NINscope, a versatile miniscope for multi-region circuit investigations
Figures
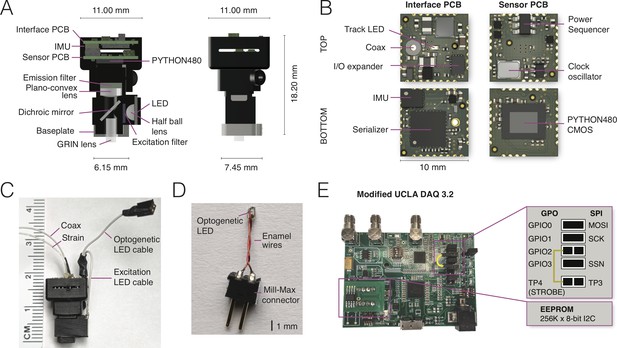
NINscope, a compact, light-weight and versatile miniscope.
(A) Schematics of the NINscope with dimensions in mm. Two 10 by 10 mm HDI printed circuit boards (PCBs), one for interfacing with the data acquisition box (Interface PCB), the other containing the CMOS imaging sensor (Sensor PCB), are stacked and mounted in a 3D printed enclosure. Excitation light from an LED is collimated with a half ball lens, passes through an excitation filter and is reflected by the dichroic mirror onto the specimen. The emitted fluorescence is collected through the GRIN objective lens, and passes the dichroic and a plano-convex lens, which focuses an image onto the CMOS sensor. An emission filter is glued onto the plano-convex lens with optical bonding glue. (B) KiCad renders of the custom-built interface and sensor PCBs with top and bottom views. The interface PCB contains an inertial measurement unit (IMU) for measuring head acceleration and orientation, three LED drivers including one for optogenetic (strobe) control and two for excitation LEDs (one used), as well as a red tracking LED, the serializer, and the IO expander. The sensor PCB contains the PYTHON480 CMOS sensor, clock oscillator and a power sequencer, which provides the image sensor with the necessary voltages as well as their timing and sequence. (C) Photograph of NINscope with coax and strain cables, excitation LED and optogenetic LED cables. (D) Custom-built implantable LED probe for optogenetic stimulation that is connected to NINscope using the optogenetic LED cable. (E) The UCLA Miniscope DAQ card v3.2 was used with minor modifications including a 256 kB x 8-bit I2C EEPROM (STMicroelectronics) and a wired connection bridging general purpose input/output 2 (GPIO2) with test point 4 (TP4). The serial peripheral interface (SPI) signals: master output slave input (MOSI), serial clk (SCK) and slave select (SSN) are connected to GPIO0, GPIO1 and GPIO3 through jumpers.
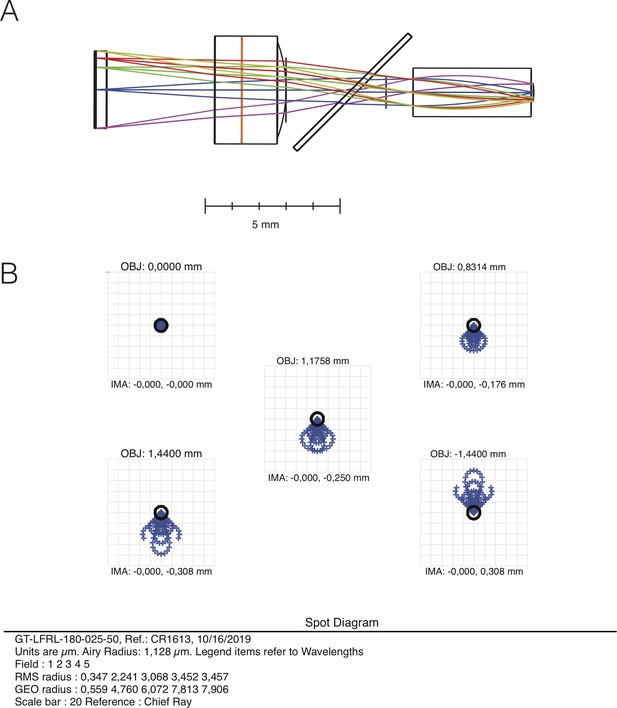
NINscope optical design.
(A) Ray tracing diagram. (B) Spot diagram. Both diagrams were obtained in Zemax Optic Studio.
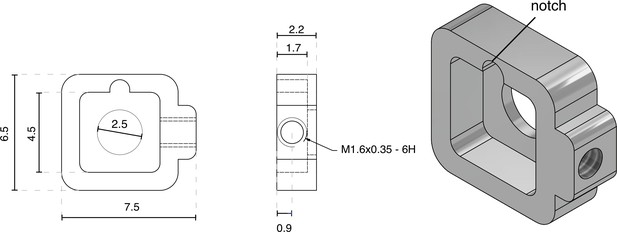
NINscope baseplate.
NINscope is secured in a custom designed baseplate using a hexagonal set screw (M1.6 × 3, 0.7 Hex, part # 913000100016003, Jeveka). The lower half of the NINscope housing has a protrusion that fits in the baseplate notch to add stability. Numbers indicated are in mm.
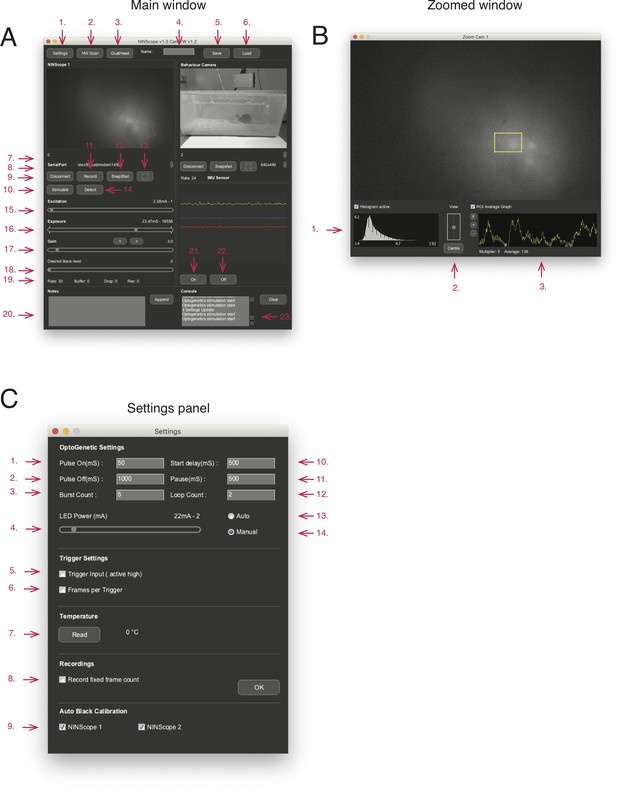
NINscope software.
(A) Acquisition and control software: 1. Access settings panel for optogenetic stimulus parameters, input trigger settings 2. Update the list of connected devices (UVC cameras and serial ports) 3. Switch to dual or single cam mode 4. Name of saved settings file 5. Save: save the current settings in a file 6. Load: load a previously stored settings file 7. Display connected device 8. Serial port selection for device connection 9. Connect or disconnect from a device 10. Initiate manual optogenetic stimulus if enabled 11. Start and stop a recording (disabled in input trigger mode) 12. Take a snapshot 13. Expand camera view 14. Detect which miniscope is connected (tracking LED blinks 5x) 15. Excitation LED current 16. CMOS sensor exposure 17. CMOS gain setting 18. Black level setting (default: auto-calibration) 19. Displays frame rate, number of buffer frames, frame drops and number of frames recorded 20. Text field to add notes, written to disk after pressing the append button. 21. Turn G-sensor logging and display on 22. Turn G-sensor logging and display off 23. Console displays hardware checks upon startup and after a HW scan (see 2.) (B) Zoomed window: opened after hitting the expand camera view icon (see 13 in A). 1. Live histogram showing the distribution of pixel intensity values. 2. Box for panning the region of the sensor to record from with re-centering button. 3. A region-of interest (ROI) can be selected (yellow box) by clicking and dragging the mouse cursor down and to the right in the zoomed window after selecting the 'ROI Average Graph' checkbox. The mean value within this ROI can be plotted live. (C) Settings panel: 1. Optogenetic pulse on duration 2. Optogenetic pulse off duration 3. Optogenetic burst count 4. Optogenetic pulse start delay 5. Optogenetic pulse pause time 6. Optogenetic pulse loop count 7. Start optogenetic stimulation automatically upon record 8. Start optogenetic stimulation upon manual trigger (see 10.) 9. Set current to optogenetic LED 10. Turn on/off trigger input (active high 3.3 V) 11. Set number of frames to record for a trigger input (if not set hold voltage at 3.3 V to keep recording) 12. Read CMOS sensor temperature 13. Record a fixed number of frames for each recording (overrules setting 10). For more information please visit https://github.com/ninscope/Software/wiki/Software-Usage, which has videos demonstrating software usage.
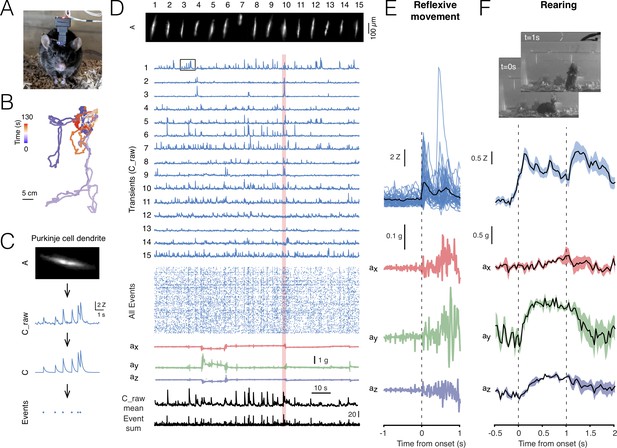
Cerebellar imaging with NINscope.
(A) A mouse wearing a single NINscope mounted over lobule V of cerebellum, where Purkinje cells were selectively transduced with GCaMP6f. (B) Animal position can be extracted from the tracking LED when combined with concurrent webcam recordings. Colors represent the time from the start of recording and the track represents cage exploration over the course of ~2 min. (C) Spatial footprints of Purkinje cell dendrites (A) raw (C_raw) and deconvoluted signals (C) were extracted using CNMF-E (Zhou et al., 2018) after motion correction using NoRMCorre (Pnevmatikakis and Giovannucci, 2017). Event times (Events) were extracted from the deconvolved signals. Signal shown is depicted in the boxed region in D. (D) Spatial footprints of 15 Purkinje cell dendrite arbors (A) and their corresponding calcium transients (C_raw), event raster across all (168) extracted signals, the x, y, z accelerometer channels, as well as the mean raw signal (C_raw mean) and sum of all events (Event sum). (E) Purkinje cell dendrite transients were aligned to acceleration onset in the red shaded area in D. In this example a reflexive movement (twitch) was triggered by a loud clap. (F) Spontaneous behaviors monitored with a webcam can be associated with distinct signatures in the accelerometer data such as during rearing. In this recording a subset of Purkinje cells showed a significant response during animal rearing with transient elevations associated with the lifting and falling phase of the mouse (average of 4 rearings, mean ± SEM, N = 1 mouse). The dashed lines indicate the time points at which the webcam images were captured.
-
Figure 2—source data 1
Cerebellar imaging with NINscope.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig2-data1-v2.zip
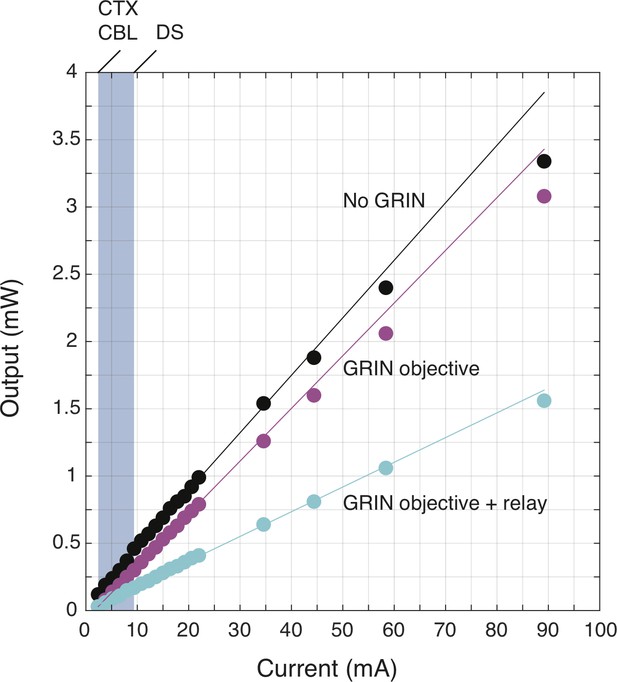
Excitation LED light power as a function of current supplied before and after the GRIN objective and relay lenses.
Excitation LED light power measured as a function of supplied current without or with GRIN lenses. measured directly below the miniscope without a lens (No GRIN), after the GRIN objective (GRIN objective) or after a GRIN objective and a relay lens (GRIN objective + relay). Imaging was performed at current settings ranging from 2.38 to 9.38 mA, which resulted in light output powers of 100–200 µW for regions imaged with a GRIN objective only such as cerebellum (CBL) and cortex (CTX) or with both a GRIN objective and relay such as dorsal striatum (DS).
-
Figure 2—figure supplement 1—source data 1
Excitation LED light power as a function of current supplied measured before and after the GRIN objective and relay lenses.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig2-figsupp1-data1-v2.zip
Cerebellar imaging with NINscope.
Purkinje cell dendrites of cerebellar lobule V were transduced with GCaMP6f and imaged with NINscope. The raw data after motion correction (NoRMCorrre) are shown together with the demixed signal (A*C), which represents activity in spatial footprints of Purkinje cell dendrites (blue) obtained after segmentation with CNMF-E. Simultaneously acquired webcam data are also shown. Acquisition rate 30 Hz, video speed 3x.
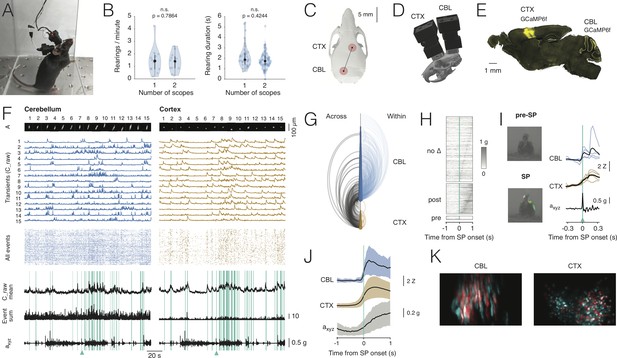
Dual-region imaging with NINscope.
(A) A mouse with two NINscopes mounted over cerebellum and cortex. (B) Behavior was unimpaired as quantified by counting the number of rearings and their duration in mice wearing single or dual miniscopes (rearings/minute, p=0.7864, n.s.; rearing duration, p=0.4244, n.s.; N = 4 mice). (C) Mouse skull with red circles indicating the recording configuration (craniotomy positions). For other possible configurations see Figure 3—figure supplement 1 and Figure 3—figure supplement 2. (D) CAD rendering showing the rostro-caudal placement of two NINscopes to image from cerebellum an cortex concurrently (~8 mm inter-baseplate distance at angles of 15–20°). (E) GCaMP6f was transduced selectively in cerebellar Purkinje cells (lobule VI or simplex lobule) and in neurons of motor cortex. (F) Dual site recordings from cerebellar lobule VI and motor cortex showing responses of segmented neurons (spatial footprints, A) in each region with the Z-mean scored signal, number of co-active Purkinje cell dendrites and compound acceleration signal (axyz, √(x2+y2+z2)). Cyan lines represent epochs where synchronous patterns (SPs) were found across cerebellum and cortex. (G) Combined arc plots for this dataset visualizing intra-cerebellar, intra-cerebral (within) and cerebello-cerebral (across) SPs. Node radii scale by the number of cells that a node connects to. In this example cerebellar neurons with high within SPs also displayed significant SPs across regions. (H) SPs were used to trigger the compound acceleration signal. Behavioral acceleration could be assigned to four categories consisting of no change (no Δ, 64%, sorted by peak response), behavioral acceleration post-SP (31%), pre-SP (4%), or around-SP (1%, not shown). (I) Across-regions SPs are associated with significant deviations from baseline in the accelerometer compound signal. Responsive cells are shown in cerebellum (lobule VI) and cortex, triggered off of the SP. A mouse resting prior to an SP that made a (left, upward) movement around SP onset. Animal movement visualized with optic flow is color-coded. (J) Population averaged responses triggered around detected SPs reveal responses in cerebellum and cortex during accelerometer upslope. (K) Example showing neurons in the cerebellum and cortex that participated in an SP (red) and cells that did not (cyan).
-
Figure 3—source data 1
Dual-region imaging with NINscope.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig3-data1-v2.zip
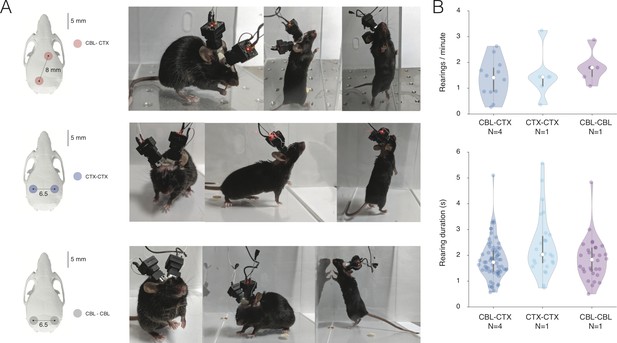
Tested dual NINscope configurations.
(A) Different mice were fitted with two miniscopes above cerebellum and cortex (CBL-CTX, N = 4 mice), two over left and right cortex (CTX-CTX, N = 1 mouse) or two above the left and right (CBL-CBL, N = 1) cerebellar hemispheres. Colored circles denote craniotomy positions for the two miniscopes and the center-to-center distance between the locations (in mm) is indicated. Photographs are shown of mice wearing dual miniscopes in the three conditions. Mice were grooming, eating and rearing. (B) To assess the impact of dual scope recordings on rearing for the (CBL-CTX, CTX-CTX and CBL-CBL) configurations shown, we counted the number and duration of rearings. A comparable number of rearings and rear durations were found for all three conditions.
-
Figure 3—figure supplement 1—source data 1
Data pertaining to rearing behavior with various configurations of NINscope.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig3-figsupp1-data1-v2.zip
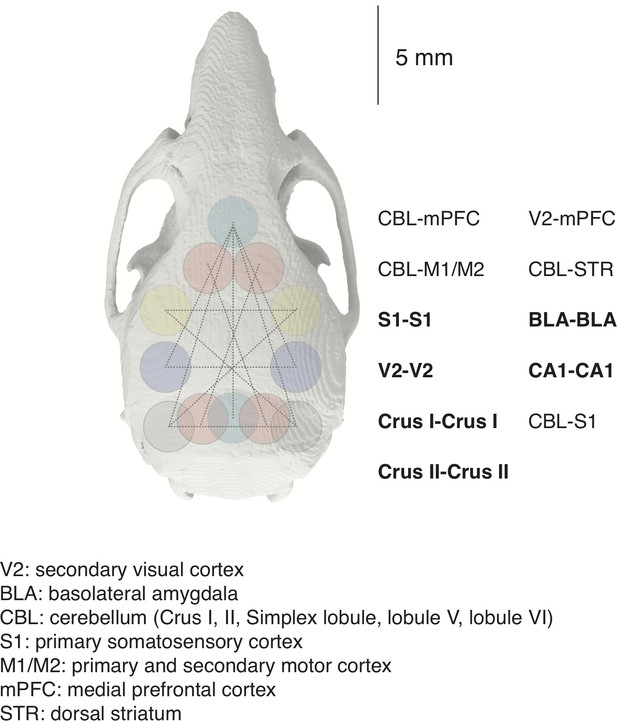
Possible dual NINscope configurations.
Colored circles (diameter of 1.8 mm GRIN objective) denote craniotomy positions. Connecting lines indicate a selection of possible rostro-caudal and bihemispheric (bold text) configurations.
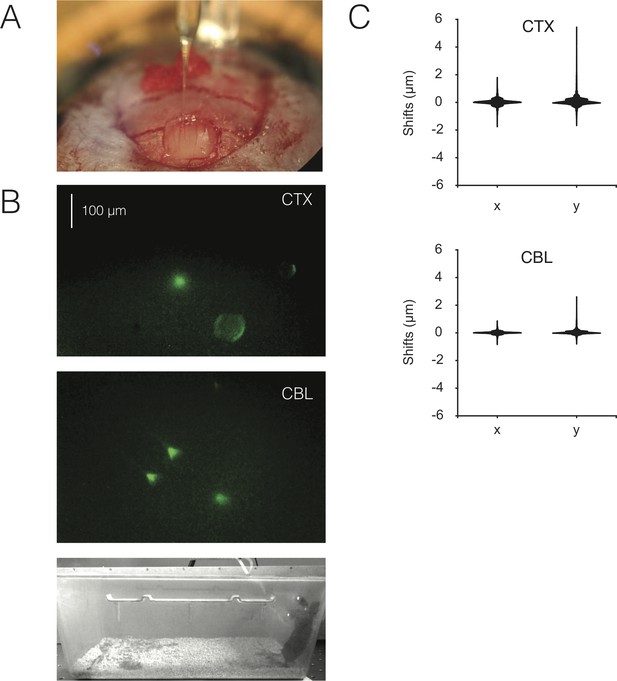
Bead imaging to assess motion artifacts with dual miniscopes.
(A) Injection of 6 µm diameter fluorescent beads into the cerebellum using a Nanoject II. (B) Imaged fields with beads in cortex (CTX) and cerebellum (CBL) in a mouse wearing two NINscopes one mounted above CTX, one above CBL. (C) Violin plots showing the distribution of x and y-shifts in µm after running NorMCorre on the data (N = 2 mice). Figure 3—video 2 shows fluorescent beads imaged in the two regions before and after motion correction.
-
Figure 3—figure supplement 3—source data 1
Data related to x and y shifts for beads injected in cerebellum and cortex.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig3-figsupp3-data1-v2.zip
A jumping mouse wearing two NINscopes.
A jumping mouse wearing two NINscopes.. Acquisition rate 30 Hz, video speed 0.33x.
Motion correction and segmentation in mice wearing dual miniscopes.
Raw data, motion corrected (NoRMCorre) data and the demixed signal (A*C obtained after running CNMF-E) representing activity in spatial footprints of Purkinje cell dendrites are shown together with video and accelerometer data. Acquisition rate 30 Hz, video speed 1x.
Bead imaging with dual NINscopes to assess motion artifacts.
We injected 6 µm diameter fluorescent beads in the cerebellum (CBL) and cerebral cortex (CTX) and imaged them using two mounted NINscopes. We applied motion correction (NoRMCorre) to the two datasets (CBL mc, CTX mc). Raw and motion corrected data are shown together with a concurrent webcam recording of a mouse in its home cage and the accelerometer signals acquired during the same session. Acquisition rate 30 Hz, video speed 1x.
Dual-region imaging with NINscope.
Purkinje cells in the cerebellar cortex (lobule VI) and neurons in the motor cortex were transduced with GCaMP6f and concurrently imaged with dual miniscopes. The video shows the (NorMCorre) motion-corrected miniscope data, demixed signals (top: cerebellum; bottom: cortex) and webcam video data with deconvoluted calcium transients of a subset of cells from cerebellum (blue) and cortex (ocre) plotted over the course of the recording. Acquisition rate 30 Hz, video speed 3x.
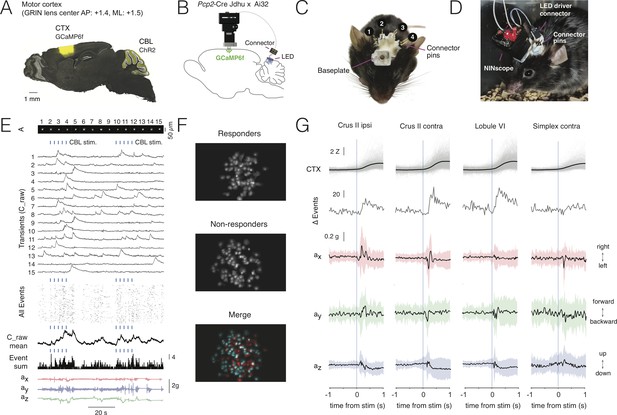
Multi-site optogenetic stimulation with NINscope.
(A) Pcp2-Cre Jdhu mice were crossed with Ai32 mice to obtain selective expression of ChR2(H134R) in cerebellar Purkinje cells. Neurons of the motor cortex were transduced in these mice with GCaMP6f. (B) Experimental configuration to combine optogenetic stimulation of cerebellum with imaging in cortex. (C) Mouse with a baseplate above the cortex and four LED probes mounted above the cerebellum (1: Crus II ipsi, 2: lobule VI, 3: Crus II contra, 4: Simplex lobule). The connector pins were used to connect the NINscope LED driver to each of the four probes. (D) Mouse with a NINscope and connection to one of the four stimulation sites. (E) Optogenetic stimulation of Purkinje cells (50 ms, 22 mA, 2.3 mW) evoked clearly discernible increases in both mean response (C_raw mean) and number of co-active cells (spatial footprints, A) in motor cortex (All Events, Event Sum). Repeated stimulation induced ramp-like activity in cortex. (F) In this example a large fraction of cells responded (responders) to optogenetic stimulation of the contralateral cerebellar hemisphere (Crus II). Color merge shows spatial localization of responders (red) and non-responders (cyan). Responsive cells were selected using the criterion that the post-stimulus signal had to exceed mean+2σ of the pre-stimulus baseline. (G) Calcium transients (gray: individual transients, black: mean) and change of summed events (Δ Events) triggered to stimulus onset at four different locations over the cerebellar surface and corresponding x (red), y (green) and z (blue) channel accelerometer data. Stimulation of the cerebellar hemispheres reveals lateralization of the behavioral response with stimulation on the left or right eliciting leftward and rightward head movements, respectively. Evoked behavioral reflexes generally commenced prior to calcium transient onsets in the cerebral cortex.
-
Figure 4—source data 1
Multi-site optogenetic stimulation with NINscope.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig4-data1-v2.zip
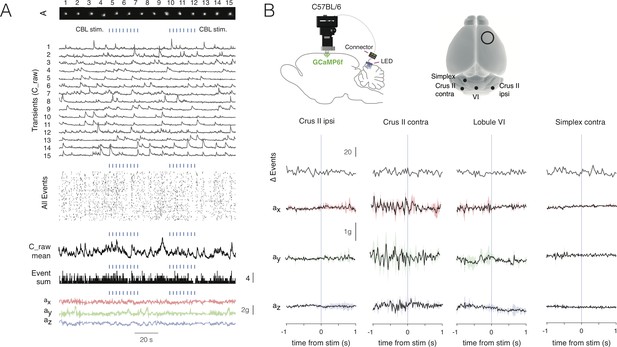
Light stimulation in absence of ChR2 neither evokes cerebral cortical nor behavioral responses.
(A) Extracted spatial components (A) and corresponding transients (C_raw), all onset events (All Events) extracted from the deconvoluted transients (C), the mean response of C_raw (C_raw mean), the sum of event onsets (Event sum) and the x, y and z accelerometer channels (ax, ay, az). Note the apparent lack of modulation of cortical activity by stimulating with light (blue dashes, CBL stim.) in the absence of ChR2. B, Schematic showing the stimulus configuration with NINscope and LED probe as well as the stimulation locations used for stimulation in a C57BlL/6 wildtype mouse (N = 1 mouse). No change was observed for the number of events triggered off of the stimulation (Δ Events). The accelerometer channels lacked stimulus-triggered deflections.
-
Figure 4—figure supplement 1—source data 1
Control experiment data in which optogenetic stimulation and imaging was performed in a wildtype mouse lacking ChR2(H134R).
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig4-figsupp1-data1-v2.zip
Combining remote optogenetic stimulation with cortical imaging using NINscope.
Neurons of motor cortex were transduced with GCaMP6f in transgenic mice that were obtained after crossing the Pcp2-Cre Jdhu and the Ai32 mouse lines to drive expression of ChR2(H134R) in cerebellar Purkinje cells. The video shows the demixed signal representing activity in spatial footprints of cortical neurons during optogenetic stimulation of the cerebellum (here 50 ms stimulation of contralateral Crus II) indicated by a blue bar together with the video and accelerometer data. Acquisition rate 30 Hz, video speed 1x.
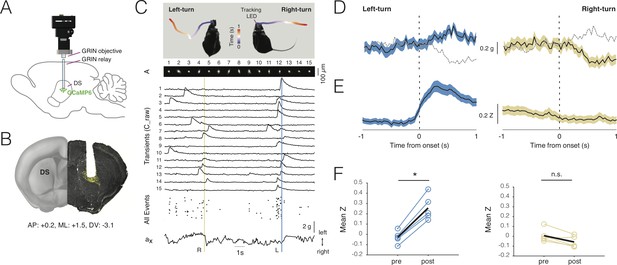
Deep-brain imaging and behavioral parsing with NINscope.
(A) Schematic of the NINscope configuration, which combines a GRIN objective with a GRIN relay lens (600 µm) to image from the dorsal striatum (DS) of the right hemisphere. (B) Coronal section showing the GRIN relay lens track and neurons in right DS expressing GCaMP6f (yellow). (C) Left- and right-turns were quantified by combining video observations or tracking LED with analysis of G-sensor data. Shown on top are examples of a mouse making a left and right turn in an open-field arena and the path obtained from the NINscope tracking LED where time progression over a duration of one second is color-coded. Below this the spatial components (A) and transients (C_raw) of 15 neurons extracted with CNMF-E are shown, the onset times of all events extracted from C as well as the x channel of the accelerometer. The vertical bars indicate that the animal first turned right (yellow) and then left (blue), with activity modulation in the right DS coinciding with contraversive movements. (D) Accelerometer data showing mean left and right acceleration of the x channel around movement onset (mean ± SEM). Dashed gray lines represent contraversive acceleration. (E) Mean calcium transient responses one second before and after movement onset (N = 5 mice) reveal a clear modulation of activity during left turns (mean ± SEM) when imaging right DS. (F) Quantification of calcium transient responses before and after movement onset for left and right turns, respectively. Right DS only displayed a significant calcium-transient increase for left turns (p<0.05).
-
Figure 5—source data 1
Deep-brain imaging and behavioral parsing with NINscope.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig5-data1-v2.zip
Deep imaging with NINscope.
Neurons in dorsal striatum were transduced with GCaMP6f and imaged with NINscope through a GRIN objective and relay lens. Shown are the demixed signals representing activity in neurons of dorsal striatum together with webcam recording of the animal in an open-field and the accelerometer signals. Acquisition rate 30 Hz, video speed 1x.
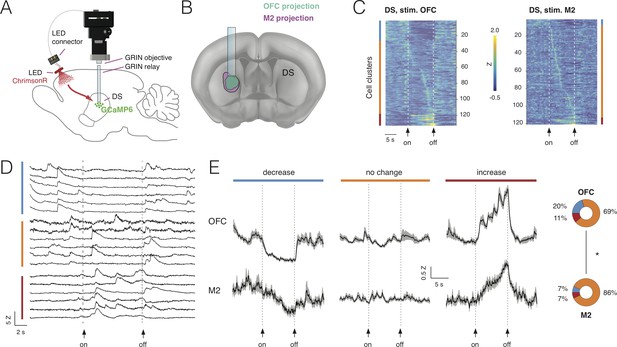
Combining deep-brain imaging with optogenetic stimulation using NINscope.
(A) Schematic showing placement of the NINscope with GRIN objective and GRIN relay lens to record from dorsal striatum (DS) as well as location of the optogenetic LED probe driven by the integrated LED driver. Viral vectors were injected either in orbitofrontal cortex (OFC) or secondary motor cortex (M2) to transduce neurons with ChrimsonR, or in DS to transduce neurons with GCaMP6f for calcium imaging. (B) Terminal fields of OFC and M2 mapped onto an Allen Brain Atlas template show their overlap in DS underneath the GRIN relay lens. (C) Responses for each neuron averaged over 10 trials for 254 cells in four mice. Different types of responses are found in DS when either OFC or M2 are optogenetically stimulated. Neurons exhibited decreases of activity (blue cluster), no apparent change (orange cluster), or increased responses (red cluster). (D) Z-scored calcium transients during OFC (left, N = 2 mice) and M2 (right, N = 2 mice) stimulation (10 s pulse, 20 Hz). For each neuron, 10 trials were averaged. (E) Responses averaged over all DS neurons for all 10 trials revealed similar types of modulation during stimulation of OFC and M2 (mean ± SEM). The circular insets denote the fraction of cells that showed suppression, no change, or increased responses.
-
Figure 6—source data 1
Combining deep-brain imaging with optogenetic stimulation using NINscope.
- https://cdn.elifesciences.org/articles/49987/elife-49987-fig6-data1-v2.zip
Tables
Features of NINscope and other miniscopes.
An overview of the features of currently released open-source and commercially available (wide-field illumination) miniscopes. Opto-remote: ability to optogenetically stimulate outside of the imaging field. Opto in-field: ability to optogenetically stimulate in the imaging field. IMU: inertial measurement unit. eFocus: electric focusing using an electrowetting lens (EWL). 1: (Cai et al., 2016), 2: (Shuman et al., 2020), 3: (Leman et al., 2018), 4: (Jacob et al., 2018), 5: (Zhang et al., 2019), 6: (Barbera et al., 2019), 7: (Gonzalez et al., 2019). n.a. = data not available. *nVoke system. **Optogenetically Synchronized Fluorescence Microscope.
| Miniscope | Weight (g) | 3D printed | Cross platform? | Two scopes? | Opto-remote | Opto -in field | Microphone | IMU | eFocus |
|---|---|---|---|---|---|---|---|---|---|
| NINscope | 1.6 | yes | yes | yes | yes | no | no | yes | no |
| UCLA Miniscope v31 | 3.2 | no | no | no | no | no | no | no | no |
| UCLA Miniscope wireless2 | 4. 5 | no | no | no | no | no | no | no | no |
| FinchScope3 | 1.8 | no | no | n.a. | no | no | yes | no | no |
| CHEndoscope4 | 4.5 | yes | no | no | no | no | no | no | no |
| miniScope Lin5 | 2.4 | yes | yes | no | no | no | no | no | no |
| miniScope Lin Wireless6 | scope: 3.9 battery:2.2–5 | yes | no | no | no | no | no | no | no |
| Miniscope Gonzalez et al.7 | n.a. | no | n.a. | yes | no | no | no | no | no |
| Inscopix (commercial) | 1.8 and up | no | yes | n.a. | no | yes* | no | no | yes |
| Doric lenses (commercial) | 2.2, 3.0 excl. canula | no | no | no | no | yes** | no | no | yes |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | B6.Cg-Tg(Pcp2-cre)3555Jdhu/J | PMID:15354293 | IMSR Cat# JAX:010536, RRID:IMSR_JAX:010536 | https://www.jax.org/strain/010536 |
| Genetic reagent (M. musculus) | Ai32 | PMID: 22446880 | IMSR Cat# JAX:012569, RRID:IMSR_JAX:012569 | https://www.jax.org/strain/012569 |
| Recombinant DNA reagent | AAV1.CAG.flex.GCAMP6f.WPRE.SV40 | Addgene | RRID:Addgene_100835 | https://www.addgene.org/100835/ |
| Recombinant DNA reagent | AAV1.CMV.PI.Cre.rBG | Addgene | RRID:Addgene_105530 | https://www.addgene.org/105530/ |
| Recombinant DNA reagent | AAV.Syn.GCaMP6f.WPRE.SV40 | Addgene | RRID:Addgene_100837 | https://www.addgene.org/100837/ |
| Recombinant DNA reagent | AAV.Syn.GCaMP6f.WPRE.SV40 | Addgene | RRID:Addgene_100843 | https://www.addgene.org/100843/ |
| Other | NINscope | This paper | RRID:SCR_017628 | https://github.com/ninscope |
| Software, algorithm | FIJI | PMID: 22743772 | RRID: SCR_002285 | https://imagej.net/Fiji/Downloads |
| Software, algorithm | MATLAB | MathWorks | RRID: SCR_001622 | https://uk.mathworks.com/products/matlab.html |
| Software, algorithm | Bonsai | PMID: 25904861 | RRID:SCR_017218 | https://bonsai-rx.org/ |
| Software, algorithm | R | R project | RRID:SCR_001905 | http://www.r-project.org/ |
| Software, algorithm | Python | Python | RRID:SCR_008394 | http://www.python.org/ |
| Software, algorithm | SPSS | IBM | RRID:SCR_002865 | http://www-01.ibm.com/software/uk/analytics/spss/ |
| Software, algorithm | Prism | Graphpad | RRID:SCR_002798 | http://www.graphpad.com/ |
| Software, algorithm | Custom Python scripts | This paper | https://github.com/Romanovg185/sps-continuous-time-data |





