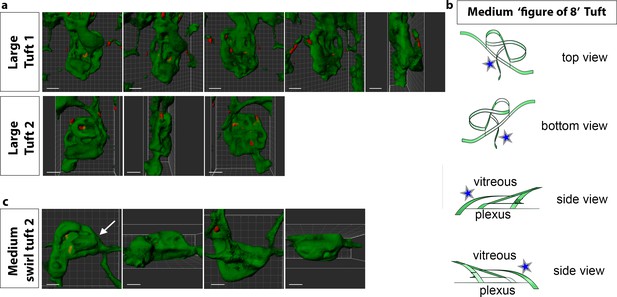
LSFM reveals knotted morphologies in vascular tufts.
(a) extended rotational views to see knotted structure from different angles of tuft 1 and 2 in Figure 6a taken from Videos 14 and 15. Scale bar = 50 μm. (b) 3D illustration (made with uMake software) while viewing the tuft with curved nuclei (from Figure 6c and Video 13) from top to bottom with clipping planes to better understand how the knot topology changes through the tuft from the vitreous to the plexus side. Blue star indicates the unexpected vessel joining the top (vitreous facing) surface of the tuft to the plexus below. (c) a second example of a medium tuft with a swirl structure and clear protrusive sprout-like morphology to the upper vitreous facing end point of the swirl. Scale bar = 30 μm.
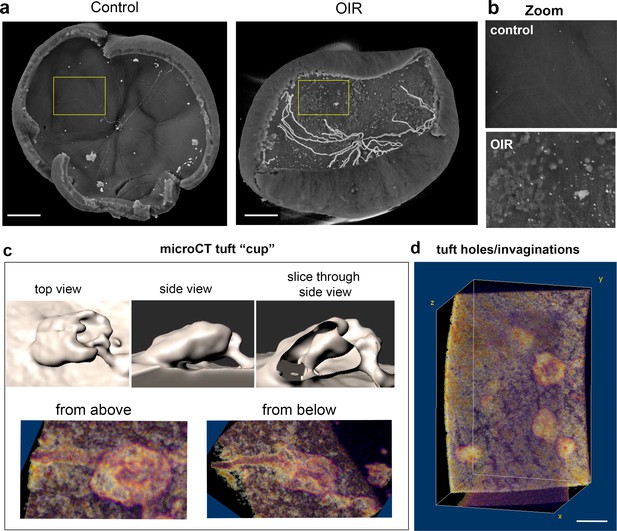
MicroCT of OIR retinas shows tuft appearing to undergo invagination.
(a) Representative overview microCT images showing vitreous surface is smooth in control retina (P15), whereas epiretinal tufts protrude clearly into the vitreous space in OIR retina (P15). Scale bar = 500 μm for both images. Hyaloid vessels tend to persist during OIR protocol at P15, contrary to normal development, and are shown centrally, but do not connect to the retina surface, making them easily segmented out when studying tufts. (b) zoom regions of a (yellow boxes). (c,d) Higher resolution microCT analysis (see Materials and methods for details) of individual tufts on OIR retina surface show tufts with holes/invaginations through them as characterised with LSFM in Figure 6. Surfaces rendered in Imaris, depth volumes (lower panels) rendered in FIJI with the volume viewer plugin reveal the long tubular vessel in the plexus layer beneath this tuft. Scale bar = 30 μm. (d) a set of three neighbouring tufts appeared to have holes, depressions or invaginations (scale bar = 100 μm).
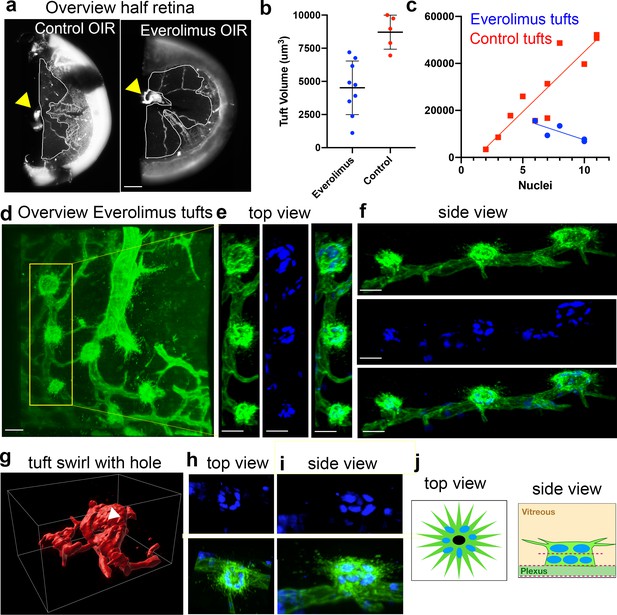
Everolimus-treated tufts exhibit highly active filopodia and cup morphology.
(a) overview image of control (vehicle-treated OIR) and Everolimus-treated OIR retinas (from n = 2 control and n = 2 drug-treated retinas). Avascular areas were visibly larger in Everolimus-treated retinas (marked with white line and adjacent to the optic nerve, yellow arrow) as previously reported. Scale bar, 300 μm. (b) tuft volume comparison n = 2 control OIR and n = 2 Everolimus-treated OIR, (c) quantification of tuft volume vs nuclei count, with and without drug treatment. n = 5 tufts from one drug-treated retina compared to comparable (medium) tuft volume data in Figure 5b. (d) Overview of Everolimus-treated OIR retina with many small tufts demonstrating highly active filopodia. Vessels were visualised with CD31, Scale bar, 20 μm. (e–f) Overhead view of three active adjacent tufts with similar morphology to d, from the top (e) and side (f). Vessels were visualised with CD31, nuclei were visualised with ERG. Scale bar, 20 μm. (g) Surface render (Imaris) of rapamycin-treated OIR tuft, which shows a swirl structure, as well as a visible hole through the centre of the tuft (white arrow). (h) Top and (i) side view of a Everolimus-treated tuft with a ring-like, layered arrangement of cells, and splayed filopodia representative of others observed in these retinas. Tufts were visualised using CD31 (green), while nuclei were visualised using ERG (blue). (j) Schematic of Everolimus treated tuft in c: viewing from the top, it is possible to see a hole or invagination (black circle) to the tuft and/or nuclei circled around it, while from the side, there are two layers of nuclei within the tuft, similar to the layered swirl of a control tuft from an LSFM imaged OIR retina in Figure 6d and microCT imaged OIR retina in Figure 6—figure supplement 2c,d.
-
Figure 6—figure supplement 3—source data 1
Excel file containing source data pertaining to Figure 6—figure supplement 3h and i.
- https://cdn.elifesciences.org/articles/49779/elife-49779-fig6-figsupp3-data1-v3.xlsx
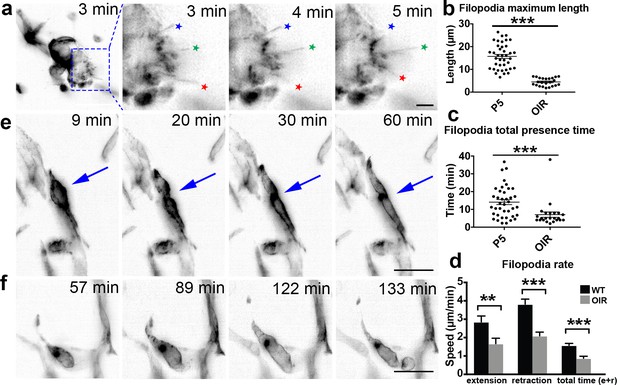
OIR live imaging.
(a) Maximum intensity projections (MIPs) of a time lapse Video of a retinal tuft of a mouse in the oxygen-induced retinopathy (OIR) model visualise short, rapidly extending and retracting filopodia as compared to control retinas (stars). (b) The maximum length that each filopodia reached was measured for each filopodia over time in P5 and OIR conditions. (c) The total time that each filopodia was present during the experiment; this time is calculated from when one filopodia appeared and then disappeared. (d) Speed of extension and retraction of filopodia were calculated for P5 and OIR conditions. Total n = 67 and 23 filopodia in 8 and 3 cropped Videos from three independent P5 and 1 OIR experiment. (e) MIPs of a time lapse Video of mouse retinal vasculature in the OIR model reveal cell shuffling in real-time (arrow). (f) MIPs of a time lapse Video of mouse retinal vasculature in the OIR model reveal abnormal vessel growth in real-time. Scale bar, 20 µm.
-
Figure 7—source data 1
Excel file containing source data pertaining to Figure 7d–f.
- https://cdn.elifesciences.org/articles/49779/elife-49779-fig7-data1-v3.xlsx
Videos
3D-rendered LSFM z-stack of retinal eye cups expressing yellow fluorescent protein, YFP (green) harvest from Thy1-YFP mice and stained with Isolectin IB4 (red).
3D-rendered LSFM z-stack of all nuclei (Draq5, magenta) visualises the inner nuclear layer and outer nuclear layer of the adult mouse retina.
Vessels were visualised by IsolectinB4 staining (green). The retinal pigment epithelium emits green autofluorescence.
3D-rendered LSFM z-stack of an adult mouse retina stained for Tuj1 (green) and horizontal cells (calbindin, red) visualising the ganglion cells and horizontal cells, respectively.
3D-rendered LSFM z-stack of an adult mouse retina stained for Smooth Muscle Actin (SMA, red) and CollagenIV staining (CollIV, green) visualises the three vascular layers and smooth muscle cells in the mouse retina.
An hour time lapse LSFM Video showing long, slender filopodia, and rapid ‘kiss and run’ fusion and disconnection of tip cells at the vascular front of mT/mG x cdh5 (PAC) CreERT2 mice.
Frame rate: one image/45 s.
A 9 hr time lapse LSFM Video showing a connection between two branches in the capillary plexus of mT/mG x cdh5 (PAC) CreERT2 mice.
Frame rate: one image/20 min.
Representative tracking of a short-lived actin-rich bundle in life-Act-EGFP retina mice imaged with LSFM (7 min).
The tracking was performed manually using Manual Tracking plugin in Fiji/ImageJ.
Representative tracking of a longer-lived actin-rich bundle in life-Act-EGFP retina mice imaged with LSFM (30 min).
The tracking was performed manually using Manual Tracking plugin in Fiji/ImageJ.
Representative tracking of a long-lived actin-rich bundle track in life-Act-EGFP retina mice imaged with LSFM (40 min).
The tracking was performed manually using Manual Tracking plugin in Fiji/ImageJ.
3D-rendered LSFM z-stack of a ‘small tuft’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (magenta).
The z-stack was rendered in Imaris.
3D-rendered LSFM z-stack of a ‘medium tuft’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (magenta).
The z-stack was rendered in Imaris.
3D-rendered LSFM z-stack of a ‘large tuft’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (magenta).
The z-stack was rendered in Imaris.
3D-rendered z-stack showed a curved nuclei in a ‘medium tuft’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (magenta).
The z-stack was rendered in Imaris.
3D-rendered LSFM z-stack deconvolved in Huygens then reconstructed with surface rendering in Imaris of ‘Large tuft 1’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (red).
3D-rendered LSFM z-stack deconvolved in Huygens then reconstructed with surface rendering in Imaris of ‘Large tuft 2’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (red).
3D-rendered LSFM z-stack deconvolved in Huygens then reconstructed with surface rendering in Imaris of a ‘small tuft’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (red).
3D-rendered LSFM z-stack deconvolved in Huygens then reconstructed with surface rendering in Imaris of ‘a Medium swirl tuft’ from an OIR mouse retina stained for blood vessels (IsolectinB4, green), and the nuclear marker ERG (red).
A 2.5 hr time lapse Video of a retinal tuft of a mouse in the OIR model using mT/mG x cdh5 (PAC) CreERT2 mice visualise short, rapidly extending and retracting filopodia as compared to control retinas.
Frame rate: one image/minute.
A 2.5 hr time lapse Video of mouse retinal vasculature in the OIR model using mT/mG x cdh5 (PAC) CreERT2 mice reveal abnormal vessel growth in real-time.
Frame rate: one image/minute.
A 2.5 hr time lapse Video of mouse retinal vasculature in the OIR model using mT/mG x cdh5 (PAC) CreERT2 mice reveal cell shuffling in real-time.
Frame rate: one image/minute.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-calretinin (Rabbit Polyclonal) | Abcam | Cat#:ab702, RRID: AB_305702 | (1:50) |
| Antibody | Anti-ERG (Rabbit Monoclonal) | Abcam | Cat#: ab92513, RRID: AB_2630401 | (1:200) |
| Antibody | Anti-calbindin (Rabbit Polyclonal) | Millipore | Cat#: AB1778 RRID: AB_2068336 | (1:300) |
| Antibody | Anti-GFAP (Rabbit Polyclonal) | Agilent, Dako | Cat#:Z0334 RRID: AB_10013382 | (1:100) |
| Antibody | Anti-collagenIV (Rabbit Polyclonal) | Bio-Rad | Cat#:2150–1470, RRID: AB_2082660 | (1:500) |
| Antibody | Neuron-specific beta-III Tubulin Biotin MAb (Clone TuJ-1) (Mouse monoclonal) | R and D Systems | Cat#: BAM1195, RRID: AB_356859 | (1:50) |
| Antibody | Anti-Smooth Muscle Actin (Mouse monoclonal) | Sigma-Aldrich | Cat#: C6198, RRID: AB_476856 | (1:1000) |
| Antibody | Anti-MGOLPH4 (Rabbit Polyclonal) | Abcam | Cat#: ab28049 RRID: AB_732692 | |
| antibody | Anti-Mouse CD31/PECAM-1 (Goat polyclonal) | R and D System | Cat#: AF3628 RRID: AB_2161028 | (1:200) |
| Commercial assay or kit | CyGEL | biostatus | CAT#: CY10500 | (1:400) |
| Software, algorithm | Arivis Vision 4D | arivis AG | N/A – (A new RRID has been requested as of paper submission). | |
| Software, algorithm | Imaris | Bitplane | RRID:SCR_007370 | |
| Software, algorithm | FIJI | Shcindelin, 2012 | RRID:SCR_002285 | StackReg plugin used |
| Software, algorithm | Zeiss Scout and Scan Control Reconstructor Software | Zeiss | N/A | |
| Software, algorithm | Drishti | Limaye, 2012 | N/A – (A new RRID has been requested as of paper submission). | |
| Software, algorithm | Zeiss Zen | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | Huygens Software | Scientific Volume Imaging | RRID:SCR_014237 | |
| Other | Alexa Fluor 568 isolectin GS-IB4 conjugate | ThermoFisher Scientific | Cat#:I21412 | Molecular probe. |
| Other | Alexa Fluor 488 isolectin GS-IB4 conjugate | ThermoFisher Scientifc | Cat#:I21411 | Molecular probe. |
| Other | Alexa Fluor 647 isolectin GS-IB4 conjugate | ThermoFisher Scientifc | Cat#: I32450 | Molecular probe. |
| Other | Draq5 | Thermo Scientific | Cat#: 62251 | Cell (DNA) stain |