Biochemical reconstitution of branching microtubule nucleation
Figures
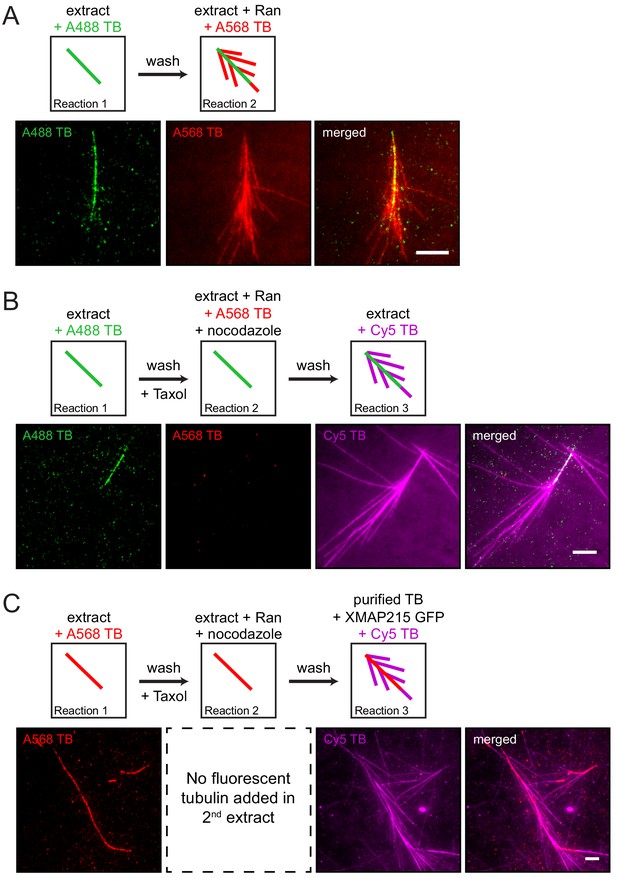
The proteins necessary for branching microtubule nucleation in Xenopus egg extract bind to a pre-existing microtubule independent of the nucleation event.
(A–C) Sequential reactions with Xenopus egg extract. (A) Single microtubules formed on the glass surface in the first extract supplemented with Alexa488 tubulin (green). A second extract supplemented with Alexa568 tubulin (red) and RanQ69L was subsequently introduced. New microtubules (red) nucleated from pre-existing microtubules (green). See Figure 1—video 1. (B) Single microtubules formed on the glass surface in the first extract supplemented with Alexa488 tubulin (green). A second extract supplemented with Alexa568 tubulin (red), RanQ69L and nocodazole was subsequently introduced, followed by a third extract supplemented with Cy5 tubulin (magenta). Branched microtubules (magenta) nucleated from pre-existing microtubules (green) via the branching factors released in the second extract, while no microtubules formed in the presence of nocodazole (red). See Figure 1—figure supplement 1 and Figure 1—figure supplement 2A. (C) Similar to (B), except that the first extract was supplemented with Alexa568 tubulin (red), the second extract contained no fluorescent tubulin, and the third extract reaction was substituted for purified Cy5 tubulin (magenta) and XMAP215. Branched microtubules (magenta) nucleated from pre-existing microtubules (red), which had been pre-loaded with branching factors in the second extract. See Figure 1—figure supplement 2B and Figure 1—video 2. For all experiments, images were collected approximately 5 min after the last solution was exchanged. Scale bars, 5 μm. The experiments were repeated three times with different Xenopus egg extracts.
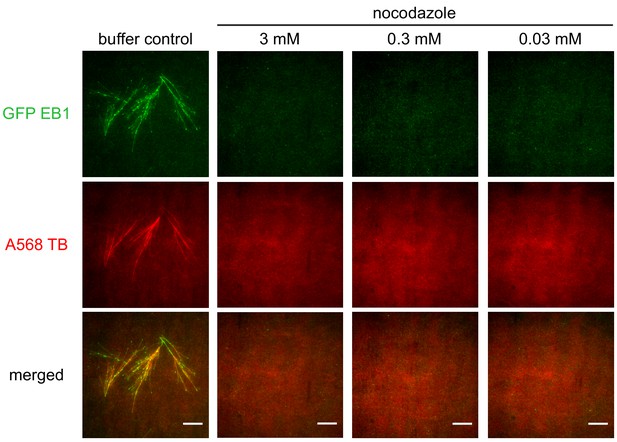
Testing the inhibitory effect of nocodazole in Xenopus egg extract.
Branching microtubule nucleation was stimulated in Xenopus egg extract with 10 μM RanQ69L in the presence of increasing concentrations of nocodazole. microtubules were labeled with Alexa568 tubulin (red) and their plus-ends with EB1-GFP (green). Scale bars, 10 μm. The experiment was repeated three times with different Xenopus egg extracts.
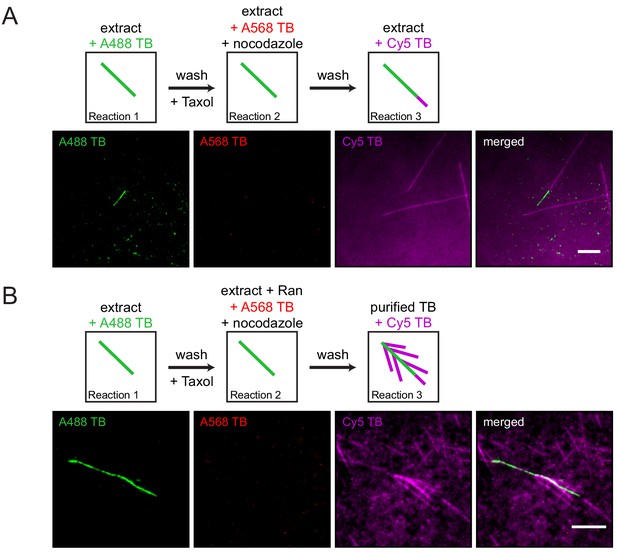
Sequential Xenopus egg extract reactions.
(A) Single microtubules formed on the glass surface in the first extract supplemented with Alexa488 tubulin (green). A second extract supplemented with Alexa568 tubulin (red) and nocodazole, but lacking RanQ69L, was subsequently introduced, followed by a third extract supplemented with Cy5 tubulin (magenta). Pre-existing microtubules (green) only extended from their plus-ends (magenta) in the third extract reaction because no branching factors were released in the second reaction step, while no microtubules formed in the presence of nocodazole (red). (B) Analogous to (A), except that the second extract was supplemented with RanQ69L, and the third extract reaction was substituted for purified Cy5 tubulin (magenta). Branched microtubules (magenta) nucleated from pre-existing microtubules (green), while no microtubules formed in the presence of nocodazole (red). For all experiments, images were collected approximately 5 min after the last solution was exchanged. Scale bars, 5 μm. The experiments were repeated three times with different Xenopus egg extracts.
Branching microtubule nucleation from a pre-existing microtubule in Xenopus egg extract (related to Figure 1A).
A single microtubule formed on the glass surface in extract supplemented with Alexa488 tubulin (green). A second extract supplemented with Alexa568 tubulin (red) and RanQ69L was subsequently introduced. Branched microtubules (red) nucleated from the pre-existing microtubule (green). The sample was imaged every 2 s. Scale bar, 10 μm.
The proteins necessary for branching microtubule nucleation in Xenopus egg extract bind to a pre-existing microtubule preceding and independent of the nucleation event (related to Figure 1C).
Single microtubules formed on the glass surface in extract supplemented with Alexa568 tubulin (green). A second extract supplemented with RanQ69L and nocodazole was subsequently introduced during which branching factors bound to the pre-existing microtubule. Finally, a mixture of purified Cy5 tubulin (red) and XMAP215 was added. Branched microtubules (red) nucleated from pre-existing microtubules (green). The sample was imaged every 2 s. Scale bar, 10 μm.
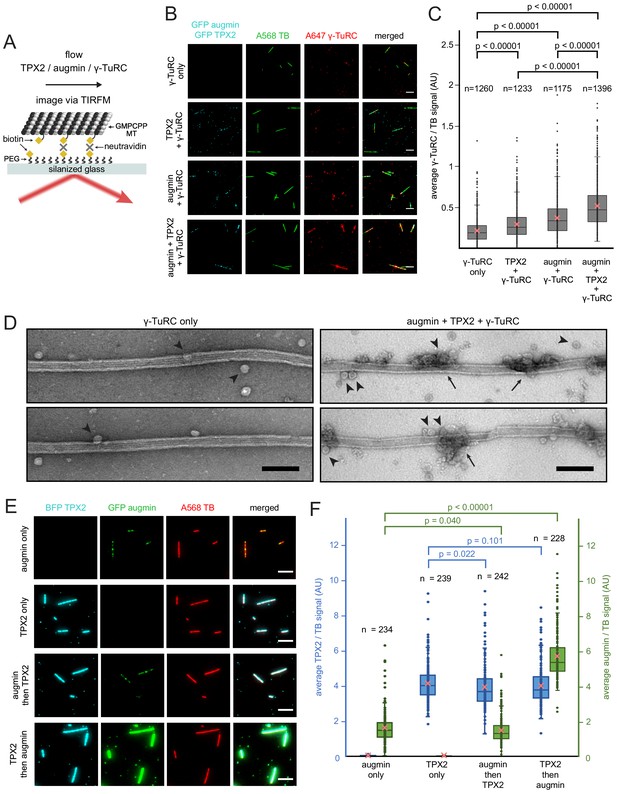
Binding of augmin, TPX2 and γ-TuRC to a template microtubule.
(A) Diagram of the experimental set-up. GMPCPP-stabilized microtubules were attached to a PEG-passivated cover glass with biotin-neutravidin links. (B) γ-TuRC (10 nM) visualized using Alexa647-labeled antibodies (red) along microtubules (green), in the absence or presence of GFP-augmin (50 nM) and GFP-TPX2 (50 nM) (cyan). Scale bars, 5 μm. (C) Boxplot of average γ-TuRC signal relative to the average tubulin signal, where each dot represents one microtubule from the experiment in (B). The number of microtubules (n) was obtained from four replicates using γ-TuRC purified from two different preps. (D) GMPCPP-stabilized microtubules incubated with γ-TuRC (10 nM) only or with augmin (50 nM), TPX2 (50 nM) and γ-TuRC (10 nM), visualized by electron microscopy after uranyl acetate staining. Ring-shaped structures that correspond to γ-TuRCs (arrowheads), and clusters of protein formed on microtubules (arrows) are visible. Scale bars, 100 nm. (E) GFP-augmin (50 nM) (green) and BFP-TPX2 (50 nM) (cyan) visualized along microtubules (red) by themselves or in sequential binding steps. Scale bars, 5 μm. (F) Boxplot of average BFP-TPX2 signal or GFP-augmin signal relative to the average tubulin signal, where each dot represents one microtubule from the experiment in (E). The number of microtubules (n) was obtained from two replicates. For (C) and (F), the boxes extend from 25th to 75th percentiles, the whiskers extend from minimum to maximum values, and the mean values are plotted as crosses. P-values were calculated from independent T-tests.
-
Figure 2—source data 1
Binding of γ-TuRC to a template microtubule.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Binding of TPX2 and augmin to a template microtubule.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig2-data2-v1.xlsx
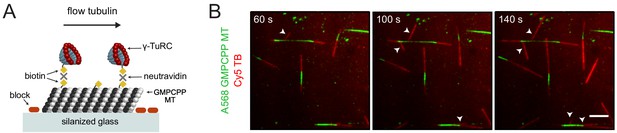
Microtubule nucleation from artificially-attached γ-TuRCs to a template microtubule.
(A) Diagram of the experimental set-up. GMPCPP-stabilized microtubules attached non-specifically to a silanized cover glass, and γ-TuRCs (10 nM) attached to the microtubules with biotin-neutravidin links. Nucleation of new microtubules was visualized using Cy5 tubulin. (B) Using the set-up in (A), the formation of artificial microtubule branches (red, arrowheads) from GMPCPP-stabilized microtubules (green) was observed. Scale bar, 5 μm. Experiment was performed once.
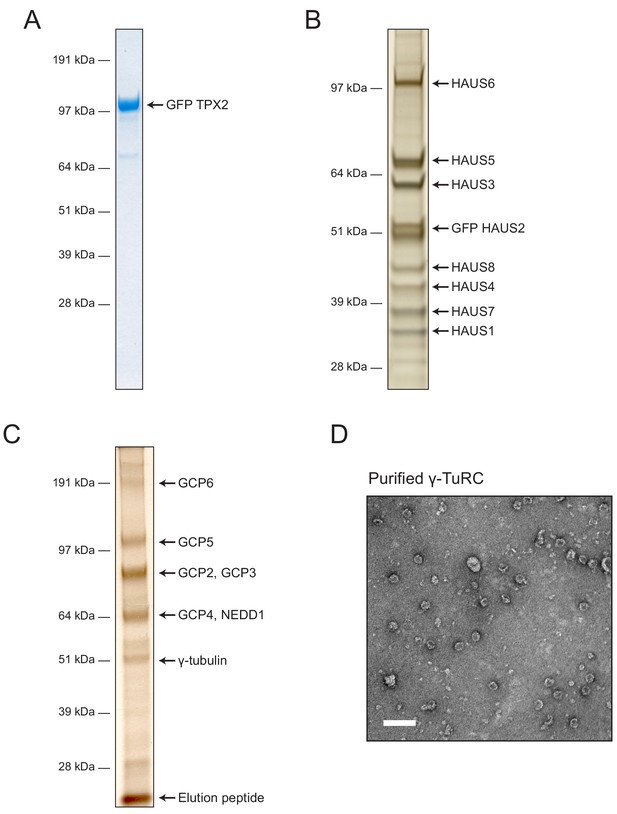
Purified TPX2, augmin and γ-TuRC.
(A) SDS-PAGE of purified GFP-TPX2 visualized by Coomassie Blue staining. (B) SDS-PAGE of purified GFP-labeled augmin holocomplex visualized by silver staining. (C) SDS-PAGE of purified γ-TuRC visualized by silver staining. (D) Purified γ-TuRC visualized using negative stain electron microscopy. Scale bar, 100 nm.
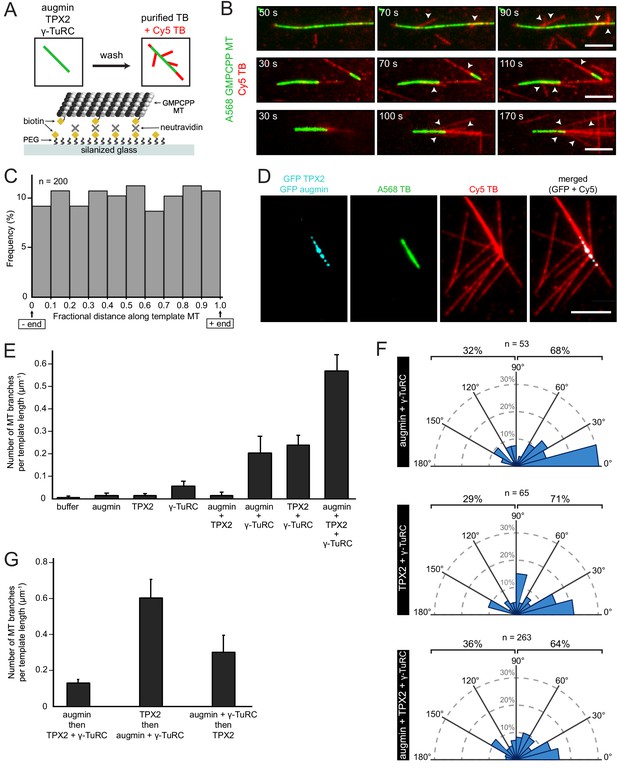
Biochemical reconstitution of branching microtubule nucleation using purified augmin, TPX2 and γ-TuRC.
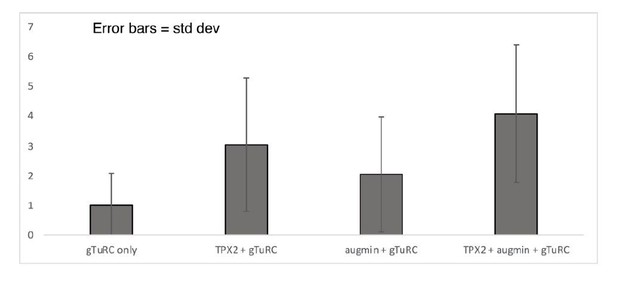
(A) Diagram of the experimental set-up. GMPCPP-stabilized microtubules were attached to a PEG-passivated cover glass with biotin-neutravidin links. Following the binding of augmin (50 nM), TPX2 (50 nM), and γ-TuRC (10 nM), nucleation of new microtubules was visualized using Cy5 tubulin. (B) Using the set-up in (A), the formation of microtubule branches (red, arrowheads) from GMPCPP-stabilized microtubules (green) was observed. Scale bars, 5 μm. See Figure 3—figure supplement 1A and Figure 3—video 1. (C) Fractional distance along the template microtubule where microtubule branches formed. The 0-point on the x-axis denotes nucleation at the minus-end of the template microtubule, while the 1-point denotes nucleation at the plus-end. The number of branching events (n) was obtained from twelve replicates using γ-TuRC purified from four different preps. (D) Same as (A), microtubule branches (red) grow from distinct GFP-augmin and GFP-TPX2 puncta (cyan) localized on GMPCPP-stabilized microtubules (green). (E) Number of microtubule branches per field of view after 4 min, normalized to the length of template microtubule available, for all the combinations of branching factors. Values are the mean of four replicates using γ-TuRC purified from one prep, and error bars represent standard error of the mean. (F) Angle of branching for three different combinations of branching factors. The number of branching events (n) was obtained from eight replicates using γ-TuRC purified from two different preps in the case of augmin + γ-TuRC and TPX2 + γ-TuRC, and from twelve replicates using γ-TuRC purified from four different preps in the case of augmin + TPX2 + γ-TuRC. (G) Number of microtubule branches per field of view after 4 min, normalized to the length of template microtubule available, for different binding sequences. Values are the mean of four replicates using γ-TuRC purified from one prep, and error bars represent standard error of the mean.
-
Figure 3—source data 1
Position of microtubule branches along the template microtubule.
Numerical data obtained from the experiment in Figure 3A–B, and represented graphically in Figure 3C.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Quantification of branched microtubules.
Numerical data obtained from the experiment in Figure 3A–B and Figure 3—figure supplement 2, and represented graphically in Figure 3E.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Angles of branching microtubule nucleation.
Numerical data obtained from the experiment in Figure 3A–B and Figure 3—figure supplement 2, and represented graphically in Figure 3F.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Quantification of non-branched microtubules.
Numerical data obtained from the experiment in Figure 3A–B and Figure 3—figure supplement 2, and represented graphically in Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig3-data4-v1.xlsx
-
Figure 3—source data 5
Quantification of branched microtubules.
Numerical data represented graphically in Figure 3G.
- https://cdn.elifesciences.org/articles/49797/elife-49797-fig3-data5-v1.xlsx
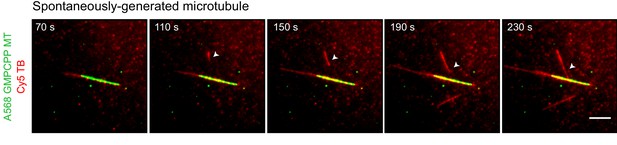
Microtubules can spontaneously form in solution and subsequently interact with the template GMPCPP-stabilized microtubule.
Time-lapse images from the experiment in Figure 3B showing an example of a microtubule (red, arrowhead) that is spontaneously nucleated in solution and contacts the GMPCPP-stabilized template microtubule (green) afterwards. Scale bar, 5 μm.
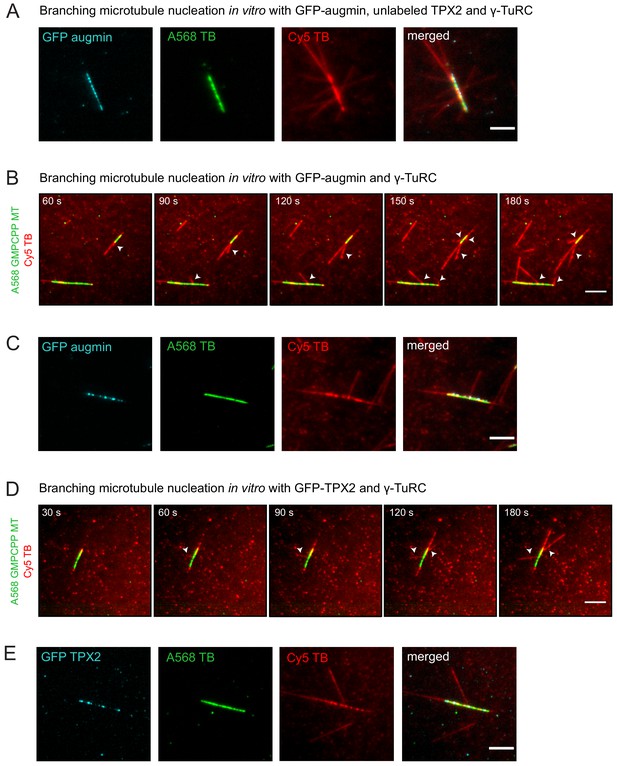
Branching microtubule nucleation with augmin + γ-TuRC and TPX2 + γ-TuRC.
(A) Similar to the experiment in Figure 3A–D, but using an unlabeled version of TPX2. GFP-augmin is present in the observed puncta on template microtubules. (B) Similar to the experiment in Figure 3A–B, but only augmin and γ-TuRC were bound to the GMPCPP-stabilized microtubule. The formation of some microtubule branches (red, arrowheads) from GMPCPP-stabilized microtubules (green) was observed. (C) Same as (B), microtubule branches (red) grow from distinct GFP-augmin puncta (cyan) localized on GMPCPP-stabilized microtubules (green). (D) Similar to the experiment in Figure 3A–B, but only TPX2 and γ-TuRC were bound to the GMPCPP-stabilized microtubule. The formation of some microtubule branches (red, arrowheads) from GMPCPP-stabilized microtubules (green) was also observed. (E) Same as (D), microtubule branches (red) grow from distinct GFP-TPX2 puncta (cyan) localized on GMPCPP-stabilized microtubules (green). Scale bars, 5 μm.

Effect of augmin and TPX2 on de novo microtubule nucleation.
(A) Quantification of non-branched microtubules from the experiment in Figure 3E. Values are the mean of four replicates using γ-TuRC purified from one prep, and error bars represent standard error of the mean. (B) Microtubule nucleation assay in solution with purified augmin (50 nM) and γ-TuRC (5 nM). (C) Microtubule nucleation assay in solution with purified TPX2 (10 nM) and γ-TuRC (5 nM). (D) Microtubule nucleation assay in solution with purified TPX2 (50 nM) and γ-TuRC (5 nM). Scale bars, 20 μm.
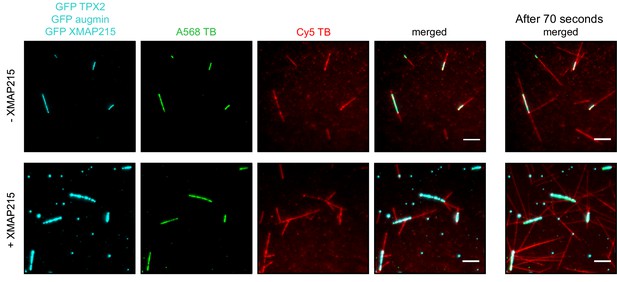
Reconstitution of branching microtubule nucleation using purified augmin, TPX2, γ-TuRC and XMAP215.
Similar to the experiment in Figure 3B, but comparing the effect of having GFP-XMAP215 in the final solution of Cy5 tubulin. The images correspond to the first frame of the time-lapse collected. The panels on the right (merged only) correspond to the same fields of view 70 s later. Scale bar, 5 μm. Experiment was performed once.
Reconstitution of branching microtubule nucleation using purified augmin, TPX2 and γ-TuRC (related to Figure 3B).
A GMPCPP-stabilized microtubule (green) with bound augmin (50 nM), TPX2 (50 nM), and γ-TuRC (10 nM), served as a template for the nucleation of branched microtubules (red). The sample was imaged every 2 s. Scale bar, 5 μm.
Tables
| Average Binding | Std Dev | Average Branching | Std Dev | |
|---|---|---|---|---|
| gTuRC only | 0.21294 | 0.14638 | 0.05715 | 0.04711 |
| TPX2 + gTuRC | 0.29349 | 0.18580 | 0.23863 | 0.09174 |
| augmin + gTuRC | 0.37321 | 0.22619 | 0.20415 | 0.14939 |
| TPX2 + augmin + gTuRC | 0.51905 | 0.26268 | 0.56789 | 0.14644 |
| Branching / Binding | Normalized Branching/Binding | Std Dev | ||
| gTuRC only | 0.2684 | 1.0000 | 1.0733 | |
| TPX2 + gTuRC | 0.8131 | 3.0293 | 2.2437 | |
| augmin + gTuRC | 0.5470 | 2.0381 | 1.9365 | |
| TPX2 + augmin + gTuRC | 1.0941 | 4.0763 | 2.3154 |
| Individual branching measurements | Individual branching measurements divided by the average binding | Average of the individual branching/ binding ratios | |
|---|---|---|---|
| γ-TuRC | 0 | 0 | 0.268392398 |
| 0.074895147 | 0.51719484 | ||
| 0.042944258 | 0.201673044 | ||
| 0.110766504 | 0.520177064 | ||
| TPX2 + γ-TuRC | 0.281327868 | 0.958560319 | 0.813061147 |
| 0.159515074 | 0.543511104 | ||
| 0.165993471 | 0.565584759 | ||
| 0.347664851 | 1.185488405 | ||
| augmin + γ-TuRC | 0.139938427 | 0.374958943 | 0.547017648 |
| 0.039399551 | 0.105569387 | ||
| 0.387446726 | 1.03814669 | ||
| 0.49825122 | 1.03814669 | ||
| TPX2 + augmin + γ-TuRC | 0.627385371 | 1.208718565 | 1.094087931 |
| 0.415528909 | 0.800556611 | ||
| 0.743284806 | 1.43201003 | ||
| 0.485346276 | 0.935066548 |
| Normalized Branching / Binding | Std Dev | |||
|---|---|---|---|---|
| γ-TuRC only | 0.999963327 | 1.073272955 | ||
| TPX2 + γ-TuRC | 3.029284911 | 2.243660565 | ||
| augmin + γ-TuRC | 2.038064177 | 1.936445753 | ||
| TPX2 + augmin + γ-TuRC | 4.076331128 | 2.315317404 | ||
| Z-scores | ||||
| Ignoring the StdDev of gTuRC only | Ignoring the StdDev of the other sample | |||
| γ-TuRC only vs TPX2 + γ-TuRC | -0.90446907 | Not significant | 1.890778645 | significant |
| γ-TuRC only vs augmin + γ-TuRC | -0.536085686 | Not significant | 0.967229115 | Not significant |
| γ-TuRC only vs TPX2 + augmin + γ-TuRC | -1.328702404 | Not significant | 2.866342442 | significant |