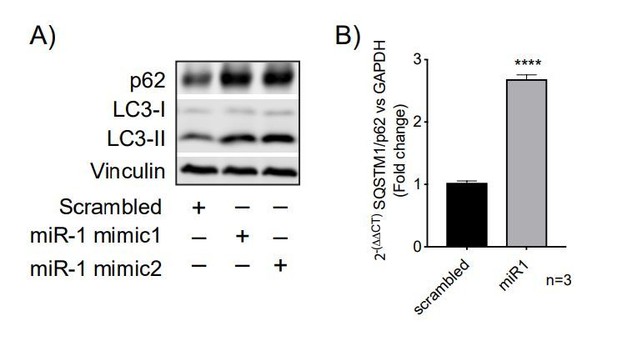
Interferon-β-induced miR-1 alleviates toxic protein accumulation by controlling autophagy
Figures
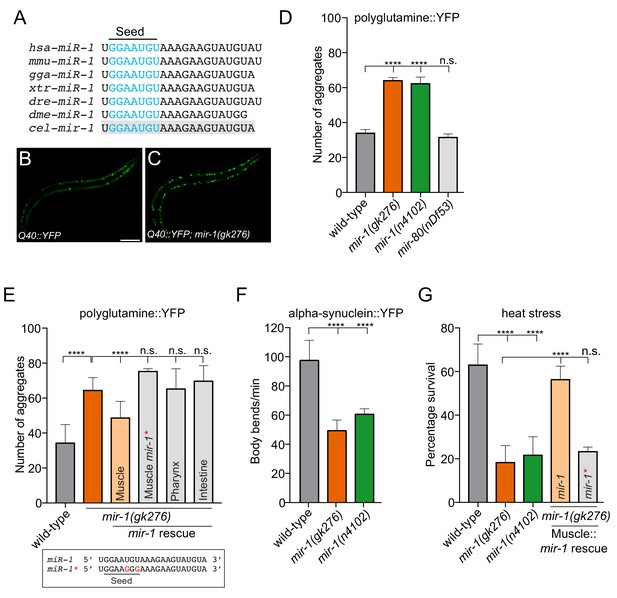
mir-1 protects against proteotoxic stress.
(A) Alignment of mature miR-1 sequences indicates deep conservation. The seed sequence of each miR-1 family member is highlighted in blue and the conservation of C. elegans mir-1 is highlighted in gray. hsa = Homo sapiens, mmu = Mus musculus, gga = Gallus gallus, xtr = Xenopus tropicalis, dre = Danio rerio, dme = Drosophila melanogaster, cel = Caenorhabditis elegans. (B–C) Visualization of Q40::YFP aggregates (green foci) in (B) wild-type and (C) mir-1(gk276) animals. Scale bar, 50 μm. (D) Quantification of Q40::YFP aggregation in wild-type, mir-1(gk276), mir-1(n4102) and mir-80(nDf53) animals. (E) Quantification of Q40::YFP aggregates in wild-type, mir-1(gk276) and mir-1(gk276) animals transgenically-expressing the mir-1 hairpin in body wall muscle (myo-3 promoter), pharynx (myo-2 promoter) or intestine (ges-1 promoter). Mutation of the mir-1 seed sequence (Muscle mir-1*) abrogates rescue from body wall muscle. Mature mir-1 sequences (wild-type mir-1 or mutated mir-1*) used for rescue experiments are shown (box). Red nucleotides indicate the mutations in the seed sequence used in mir-1* rescue experiments, which are predicted to hinder interactions with mir-1 targets. (F) Body bends in wild-type, mir-1(gk276) and mir-1(n4102) mutant animals expressing α-synuclein::YFP. (G) Survival of wild-type, mir-1(gk276) and mir-1(n4102) animals after exposure to 4 hr of 35°C heat stress. Transgenic expression of wild-type mir-1 hairpin, but not mutated mir-1*, in body wall muscle rescues mir-1(gk276) heat stress sensitivity. All experiments were performed in triplicate and at least 10 animals were scored per experiment. Error bars show standard error of the mean (SEM). ****p<0.0001, n.s. not significant to the control (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test).
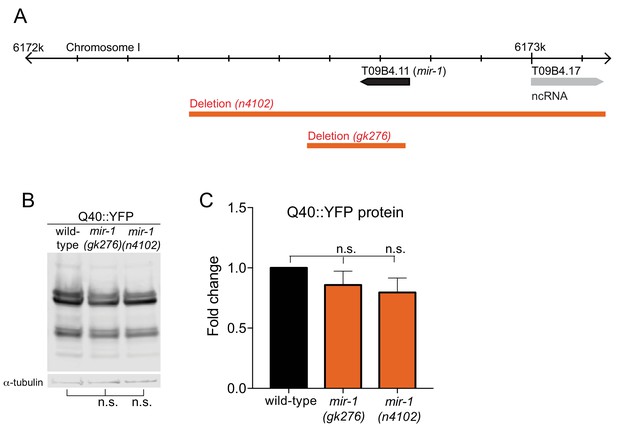
Quantification of Q40::YFP Expression.
(A) Location of mir-1(T09B4.11) on chromosome I, reverse strand of assembly; http://www.wormbase.org, WS258, showing the two deletion strains used in this study, gk276 and n4102 (orange bars). (B–C) WB analysis (B) and quantification (C) of Q40::YFP protein lysates from wild-type, mir-1(gk276) and mir-1(n4102) animals for YFP expression using an α-GFP antibody and α-tubulin antibody as a loading control (n = 3). n.s. not significant (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test).
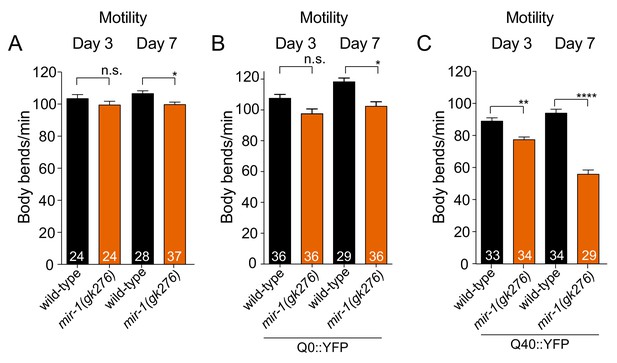
Motility Analysis.
(A–C) Quantification of body bends in wild-type and mir-1(gk276) mutant animals without a transgene (A), expressing the Q0::YFP transgene (B) or expressing the Q40::YFP transgene (C). All experiments were performed in triplicate (number of animals scored are shown in each bar). ± SEM. *p<0.05, **p<0.01, ****p<0.0001, n.s. not significant (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test).
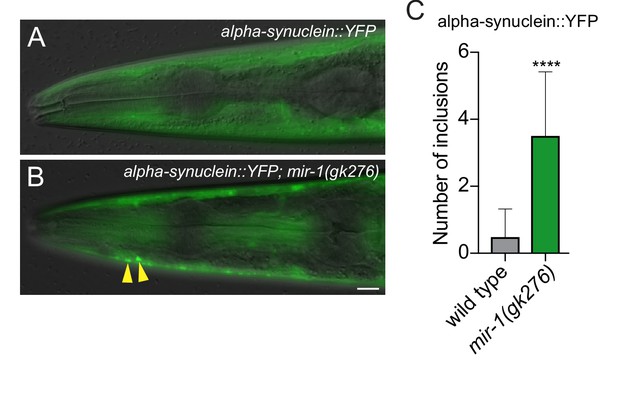
mir-1 prevents the formation of α-synuclein inclusions.
(A–B) Visualization of α-synuclein::YFP inclusions (yellow arrowheads) in (A) wild-type and (B) mir-1(gk276) animals in the first day of adulthood. Scale bar, 50 μm. (C) Quantification of α-synuclein::YFP inclusions in wild-type and mir-1(gk276) animals. n > 25. ± SEM ****p<0.0001 (Welch's t-test).
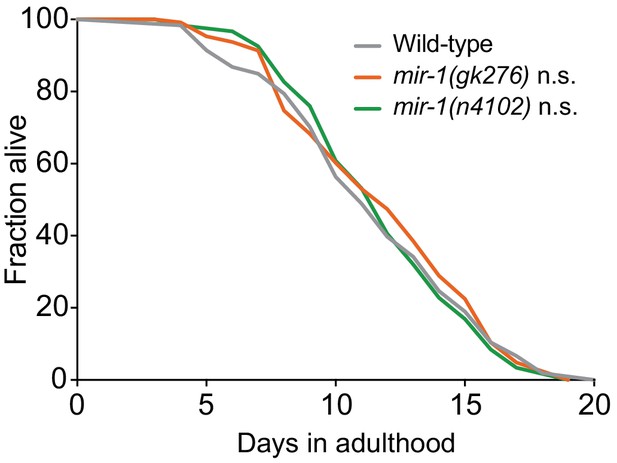
mir-1 Lifespan Analysis.
(A) Lifespan analysis of wild-type (n = 121), mir-1(gk276) (n = 131) and mir-1(n4102) (n = 125) animals. Log-rank (Mantel-Cox) test - n.s. not significant.
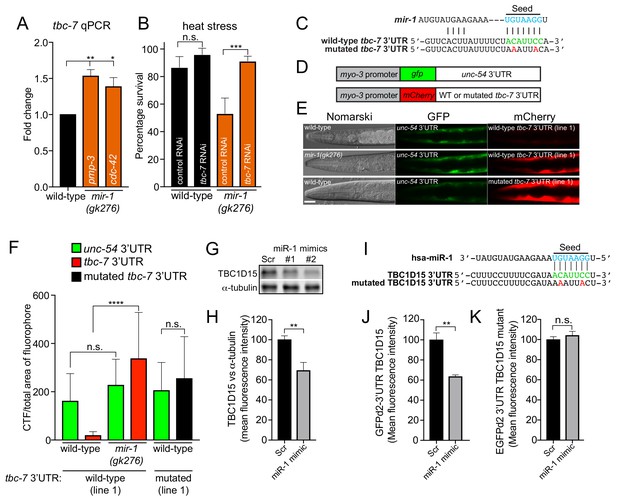
miR-1 directly regulates TBC 3′UTRs in C. elegans and mammals.
(A) Relative tbc-7 mRNA levels measured by quantitative real-time PCR in L4 larvae. Data normalized to values for wild-type worms. Two independent reference genes (pmp-3 and cdc-42) were used. Error bars show standard error of the mean (SEM) obtained from n = 3 biological replicates and three technical replicates each. **p<0.001, *p<0.005 (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test). (B) Survival of wild-type and mir-1(gk276) animals (incubated on control (L4440) or tbc-7 RNAi bacteria) after exposure to 4 hr of heat stress (35°C) (n = 30). ***p<0.001, n.s. not significant (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test). (C) Predicted mir-1 binding site on the 3′UTR of tbc-7 mRNA (green) and seed sequence in mir-1 (blue). Mutated nucleotides in the tbc-7 3′UTR for experiments (E–F) are in red. (D) Indicated DNA constructs were co-transformed as multi-copy extrachromosomal arrays for experiments in (E–F). (E) Expression of heterologous reporter transgenes for control unc-54 3′UTR (gfp) and wild-type and mutated tbc-7 3′UTR (mCherry) constructs in body wall muscle. (F) Quantification of gfp and mCherry fluorescence of transgenic animals calculated as CTF/total area of fluorophore (n = 30). ****p<0.0001, n.s. not significant (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test). (G) WB of TBC1D15 and α-tubulin and (H) quantified bands from HeLa cells transfected with scrambled (Scr) or miR-1 mimics (n = 5). Data are mean fluorescence intensities ± SEM. **p<0.01 (Students t-test). (I) Predicted miR-1 binding site on the 3′UTR of TBC1D15 mRNA (green) and seed sequence in miR-1 (blue). Mutated nucleotides in the TBC1D15 3′UTR for experiments (L–M) are in red. (J–K) Quantification of flow cytometry analysis of HeLa cells co-expressing scrambled (Scr) or miR-1 mimic together with (J) GFPd2-3′UTR TBC1D15 (n = 4) or (K) mutated GFPd2-3′UTR TBC1D15mutant (n = 5). Data are mean fluorescence intensities ± SEM, **p<0.01 (Students t-test).
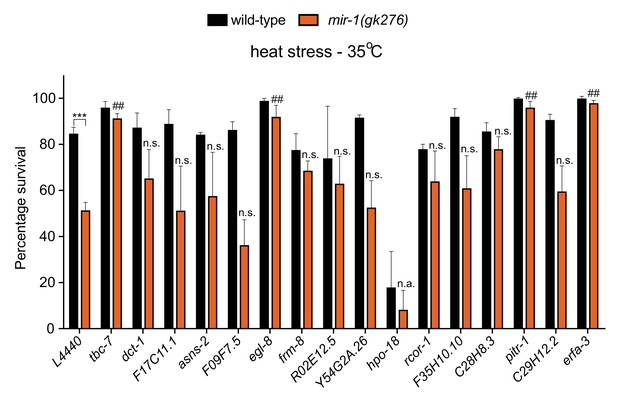
RNAi screen to identify mir-1 targets important for the heat stress response.
Survival of wild-type and mir-1(gk276) animals after exposure to 4 hr of 35°C heat stress. Animals were incubated on RNAi bacteria to reduce expression of predicted mir-1 targets (TargetScanWorm release 6.2). L4440 = control RNAi bacteria. n = 30. ***p<0.001, wild-type compared to mir-1(gk276) on control RNAi bacteria. ##p<0.001 and n.s. not significant when comparing knockdown of predicted mir-1 target to control RNAi in mir-1(gk276) animals (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test). n.a. - hpo-18 RNAi causes lethality in wild-type and mir-1(gk276) animals. All experiments were performed in triplicate.
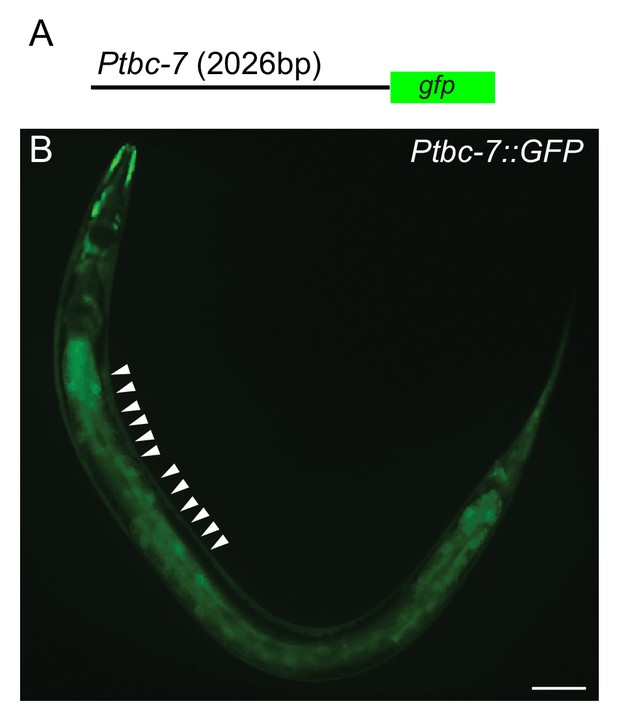
tbc-7 expression pattern.
(A) DNA construct containing a 2026 bp tbc-7 promoter upstream of gfp coding sequence was used to generate an extrachromosomal array to report tbc-7 expression. (B) Expression of gfp controlled by the tbc-7 promoter is detected in the intestine, unidentified head cells and BWM cells (white arrowheads). Scale bar, 20 μm.
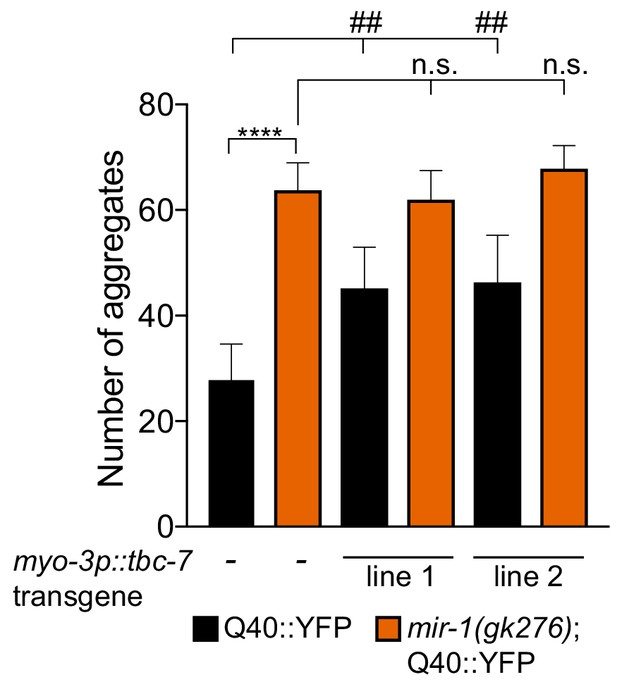
Overexpression of tbc-7 causes Q40::YFP aggregation.
Quantification of Q40::YFP aggregates in wild-type and mir-1(gk276) animals transgenically-expressing tbc-7 cDNA in body wall muscle (myo-3 promoter). Experiments were performed in triplicate (n = 30). Error bars show standard error of the mean (SEM). ****p<0.0001, ## p<0.001 compared to the control and n.s. not significant compared to mir-1(gk276) (one-way ANOVA analysis, followed by Dunnett’s multiple comparison test).

miR-1 targeting of TBC proteins is conserved.
Predicted mir-1 binding sites are found in the 3′UTRs of mRNAs that encode TBC proteins in C. elegans (tbc-7), D. melanogaster (Skywalker) and humans (TBC1D15). This conservation is found in all vertebrate species examined (Targetscan). The mir-1 seed sequences are shown in blue and the predicted tbc-7-related 3′UTRs are shown in green.
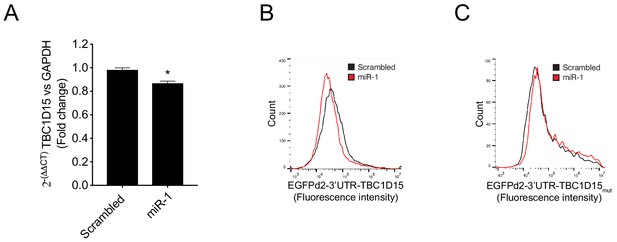
TBC1D15 3′UTR analysis.
(A) Relative TBC1D15 mRNA levels normalized to GAPDH measured by qRT-PCR from HeLa cells expressing scrambled miRNA or miR-1 mimic. Bar graph show fold changes compared to scrambled control ± SEM (n = 3). (B–C) Representative fluorescence intensity histograms from flow cytometry analysis of HeLa cells expressing scrambled miRNA (Scr) or miR-1 mimic together with (B) GFPd2-3′UTR-TBC1D15 or (C) GFPd2-3’UTR-TBC1D15 containing mutated miR-1 target sequence. Quantification of these histogram data is shown in Figure 2J–K. Wild-type hsa TBC1D15 3’UTR = 5′-CUUUCCUUUUCGAUAACAUUCCU-3′ and mutated hsa TBC1D15 3’UTR = 5′-CUUUCCUUUUCGAUAAAAUUACU-3′.
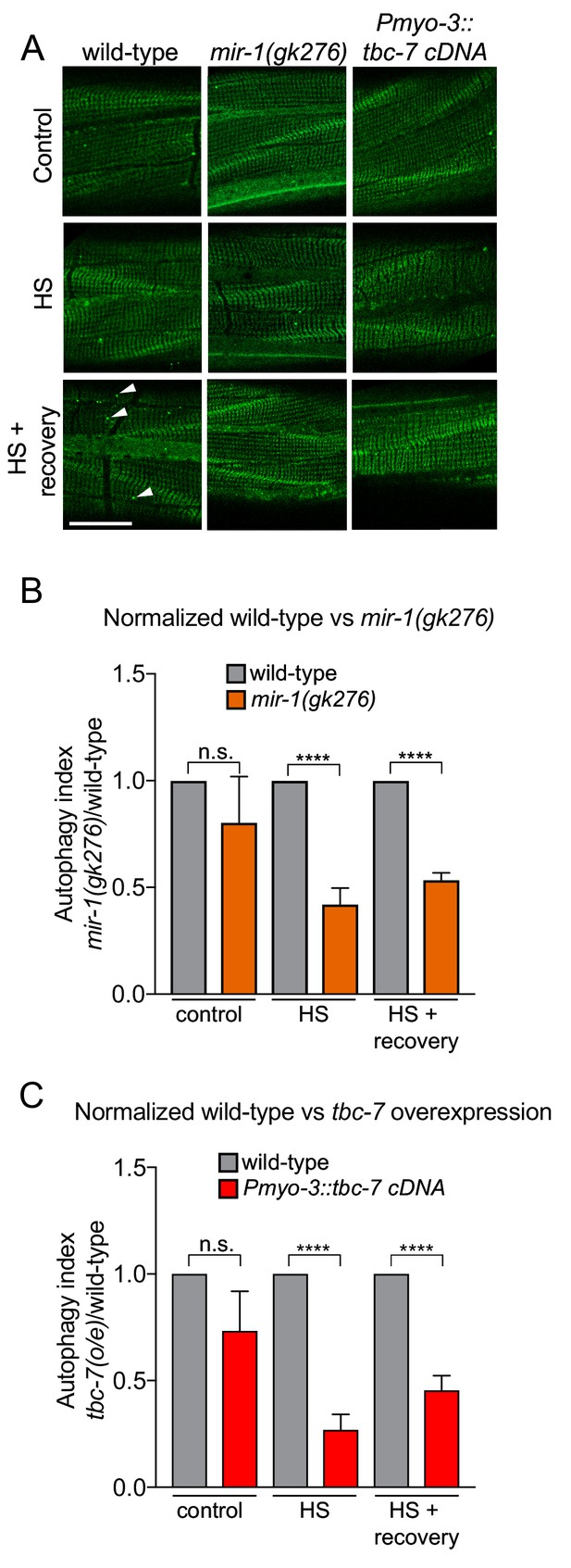
mir-1 and tbc-7 control stress-induced autophagy.
(A) Fluorescent images of BWM expressing GFP::LGG-1/Atg8 in wild-type, mir-1(gk276) and Pmyo-3::tbc-7 overexpressing animals under control conditions, immediately after heat shock for 1 hr at 35°C (HS) or 1 hr after recovery from heat shock at 15°C (HS + recovery). GFP::LGG-1 puncta = arrowheads. Scale bar, 10 μm. (B–C) Quantification of GFP::LGG-1/Atg8 puncta in BWM of animals and conditions shown in (A). The values represent the number of green puncta in mir-1(gk276) (B) and Pmyo-3::tbc-7 overexpressing (C) animals normalized to one green puncta in wild-type animals for each condition. n > 15. ± SEM ****p<0.0001, n.s. not significant (Welch's t-test).
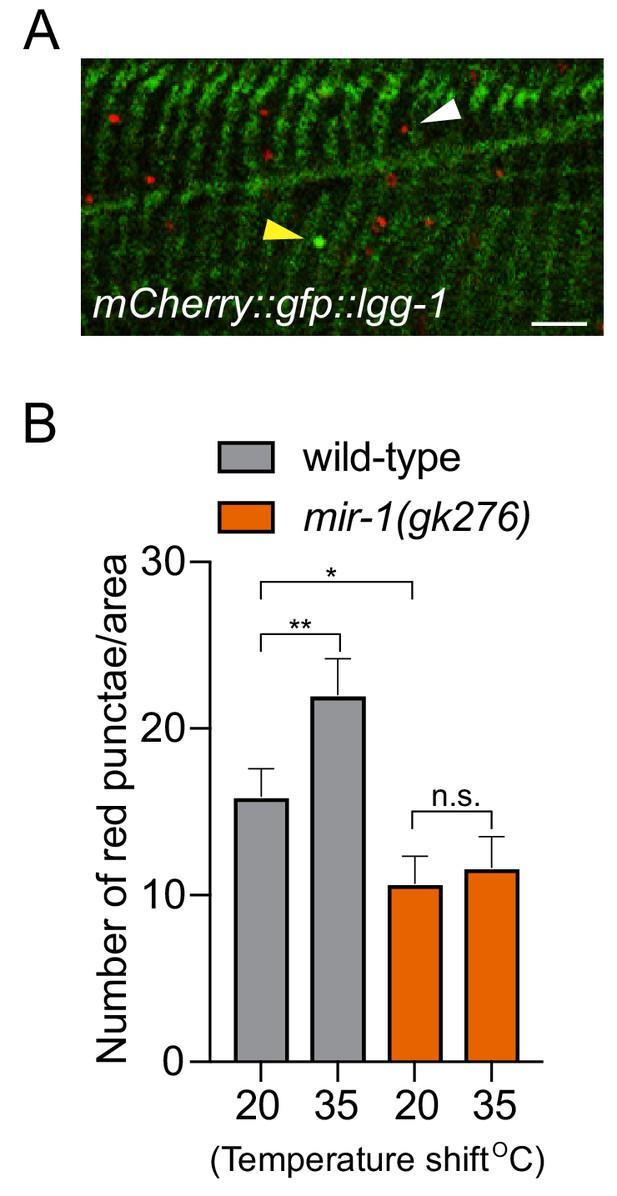
mir-1 controls stress-induced autophagy.
(A) Fluorescent image of BWM expressing mCherry::GFP::LGG-1 in wild-type animals. Yellow arrowhead = autophagosome and white arrowhead = autolysosome. Scale bar, 10 μm. (B) Quantification of autolysosomes in BWM of wild-type and mir-1(gk276) animals incubated at 20°C and then shifted for 1 hr at 35°C. The values represent the mean number of red puncta (autolysosomes). n > 15. ± SEM ****p<0.0001, n.s. not significant (Welch's t-test).
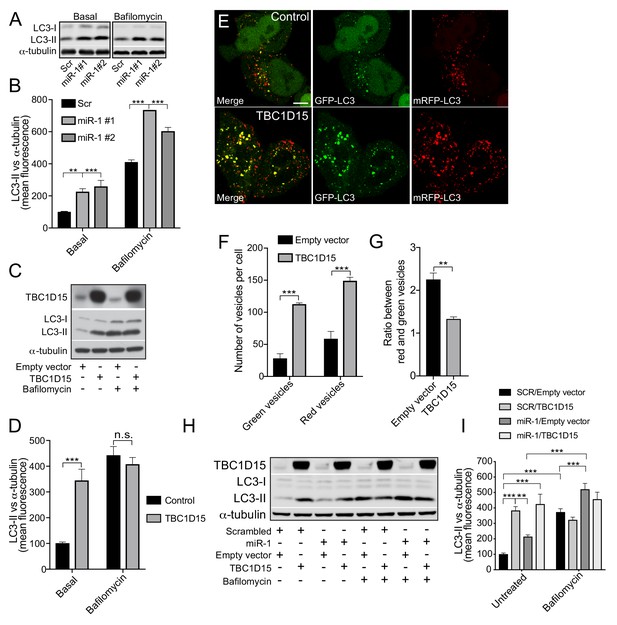
Human miR-1 regulates autophagy by controlling TBC1D15 expression.
(A) WB and (B) quantification of LC3-II normalised to α-tubulin from HeLa cells expressing Scr or miR-1 mimics +/- bafilomycin. Data are mean fluorescence intensities of bands ± SEM (n = 3–5). **p<0.01, ***p<0.001 (one-way ANOVA with Dunnett’s correction). (C) WB and (D) quantification of LC3-II normalised to α-tubulin from HeLa cells expressing empty vector (control) or TBC1D15 overexpression vector +/- bafilomycin. Data are mean fluorescence intensities of bands ± SEM normalised to α-tubulin (n = 5). n.s. not significant to the control, ***p<0.001 (two-way ANOVA with Bonferroni correction). (E) IF images of HeLa cells stably expressing mRFP-GFP-LC3 and transfected with empty vector (control) or TBC1D15 overexpression vector. Scale bar, 10 μm. (F) Quantification of green and red vesicles and (G) red/green vesicle ratio from (E) ± SEM (n = 3, 12–14 cells per replicate). **p<0.01, ***p<0.001 (Student’s t-test). (H) WB and (I) quantification of HeLa cells co-transfected with Scr or miR-1 mimic together with empty vector or TBC1D15 overexpression vector +/- bafilomycin. Data are mean fluorescence intensities of LC3-II bands normalized to α-tubulin ± SEM (n = 7). **p<0.01, ***p<0.001 (two-way ANOVA with Bonferroni correction).
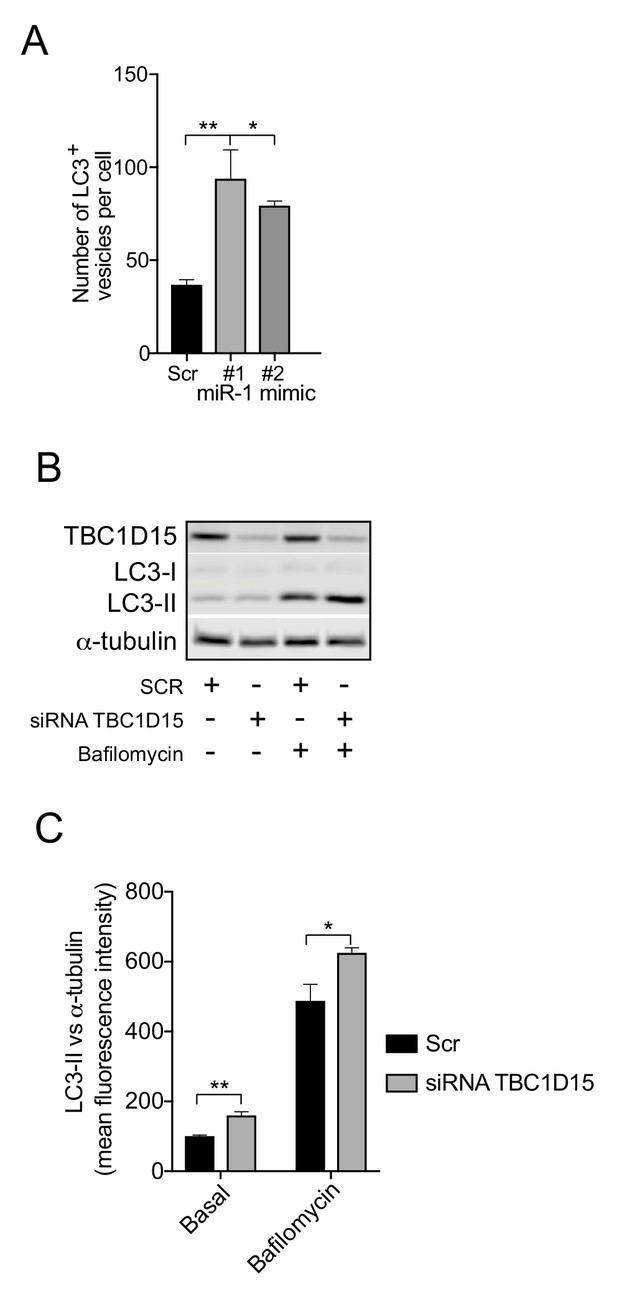
miR-1 and TBC1D15 control autophagy.
(A) Quantification of the mean number of LC3-positive vesicles per HeLa cell ± SEM expressing scrambled miRNA (Scr) or independent miR-1 mimics immunostained with antibodies against LC3 (n = 3). *p<0.05, **p<0.01 (one-way ANOVA with Dunnett’s correction). (B) WB of HeLa cells transfected with Scr siRNA or siRNA against TBC1D15 in the presence or absence of bafilomycin (400 nM for 4 hr). (C) Mean fluorescence intensities of LC3-II WB bands ± SEM normalized to α-tubulin (n = 4). *p<0.05, **p<0.01 (Student’s t-test).
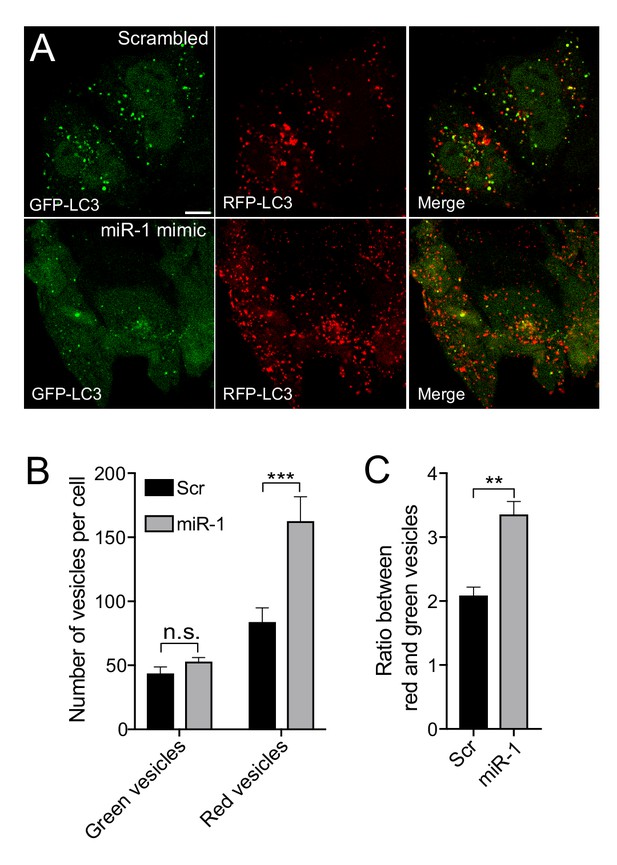
miR-1 overexpression induces autophagy flux.
(A) IF images of HeLa cells stably expressing mRFP-GFP-LC3 and transfected with scrambled miRNA (Scr) or miR-1 mimic. Scale bar, 10 μm. (B) Quantification of mean number of green and red vesicles per cell and (D) the red/green vesicle ratio ± SEM (n = 4). **p<0.01, ***p<0.001 (Student’s t-test).
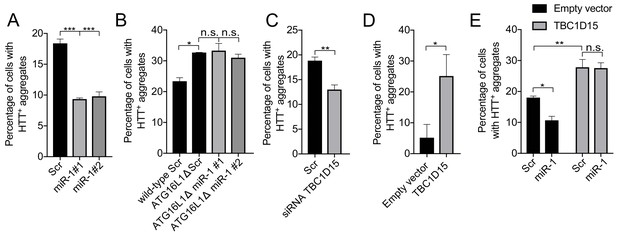
miR-1 reduces mutant Huntingtin aggregation through the autophagy pathway.
(A) Quantification of the percentage of cells containing HTT-positive aggregates co-expressing scrambled (Scr) or miR-1 mimics with EGFP-HTTQ74 for 48 hr ± SEM (n = 3, 200–400 cells per replicate). ***p<0.001 (one-way ANOVA). (B) CRISPR/Cas9 ATG16L1 knockout HeLa cells co-expressing scrambled (Scr) or miR-1 mimics with EGFP-HTTQ74 for 48 hr. Quantification of the percentage of cells containing HTT-positive aggregates ± SEM (n = 3, 200–400 cells per replicate). *p<0.05 (Student’s t-test), n.s. not significant (one-way ANOVA). (C–E) Quantification of the percentage of cells containing HTT-positive aggregates in HeLa cells co-expressing EGFP-HTTQ74 with (C) scrambled (Scr) or siRNA against TBC1D15 for 48 hr (n = 4, 200–400 cells per replicate), (D) empty or TBC1D15 overexpression vector for 24 hr (n = 3, 200–400 cells per replicate), or (E) a combination of Scr or miR-1 mimic together with empty or TBC1D15 overexpression vector for 48 hr (n = 6, 200–400 cells per replicate) ± SEM. (C–D) *p<0.05, **p<0.005, n.s. not significant (Student’s t-test) or (E) (two-way ANOVA with Dunnett’s correction).
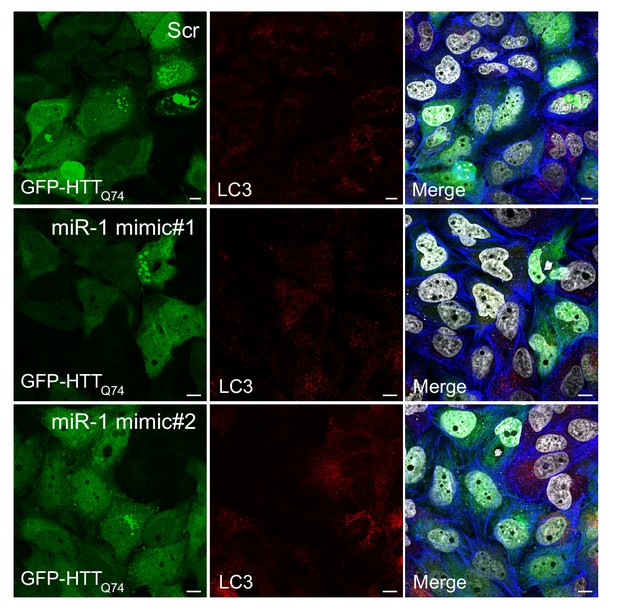
miR-1 overexpression reduces HTTQ74 accumulation IF images of HeLa cells co-expressing scrambled miRNA (Scr) or independent miR-1 mimics with EGFP-HTTQ74 stained with antibodies against LC3 (red), phalloidin (blue), and DAPI (gray).
Scale bar, 10 μm.
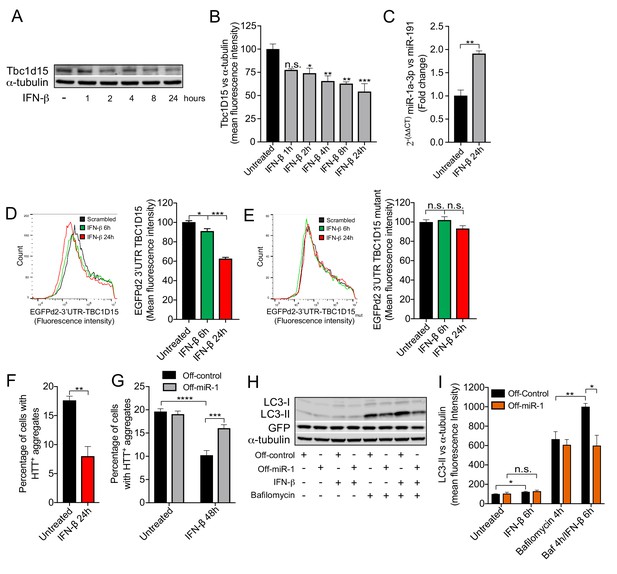
IFN-β induction of miR-1 controls mutant Huntingtin aggregation.
(A) WB and (B) quantification of Tbc1d15 normalized to α-tubulin of cortical neurons from mice treated with recombinant mouse IFN-β (100 U/ml) for 1–24 hr (n = 4). Data are mean fluorescence intensities of bands ± SEM. n.s. not significant to the control, *p<0.05, **p<0.01, ***p<0.001 (one-way ANOVA). (C) RT-PCR of miR-1a-3p normalized to miR-191 from mouse cortical neurons treated with recombinant mouse IFN-β (100 U/ml) for 24 hr (n = 3). **p<0.01 (Student’s t-test). (D–E) Flow cytometry analysis of HeLa cells expressing (D) GFPd2-3′UTR TBC1D15 (n = 4) or (E) mutated GFPd2-3′UTR TBC1D15mutant (n = 5) treated with recombinant human IFN-β (1000 U/ml) for 6 or 24 hr. Data are presented as fluorescence intensity histograms and bar graphs showing mean fluorescence intensities ± SEM. *p<0.05, ***p<0.0001 (one-way ANOVA). (F) Quantification of HTTQ74 aggregates in HeLa cells expressing EGFP-HTTQ74 treated with recombinant human IFN-β (1000 U/ml) for 24 hr. Graph shows percentage of cells containing EGFP-HTTQ74-positive aggregates (n = 4, 400 cells per replicate) ± SEM. **p<0.01 (Student’s t-test). (G) Quantification of HTTQ74 aggregates in HeLa cells expressing GFP-Off-control or GFP-Off-miR-1 (miR-1 hairpin inhibitor) with EGFP-HTTQ74 and treated with recombinant human IFN-β (1000 U/ml) for 48 hr. Graph represents percentage of cells containing EGFP-HTTQ74-positive aggregates (n = 5, 400 cells per replicate) ± SEM. ***p<0.001, ****p<0.0001 (two-way ANOVA with Bonferroni correction). (H) WB of LC3, GFP and α-tubulin and (I) quantification of LC3-II normalized to α-tubulin from HeLa cells stably expressing GFP-Off-Control and GFP-Off-miR-1 treated with recombinant human IFN-β (1000 U/ml) for 6 hr, bafilomycin (400 mM) for 4 hr or in combination (n = 4) ± SEM. *p<0.05, **p<0.01, n.s. not significant (Student’s t-test).
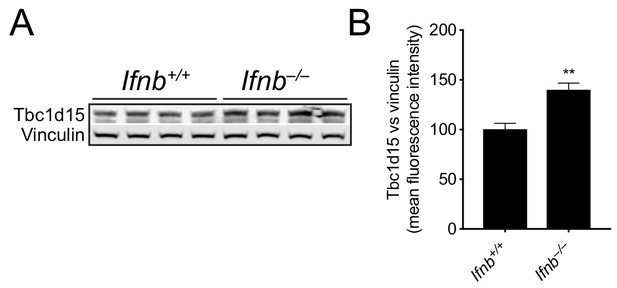
IFN-β regulates TBC1D15 expression the mouse brain.
(A) WB and (B) quantification of Tbc1d15 (normalized to vinculin) in the brain of wild-type (Ifnb+/+) and Ifnb–/– 3 month old male mice (n = 4). Data are mean fluorescence intensities of bands ± SEM. **p<0.01 (Student’s t-test).
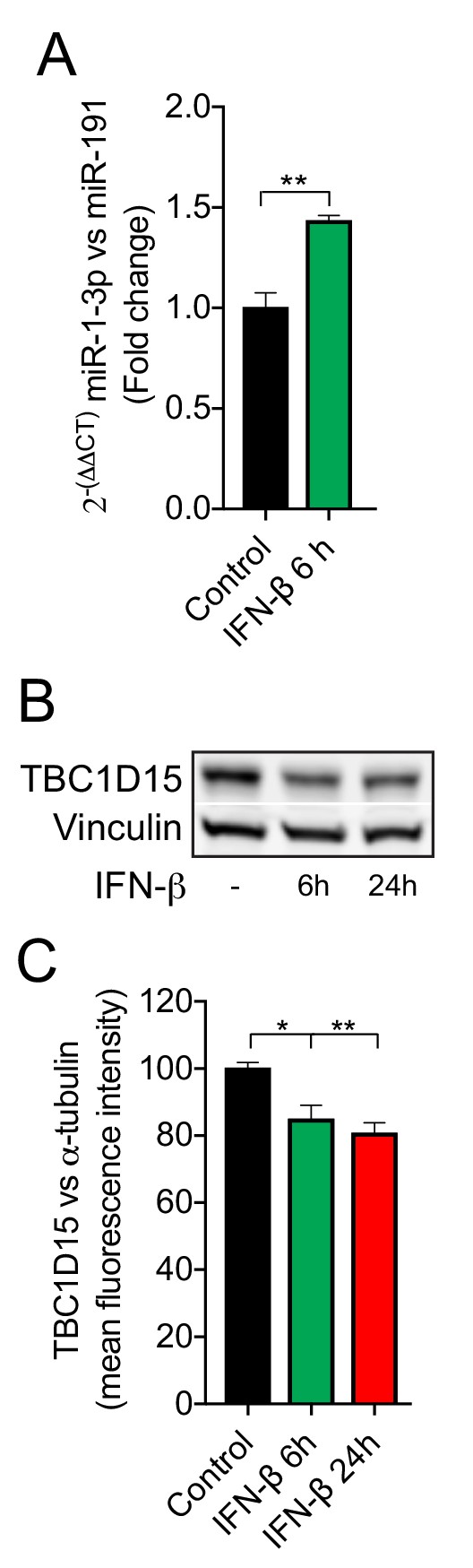
IFN-β regulates miR-1 and TBC1D15 expression in HeLa cells.
(A) RT-PCR of miR-1–3 p normalized to miR-191 from HeLa cells treated with recombinant human IFN-β (1000 U/ml) for 6 hr (n = 3). **p<0.01 (Student’s t-test). (B) WB and (C) quantification of TBC1D15 bands normalised to vinculin from HeLa cells treated with recombinant human IFN-β for 6 or 24 hr (n = 4). Data are mean fluorescence intensities ± SEM. *p<0.05, **p<0.01 (one-way ANOVA).
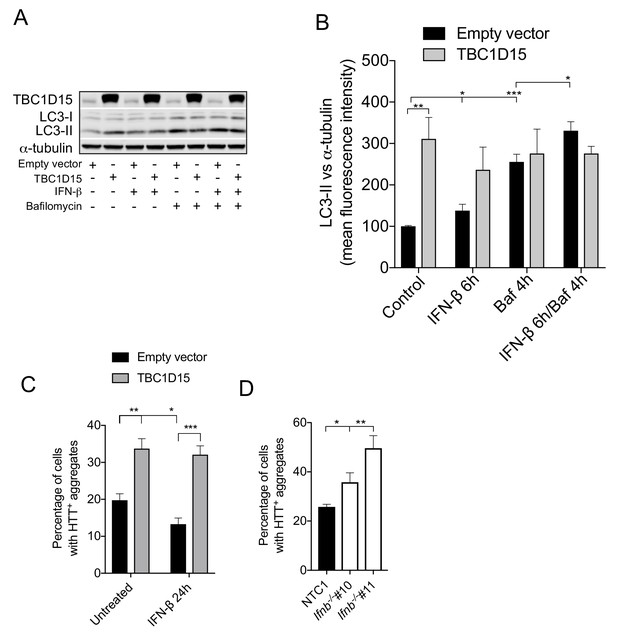
TBC1D15 overexpression abrogates IFN-β-induced reduction of HTTQ74 aggregates.
(A) WB of TBC1D15, LC3 and α-tubulin in HeLa cells expressing empty or TBC1D15 overexpression vector, treated with recombinant human IFN-β (1000 U/ml) for 6 hr, bafilomycin (400 mM) for 4 hr, or a combination of both. (B) Quantification of mean fluorescence intensities of LC3-II bands from (A) (n = 4) ± SEM. *p<0.05, **p<0.01 ***p<0.001 (Student’s t-test). (C) HeLa cells co-expressing EGFP-HTTQ74 with either empty or TBC1D15 overexpression vector with or without recombinant human IFN-β treatment (1000 U/ml) for 24 hr. Graph represents percentage of cells containing EGFP-HTTQ74-positive aggregates ± SEM. *p<0.05, **p<0.01 ***p<0.001 (Student’s t-test). (D) Neuronally differentiated N2A cells with non-targeting control-1 (NTC1) or Ifnb CRISPR/Cas9 knockout co-expressing EGFP-HTTQ74. Graph represents percentage of cells containing EGFP-HTTQ74-positive aggregates (n = 3) ± SEM. *p<0.01, **p<0.001 (Student’s t-test).
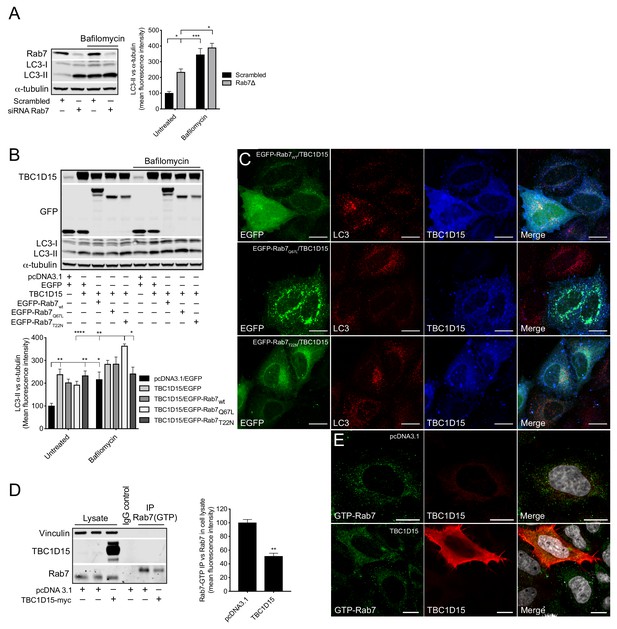
TBC1D15 reduces GTP-bound Rab7.
(A) WB and quantification of LC3-II from HeLa cells transfected with scrambled siRNA or siRNA against Rab7 (Rab7Δ) +/- bafilomycin (4 hr, 400 nM). Data are mean fluorescence intensities of bands ± SEM normalised to α-tubulin (n = 3). *p<0.05, ***p<0.001 (two-way ANOVA). (B) WB and quantification of LC3-II normalised to α-tubulin from HeLa cells co-expressing pcDNA3.1 and EGFP, or TBC1D15 with either EGFP, pIRESneo-myc--Rab7wt, EGFP-Rab7Q67L, or EGFP-Rab7T22N for 24 hr before treatment with bafilomycin (4 hr, 400 nM). Data are mean fluorescence intensities of bands ± SEM normalised to α-tubulin (n = 6). *p<0.05, **p<0.01, ***p<0.001 (two-way ANOVA). (C) Immunofluorescence (IF) images of HeLa cells co-expressing TBC1D15 with pIRESneo-myc-Rab7wt, EGFP-Rab7Q67L or EGFP-Rab7T22N stained with antibodies against LC3 and TBC1D15. Scale bars, 10 μm. (D) WB showing immunoprecipitation (IP) of GTP-bound (active) Rab7 from HeLa expressing pcDNA3.1 or TBC1D15. Data are mean fluorescence intensities of GTP-bound Rab7 (IP) normalized to the endogenous level of Rab7 (cell lysate) ± SEM (n = 3). **p<0.01 (Student’s t-test). (E)IF of HeLa cells expressing empty vector or TBC1D15 stained with antibodies against GTP-bound Rab7, TBC1D15 and DAPI. Scale bar, 10 μm.
Videos
Autophagy flux in cells expressing an empty (control) vector.
HeLa cells stably expressing mRFP-GFP-LC3 were transfected with empty vector (Video 1) or TBC1D15 overexpression vector (Video 2) and live cell imaging was conducted the following day. Notice the presence of large immobile mRFP- and GFP-positive autophagosomes in TBC1D15 overexpressing cells, implying a block in autophagosome maturation. Cells were imaged once every second for a period of 2 min and the movies are displayed at a speed of 10 frames per second.
TBC1D15 overexpression causes large stationary autophagosomes.
Additional files
-
Supplementary file 1
Predicted C. elegans mir-1 target genes.
Predicted mir-1 target genes (TargetScanWorm release 6.2) showing the number and type of putative conserved mir-1 binding sites in their 3′UTRs. Aggregate PCT = probability of conserved targeting.
- https://cdn.elifesciences.org/articles/49930/elife-49930-supp1-v2.xlsx
-
Supplementary file 2
RNA sequencing data.
Differentially expressed genes in mir-1(gk276) animals compared to wild-type for each of the three biological replicate samples. Raw counts and counts per million reads (CPM) are shown. The false discovery cut-off was set at 0.1 and absolute log fold change set to 0.3 (1.2x change in expression). Full dataset is located at NCBI - GSE128968.
- https://cdn.elifesciences.org/articles/49930/elife-49930-supp2-v2.xls
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/49930/elife-49930-transrepform-v2.docx