Developmental variability channels mouse molar evolution
Figures
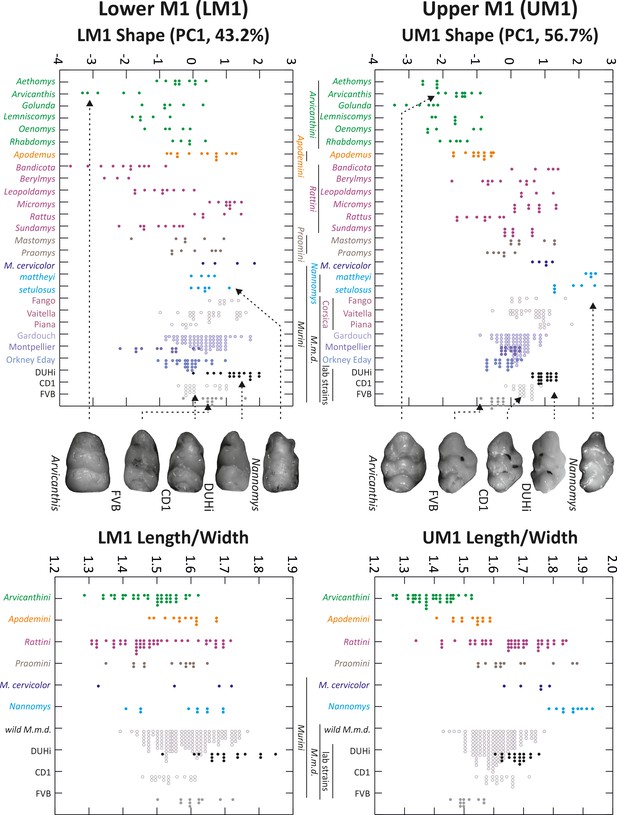
Morphological variation in murine first molar shape, based on 2D outlines.
Morphological variation of the first molar, both upper and lower, examined in individuals from several murine species, including mouse natural populations of the house mouse Mus musculus domesticus (M.m.d) and three mouse strains (Mus musculus). The left panel shows the first axis of a principal component analysis of 2D outline descriptors (Fourier coefficients). The right panel shows the length/width ratio of the molar (measures were taken on the same individuals as in the left panel, but they were grouped according to the phylogenetic groups shown in the upper left panel). Tooth images in the middle represent an example of the strain or species indicated, with arrows from each pointing to the point for that individual on each graph. UM1 – upper first molar; LM1 – lower first molar.
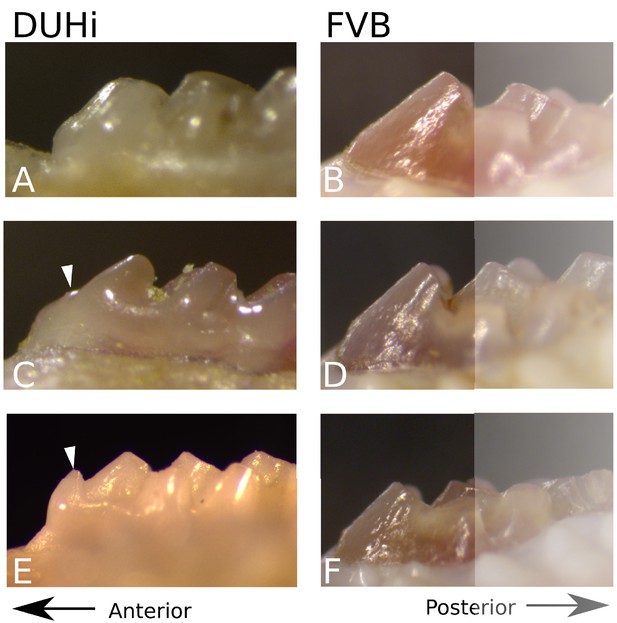
Representative examples of adult upper molar morphology in DUHi and FVB.
Side views of tooth morphology in DUHi (A, C, E) and FVB (B, D, F) mouse strains. Three individuals are shown for each strain, displaying the variation of the morphology present. The anterior part of the molar is to the left. Where present or partially present, the additional anterior small cusp is marked with a white arrow.
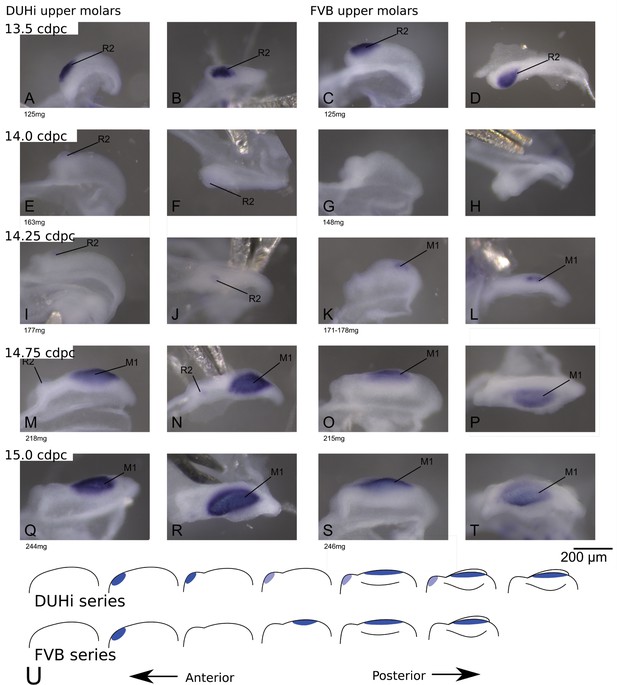
Comparative early molar tooth development in upper molar epithelia of DUHi and FVB mice.
Dissociated upper molar dental epithelia epithelia of DUHi (left-most two columns) and FVB embryos (right-most two columns), marked for Shh expression with in situ hybridisation. Samples represent a developmental series of early molar development, corresponding to 12.5dpc to 15.0dpc in FVB and 13.0dpc to 15.5dpc in DUHi. Embryo weight is noted below each sample and its equivalent in computed embryonic age is noted above each row of samples. Two images of each sample, a side and a top view, are shown.
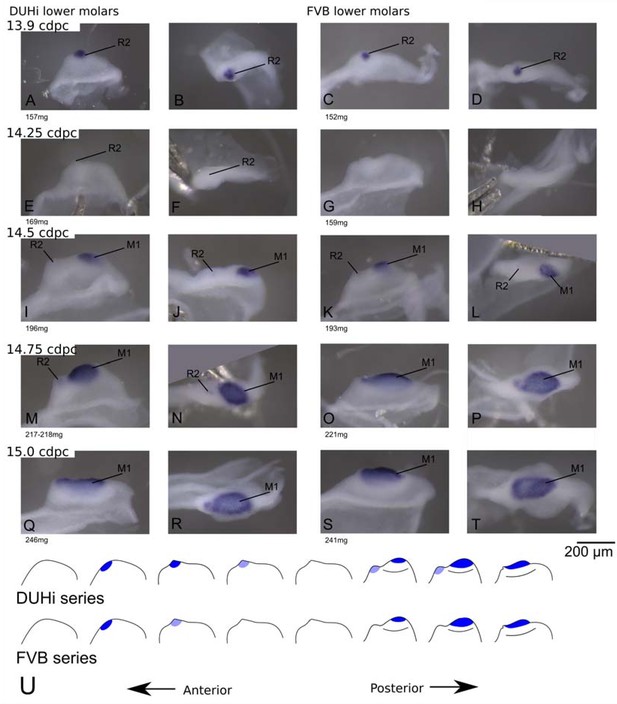
Comparative early molar tooth development in lower molar epithelia of DUHi and FVB mice.
Dissociated lower dental epithelium epithelia of DUHi (left most two columns) and FVB embryos (right-most two columns), marked forShhexpression with in situ hybridisation. Samples represent a developmental series of early molar development, corresponding to 12.5dpc to 15.0dpc in FVB. Embryo weight is noted below each sample and its equivalent in computed embryonic age is noted above each row of samples. Two images of each sample, a side and a top view, are shown. Scale bar = 200µm. Developmental progression is summarized in schematic form below (U).
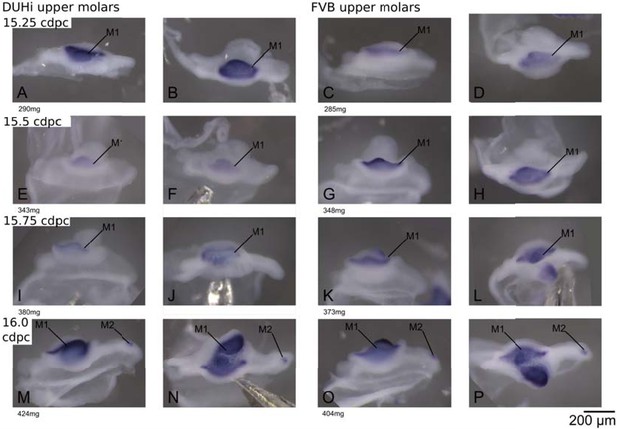
Comparative late molar tooth development in upper molar epithelia of DUHi and FVB mice.
Dissociated upper dental epithelium epithelia of DUHi (left most two columns) and FVB (right-most two columns) embryos, marked forShhexpression with in situ hybridisation. Samples represent a developmental series of later molar development, roughly corresponding to 15.0dpc to 16.5dpc in FVB. Embryo weight is noted below each sample and its equivalent in computed embryonic age is noted above each row of samples. Two images of each sample, a side and a top view, are shown. Scale bar = 200µm. The dental epithelium is oriented with anterior part to the left.
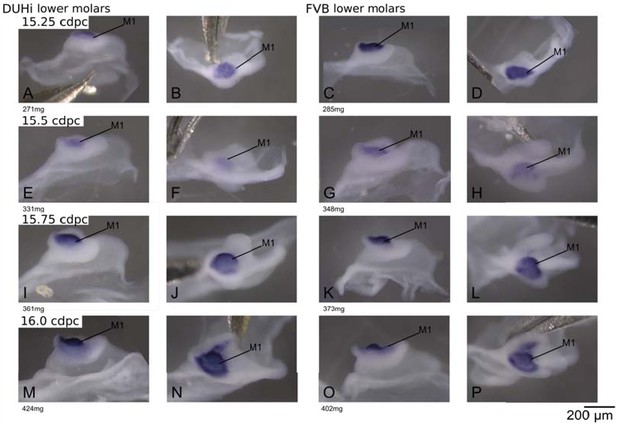
Comparative late molar tooth development in lower molar epithelia of DUHi and FVB mice.
Dissociated lower dental epithelium epithelia of DUHi (left most two columns) and FVB embryos (right-most two columns), marked forShhexpression with in situ hybridisation. Samples represent a developmental series of later molar development, roughly corresponding to 15.0dpc to 16.5dpc in FVB. Embryo weight is noted below each sample and its equivalent in computed embryonic age is noted above each row of samples. Two images of each sample, a side and a top view, are shown. Scale bar = 200µm. The dental epithelium is oriented with anterior part to the left.
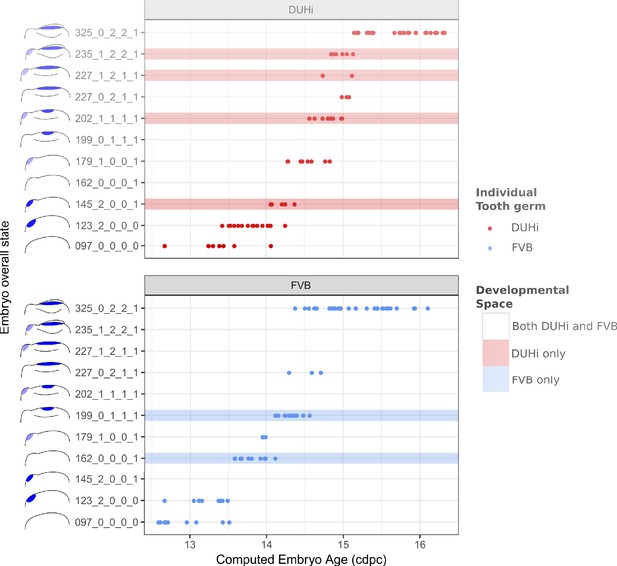
The range of possible developmental states differs between FVB and DUHi developing upper molars.
Temporal distribution of developmental state of the developing upper molar, produced by combining a value for each of the four scores for a given sample, based on criteria from Supplementary file 1. Each of the developmental states observed are schematized by a cartoon and ordered according to the average embryonic weight of the samples within that group. They are named with this weight, followed by the value of the four scores (eg. 097_0_0_0_0, means average weight 97 mg, 0 value for the four scores: R2 Shh expression, M1 Shh expression, cap transition state, R2 protuberance state). Exclusive developmental states are highlighted according to whether they are found in DUHi only (red) or in FVB only (blue). The temporal axis is given by computed embryo age (cdpc). The dental epithelium is oriented with anterior part to the left. Scale bar = 200 µm. Developmental progression is summarized in schematic form below (U).
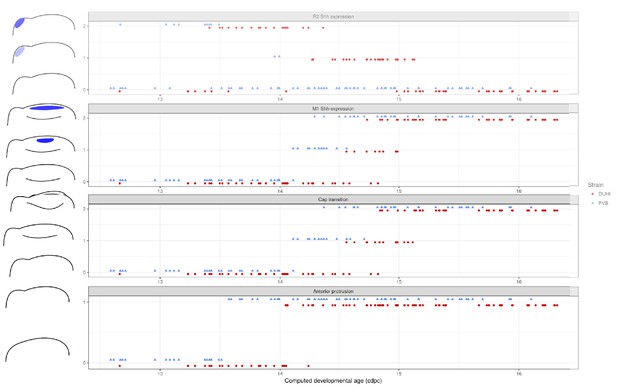
Comparative progression of scored characteristics in upper dental epithelium.
Progression of four characters (R2Shhexpression, M1Shhexpression, cap transition and anterior protrusion of the dental epithelium) is depicted in samples of upper molars from two strains (FVB and DUHi). The temporal axis is provided by computed embryonic age (cdpc). All samples were scored using the criteria provided in Supplementary file 1.
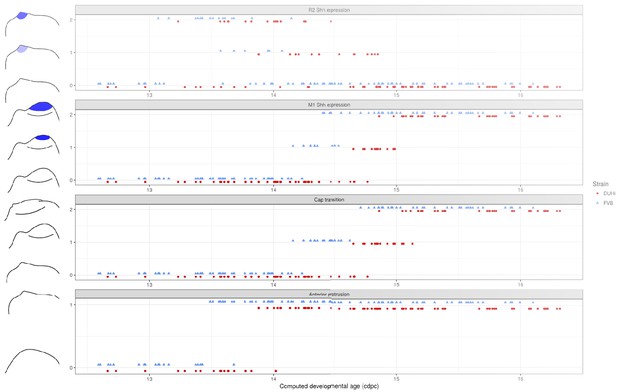
Comparative progression of scored characteristics in lower dental epithelium.
Progression of four characters (R2Shhexpression, M1Shhexpression, cap transition and anterior protrusion of the dental epithelium) is depicted in samples of lower molars from two strains (FVB and DUHi). The temporal axis is provided by computed embryonic age (cdpc). All samples were scored using the criteria provided in Supplementary file 1.
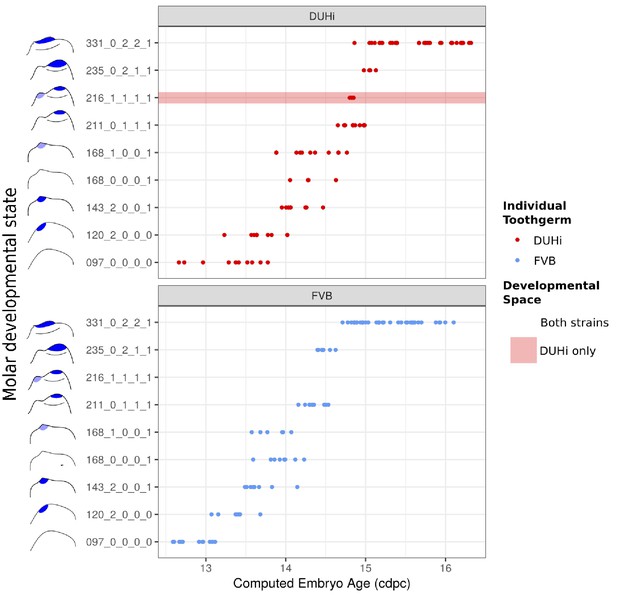
The range of possible developmental states differs between FVB and DUHi developing lower molars.
Temporal distribution of developmental state of the developing lower molar, produced by combining a value for each of the four scores for a given sample, based on criteria from Supplementary file 1. Each of the developmental states observed are shown and ordered according to the average embryonic weight of the samples within that group. Exclusive developmental states are highlighted according to whether they are found in DUHi only (red) or in FVB only (blue).The temporal axis is given by computed embryonic age (cdpc).
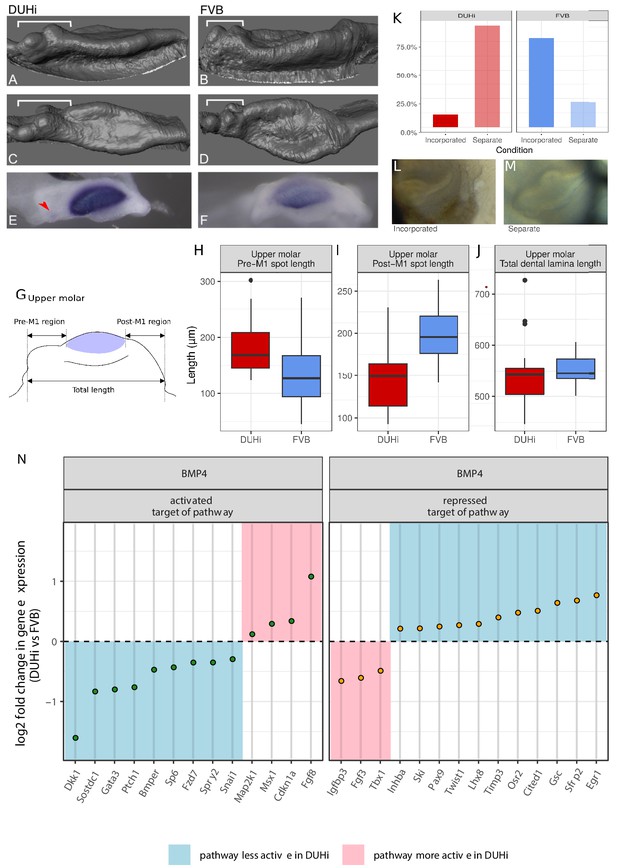
FVB and DUHi strains differ in the balance of activation-inhibition mechanisms.
(A–F) Comparative 3D morphology of the epithelial part of developing upper molars in DUHi (A, C) and FVB (B, D) strains at cap transition. A, B are side views and C, D upper views. The variable anterior region including R2 rudimentary bud is denoted by the bracket. Expression of Shh at the same timepoint is shown below for DUHi (E) and FVB (F), for comparative purposes. The red arrowhead in E points to R2 signaling center. In F, the faint staining anterior to the mature M1 signaling center might correspond to re-expression of Shh in cells that formed R2 signaling center. (G–J) panel G shows the three measurements taken from all epithelial samples of a computed age of 14.25–15 cpdc (between 180 and 250 mg weight), comparing DUHi with FVB samples. The measures were taken between the anterior or posterior limit of dental epithelium and the anterior or posterior limit of the M1 signaling center, as shown on the cartoon. Boxplots H-J show the results in upper molars (see Figure 4—figure supplement 1 for lower molars). Pre-M1 region and Post-M1 regions are significantly different in DUHi versus FVB mice (t-test; p<0.01, see Supplementary file 4). (K–M) The developing molars of DUHi and FVB mice react differently when cultured in vitro at 13.0 dpc (p=0.015 in an exact Fisher test, see Supplementary file 4): R2 bud tends to form a clear individualized bud (M) in most DUHi tooth cultures (n = 18), whereas a single developing tooth (L) is seen in most FVB tooth cultures (n = 18). (N) Target genes of the Bmp4 pathway differentially expressed between the two strains at the cap transition. Differential expression analysis was performed on both lower and upper molar samples, taking molar type into account in the statistical treatment by DEseq2. Genes were classified as targets activated or repressed by the pathway based on O'Connell et al. (2012). The log2 fold change in DUHi as compared to FVB is shown (positive: expression level increased in DUHi tooth germs; negative: expression level decreased in DUHi tooth germs). Depending if the gene is an activated or a repressed target, and is increased or decreased in DUHi tooth germs, it may suggest that the pathway is more active (pink) or less active (blue) in DUHi tooth germs.
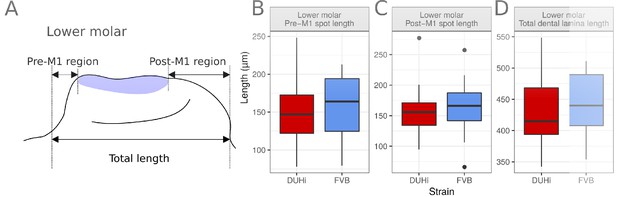
Comparative measurements in lower molar dental epithelium.
Panel A shows the three measurements taken from all epithelial samples of a computed age of 14.25-15 cpdc (between 180 and 250mg weight), comparing DUHi with FVB samples. Boxplots B-D show the results for three of them in lower molars. The measurements were taken between the anterior or posterior limit of dental epithelium and anterior or posterior limit of the M1 signaling center, respectively. Note that the pre-M1 signaling center region tends to be longer in FVB (not significant). This is due to a smoother slope of the anterior epithelium: compare E with G, I with K, M with O and Q with S, on Figure 2—figure supplement 1. Taken together with the fact that in the lower jaw, R2 signaling center is integrated in the M1 signaling center, this measure is poorly informative on R2 bud size in the lower jaw, in contrast with the upper jaw.
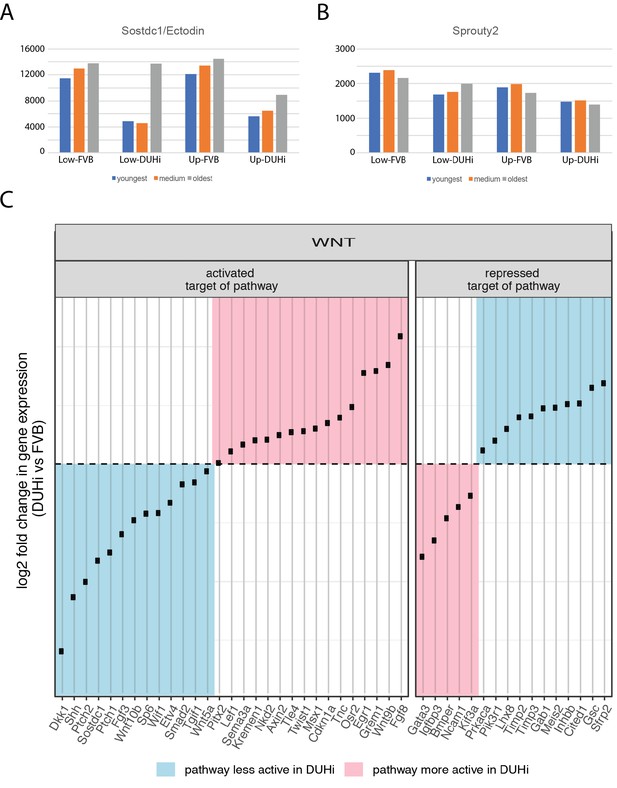
Comparative transcriptomics of DUHi versus FVB cap stage tooth germs.
(A-B) Differential expression of sostdc1 (A) and sprouty2 (B) in an RNAseq experiment for lower and upper molar germs of FVB and DUHi mice. For each strain, we sampled 3 embryos from the same litter with slightly different body weight (DUHi (embryo weight: 196, 219 and 239 mg) and FVB (195, 215 and 233 mg). This corresponds to slightly different embryonic ages, from the youngest age for the lightest embryo to the oldest age for the heaviest embryo. For each gene, the bars show normalized RNAseq counts (Basemean, DESeq2) in the lower (low) and upper (up) molar germs of these 3 DUHi and 3 FVB embryos. The test for differential expression was performed between all DUHi versus FVB samples, taking jaw into account as a factor in DEseq2.Sprouty2: fold change in DUHi=1.3, padj=2.4.E-8;Sostdc1/Ectodin: fold change in DUHi=1.8; padj=0.02. (C) A panel showing log fold change in expression level in DUHi/FVB (positive fold change: higher expression level in DUHi; negative fold change: lower expression level in DUHi) for Wnt target genes (activated or repressed by Wnt pathway), that are differentially expressed between the two strains (see supplementary text for details). In blue, targets arguing for lesser Wnt activity in DUHi mice (n=24). In pink, targets arguing for greater Wnt activity in DUHi mice (n=21).
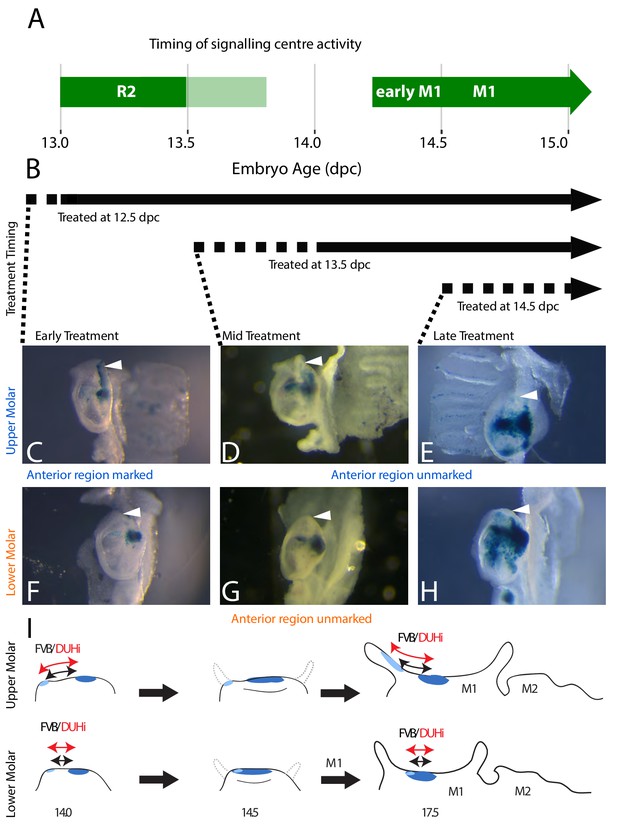
The contribution of R2 and M1 signaling centers to the anterior region of the upper and lower molar.
(A) The timing of Shh expression in R2 and M1 signaling centers is shown in green (light green: faint Shh expression in R2). (B) In this context, a tamoxifen-inducible Shh-CreERT line was used to induce the marking of Shh-expressing cells with β-galactosidase expression following tamoxifen injection. Time period of marking is indicated with black arrows, taking into account the roughly 12 hr-delay for activation (dashed line) plus the persistence of tamoxifen after injection. Early treatment (12.5 dpc injection) corresponds to activity of both R2 and M1 signaling centers, and will mark the progeny of both signaling centers during this period. Mid and late treatment (13.5dpc or 14.5dpc injections) correspond to activity of M1 signaling center only, and will mark exclusively the progeny of M1 signaling center (Supplementary file 2). (C–H) X-gal-stained epithelia of upper (C–E) or lower (F–H) molars at 17.5dpc are seen below, with the anterior region of the tooth (white arrowhead) marked in correspondence with the timing of the tamoxifen treatment. The presence of staining in the anterior part of the tooth in 12.5 dpc-treated individuals (C) with the lack of staining in later-treated individuals (D and E) indicates that R2 signaling center contributes to the anterior part of the first upper molar. The scheme in I summarizes results for lineage tracing with induction at 12.5 dpc, marking both R2 signaling center (light blue) and M1 signaling center (dark blue) descendant cells. Following M1 cap transition at 14.5 dpc, the tooth will develop anteriorly and posteriorly (shown on the 14.5 dpc scheme with dashed grey line). Only the upper R2 signaling center descendants are involved in anterior cervical loop formation. Differences in size and R2-M1 distance seen between FVB and DUHi strain (black versus red arrows) will preferentially impact the anterior part of the tooth in the upper molar only.
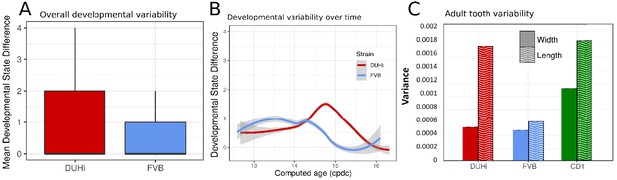
Developmental and adult variation is higher in DUHi first upper molars.
(A) A measure of developmental variation in the developing upper molars of FVB and DUHi strains. The figure shows a boxplot of developmental state differences calculated for pairs of samples with less than 0.25 difference in computed embryonic age (cdpc). Samples close in age are significantly more different in developmental state in DUHi versus FVB mice, according to a Wilcoxon test (p<0.001). See the Materials and methods for further explanation on this measure of developmental variation. (B) The mean developmental state difference between nearby samples (computed embryonic age difference <0.25) is plotted as the local regression line for both strains (standard deviation shown in grey). (C) Morphological variation in adult first molar, measured as the variance in molar width and length. Variation in length (but not width) is much greater in DUHi than that in FVB (both are inbred strains; p=0.095), and comparable to the outbred CD1 strain.
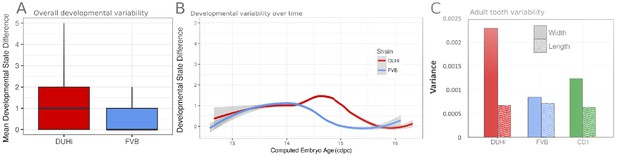
Developmental and adult variation in FVB and DUHi first lower molars.
(A) A measure of developmental variation in the developing lower molars of FVB and DUHi strains. The figure shows a boxplot of developmental state differences calculated for pairs of samples with less than 0.25 difference in computed embryonic age (cdpc). Samples close in age are significantly more different in developmental state in DUHi versus FVB mice, according to a Wilcoxon test (p<0.00001). See the material and methods for further explanation on this measure of developmental variation. (B) The mean developmental distance between nearby samples (computed embryonic age difference < 0.25) is plotted as the local regression line for both strains (standard deviation shown in grey). (C) Morphological variation in the adult first molar, measured as the variance in molar width and length. In contrast with the situation in the upper molar, the extent of variation in molar length is comparable in the lower first molars for the three strains (variation in lower molar width is however higher in DUHi and the outbred CD1 strain).
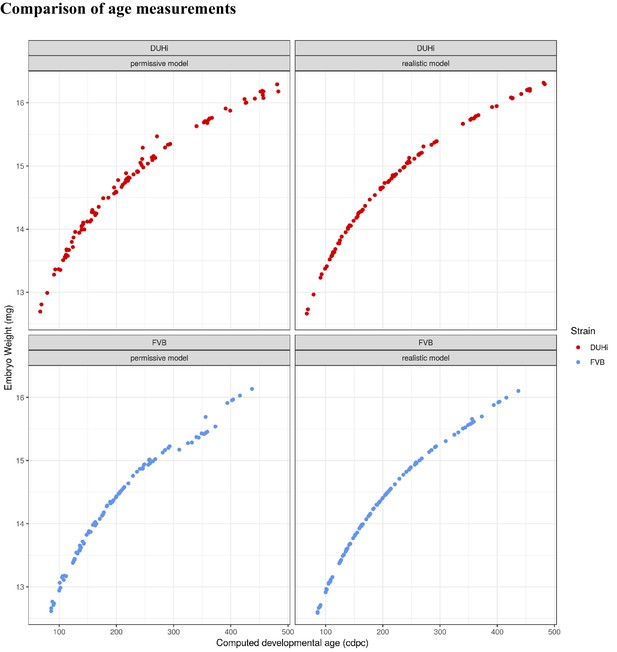
Comparison between two versions of computed embryonic age (realistic versus permissive), as plotted against embryo weight (mg).
DUHi (red) and FVB (blue) samples are shown in separate panels.
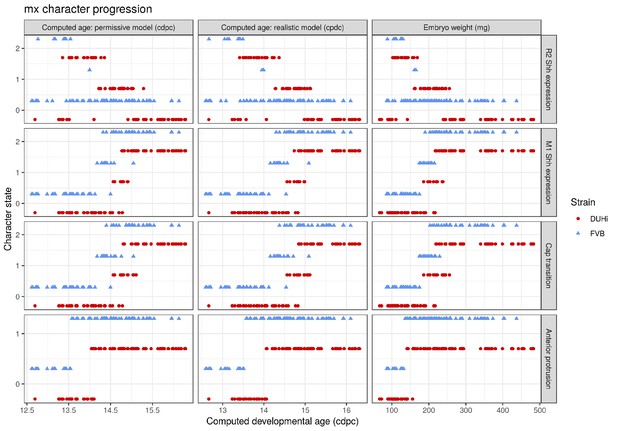
Comparative progression of scored characteristics in upper molar epithelia under different measures of age.
Progression of four characters (R2 Shh expression, M1 Shh expression, cap transition and anterior protrusion of the dental epithelium) is depicted in samples of upper molars from two strains (FVB and DUHi). In order to allow the comparison of age measurements, the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters and by embryo weight as an age proxy. All samples were scored using the criteria provided in Supplementary file 1.
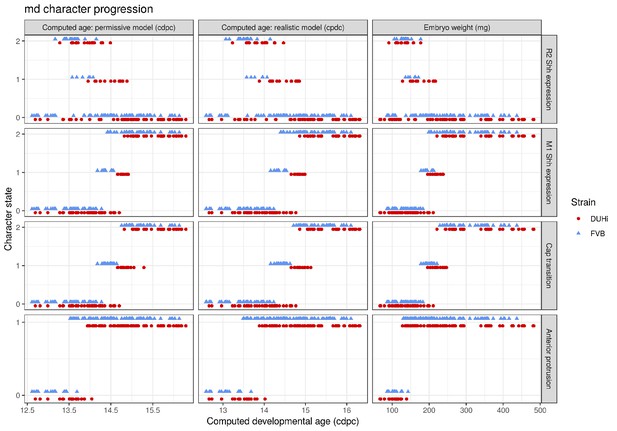
Comparative progression of scored characteristics in lower molar epithelia under different measures of age.
Progression of four characters (R2 Shh expression, M1 Shh expression, cap transition and anterior protrusion of the dental epithelium) is depicted in samples of lower molars from two strains (FVB and DUHi). In order to allow the comparison of age measurements, the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters and by embryo weight as an age proxy. All samples were scored using the criteria provided in Supplementary file 1.
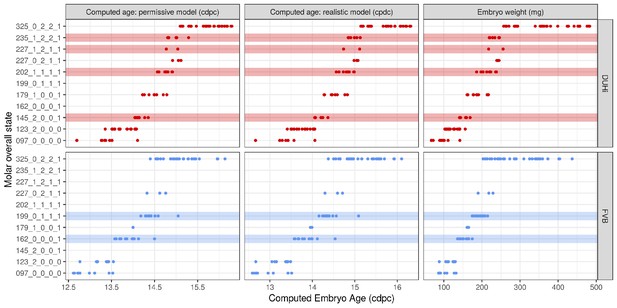
Total state of embryonic upper molars under different measures of age.
Temporal distribution of developmental state of the developing upper molar, produced by combining a value for each of the four scores for a given sample, based on criteria from Supplementary file 1. Each of the developmental states observed are shown and are ordered according to the average embryonic weight of the samples within that group. Each state present is coloured according to whether it is found in DUHi only (red), in FVB only (blue), or in both DUHi and FVB (white) samples. In order to allow the comparison of age measurements, the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters and by embryo weight as an age proxy.
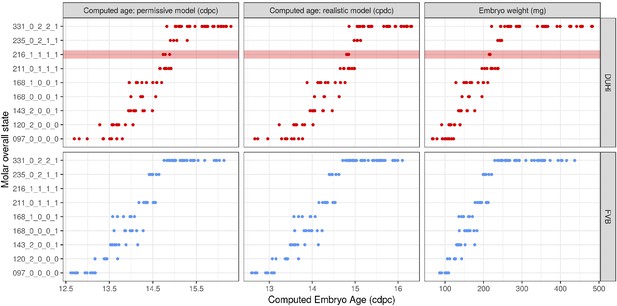
Developmental state of embryonic lower molars under different measures of age.
Temporal distribution of developmental state of the developing lower molar, produced by combining a value for each of the four scores for a given sample, based on criteria from Supplementary file 1. Each of the developmental states observed are shown and are ordered according to the average embryonic weight of the samples within that group. Each state present is coloured according to whether it is found in DUHi only (red) or in both DUHi and FVB (grey) samples. In order to allow the comparison of age measurements, the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters and by embryo weight as an age proxy.
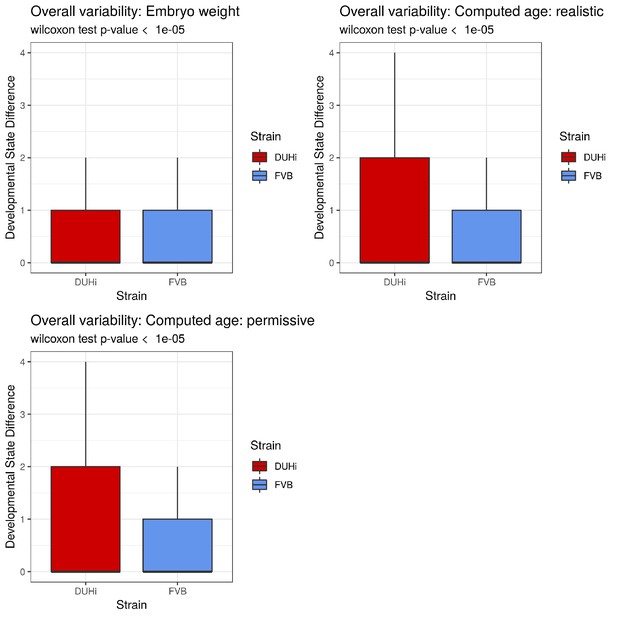
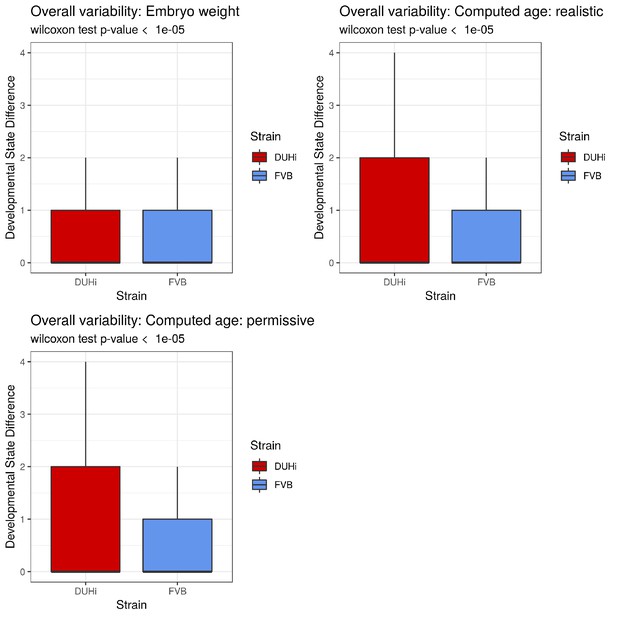
Developmental variability in the developing upper molars of DUHi and FVB mice embryos under different measures of age.
Variability in the developing upper molar is represented as a boxplot of developmental state differences calculated for pairs of samples with less than 0.25 difference in computed embryonic age (cdpc). Samples close in age are significantly more different in developmental state in DUHi versus FVB mice when the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters and when embryo weight is used as an age proxy.
Developmental variation in the developing lower molars of DUHi and FVB mice embryos under different measures of age.
Variation in the developing lower molar is represented as a boxplot of developmental state differences calculated for pairs of samples with less than 0.25 difference in computed embryonic age (cdpc). Samples close in age are significantly more different in developmental state in DUHi versus FVB mice when the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters.
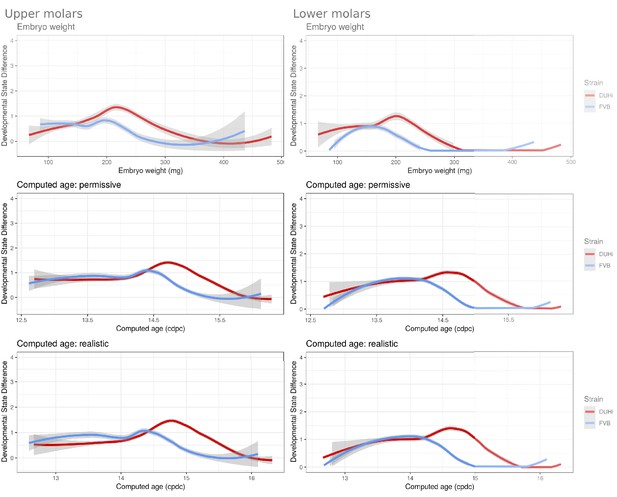
Developmental variation over time in the upper and lower molars of DUHi and FVB mice embryos under different measures of age.
The mean developmental distance between nearby samples (embryonic age difference < 0.25 d) is plotted as the local regression line (LOESS smoothing) for both strains (standard deviation shown in grey). In order to allow the comparison of age measurements, the temporal axis is provided by computed embryonic age (cdpc) under the realistic and permissive parameters and by embryo weight as an age proxy. Developmental variation is higher in DUHi under all measures of age, and peaks at time of R2 and M1 signaling center co-existence.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | FVB | Charles River, France | inbred | |
| Strain, strain background (Mus musculus) | CD1 | Charles River, France | outbred | |
| Strain, strain background (Mus musculus) | DUHi | MRC Mary Lyon centre, Oxfordshire, UK | inbred | |
| Strain, strain background (Mus musculus) | B6.129S6-Shh < tm2(cre/ERT2)Cjt>/J | Jackson Laboratory, Maine, USA | Originally in C57Bl6 background. Backcrossed to CD1 | |
| Strain, strain background (Mus musculus) | B6.129S4-Gt(ROSA)26Sortm1LacZSor/J | Jackson Laboratory, Maine, USA | Originally in C57Bl6 background. Backcrossed to CD1 | |
| Commercial assay or kit | TruSeq RNA Sample Prep Kit v2 | Illumina | RS-122–2001 | Non stranded protocol |
| Commercial assay or kit | DISPASE II (NEUTRAL PROTEASE, GRADE II) | ROCHE/SIGMA | 4942078001 | Used at 10 mg/ml in Hepes/KOH 50mM ph7.7 ; NaCl 150 mM. Incubation at 37°C from 45 min to 1h15 depending on stage. |
| Antibody | ANTI-DIGOXIGENIN AP-CONJUGATE, from sheep | ROCHE/SIGMA | 11093274910 | Fab fragments from polyclonal anti-digoxigenin antibodies (from sheep), conjugated to alkaline phosphatase. Used 1/1200ie |
| Commercial assay or kit | BM PURPLE AP SUBSTRATE, PRECIPITATING | ROCHE/SIGMA | 11442074001 | |
| Commercial assay or kit | DIG RNA Labeling Mixture, 10x | ROCHE/SIGMA | 11277073910 |
Medians and 95% credibility intervals for the parameters for each strain, and each scenario (termed realistic for sd.litter.preg = 0.05, and permissive for sd.litter.preg = 0.1 in the text).
| A: DUHi, permissive model | |||
|---|---|---|---|
| DUHi | sd.litter.preg=0.1 | ||
| 2.5% | 50% (median) | 97.5% | |
| a | 5.3937 | 5.4742 | 5.5531 |
| b | 0.4816 | 0.5402 | 0.6065 |
| sd.embryo.dev | 0.1556 | 0.1695 | 0.1855 |
| sd.litter.dev | 0.1376 | 0.1997 | 0.2889 |
| B: DUHi, realistic model | |||
| DUHi | sd.litter.preg=0.05 | ||
| 2.5% | 50% | 97.5% | |
| a | 5.3928 | 5.4732 | 5.5555 |
| b | 0.4809 | 0.5391 | 0.6050 |
| sd.embryo.dev | 0.1557 | 0.1694 | 0.1854 |
| sd.litter.dev | 0.1632 | 0.2188 | 0.3003 |
| C: FVB, permissive model | |||
| FVB | sd.litter.preg=0.01 | ||
| 2.5% | 50% | 97.5% | |
| a | 5.5019 | 5.5697 | 5.6378 |
| b | 0.4057 | 0.4622 | 0.5173 |
| sd.embryo.dev | 0.0812 | 0.0875 | 0.0946 |
| sd.litter.dev | 0.1549 | 0.2032 | 0.2683 |
| D : FVB, realistic model | |||
| FVB | sd.litter.preg=0.01 | ||
| 2.5% | 50% | 97.5% | |
| a | 5.5019 | 5.5697 | 5.6378 |
| b | 0.4060 | 0.4611 | 0.5175 |
| sd.embryo.dev | 0.0813 | 0.0875 | 0.0944 |
| sd.litter.dev | 0.1774 | 0.2211 | 0.2817 |
Additional files
-
Supplementary file 1
Table with scoring criteria used to assess embryonic dental epithelia.
- https://cdn.elifesciences.org/articles/50103/elife-50103-supp1-v2.docx
-
Supplementary file 2
An excel file for transcriptomic analysis: normalized basemean for all genes with statistical support in our DE analysis, list of DE genes for padj < 0.05, list of BMP4 pathway target genes extracted from O'Connell et al. (2012) and their classification as activated/repressed target of BMP4 pathway, list of BMP4 target genes DE in DUHi/FVB, list of Wnt pathway target genes extracted from O'Connell et al. (2012), and their classification as activated/repressed targets; list of Wnt target genes DE in DUHi/FVB.
- https://cdn.elifesciences.org/articles/50103/elife-50103-supp2-v2.xlsx
-
Supplementary file 3
Table summarizing lineage tracing experiments.
- https://cdn.elifesciences.org/articles/50103/elife-50103-supp3-v2.docx
-
Supplementary file 4
Tables for key statistical tests performed for this study.
- https://cdn.elifesciences.org/articles/50103/elife-50103-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50103/elife-50103-transrepform-v2.pdf





