Genome-wide CRISPR screening reveals genetic modifiers of mutant EGFR dependence in human NSCLC
Figures
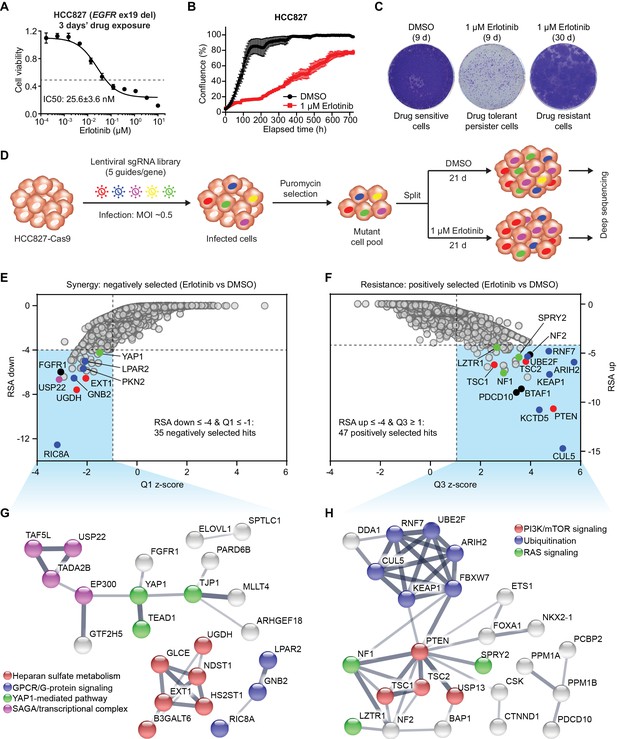
Genome-wide CRISPR-Cas9 screening identifies determinants of EGFR-TKI sensitivity in EGFR-mutant NSCLC.
(A) Cell viability assessment by CellTiter-Glo assay of HCC827 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± standard deviation (SD); n = 4. (B) Kinetic cell proliferation assay monitored by IncuCyte for HCC827 cells cultured in the presence of DMSO control or 1 µM erlotinib over a 30 day period. (C) Crystal violet staining colony formation assay of HCC827 cells treated with DMSO or 1 µM erlotinib for the indicated days. (D) Schematic outline of the genome-wide CRISPR-Cas9 screening workflow in HCC827 cells. (E) Scatterplot depicting gene level results for erlotinib negatively selected hits in the CRISPR screen. A number of representative hits are shown in color. (F) Scatterplot depicting gene level results for erlotinib positively selected hits in the CRISPR screen. A number of representative hits are shown in color. (G) STRING protein network of the 35 negatively selected hits as defined in (E). The nodes represent indicated proteins, and colored nodes highlight proteins enriched in certain signaling pathways. The edges represent protein-protein associations, and the line thickness indicates the strength of data support. The minimum required interaction score was set to default medium confidence (0.4), and the disconnected nodes were removed from the network. (H) STRING protein network of the 47 positively selected hits as defined in (F).
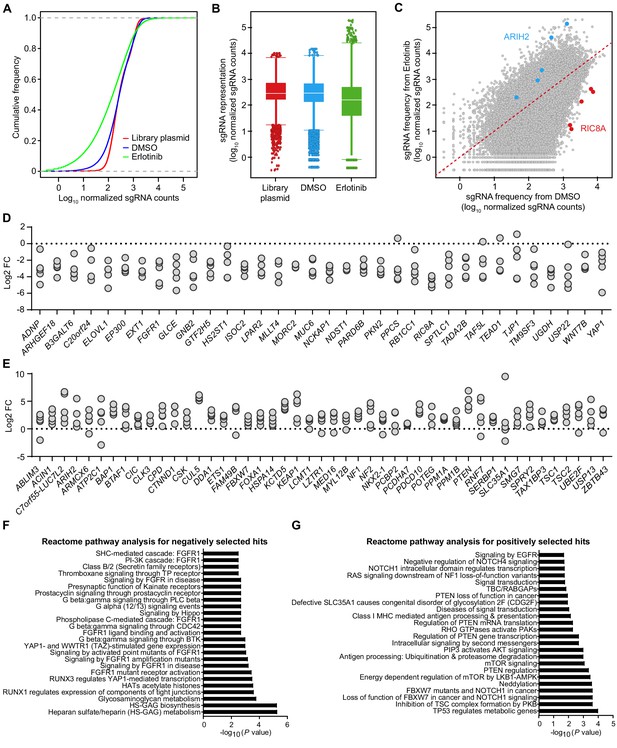
CRISPR-Cas9 screening reveals genetic determinants of EGFR-TKI sensitivity.
(A) Cumulative frequency of sgRNAs in the library plasmid and after 21 days of DMSO or erlotinib treatment in HCC827 cells. (B) Box plot showing the distribution of sgRNA representations in the library plasmid and after 21 days of DMSO or erlotinib treatment in HCC827 cells. (C) Scatterplot showing the comparison of sgRNA frequency between DMSO and erlotinib treated HCC827 cells. (D) Dot plot showing the distribution of individual sgRNAs targeting erlotinib negatively selected hits in the CRISPR screen. Data are presented as log2 fold change of each sgRNA sequence based on the abundance in the erlotinib-treated versus DMSO-treated cell population. (E) Dot plot showing the distribution of individual sgRNAs targeting erlotinib positively selected hits in the CRISPR screen. Data are presented as log2 fold change of each sgRNA sequence based on the abundance in the erlotinib-treated versus DMSO-treated cell population. (F) Reactome pathway analysis for erlotinib negatively selected hits in the CRISPR screen. (G) Reactome pathway analysis for erlotinib positively selected hits in the CRISPR screen.
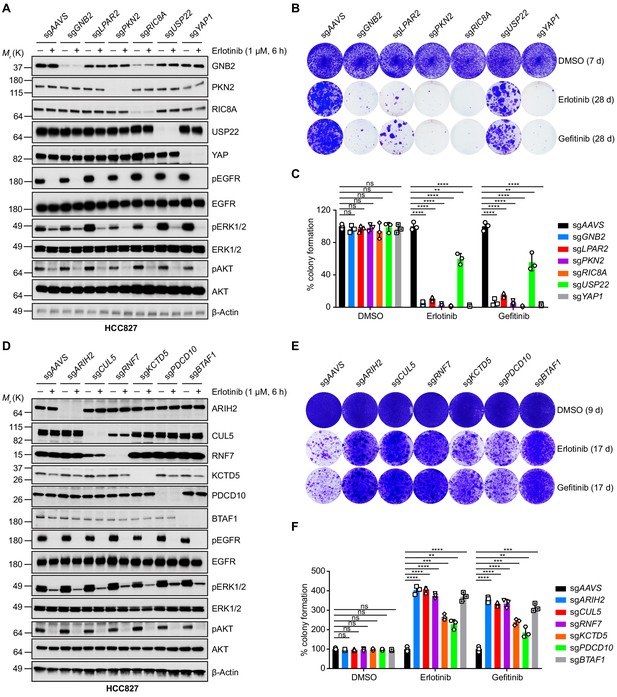
Validation of selected hits by individual knockout in HCC827 cells.
(A) Immunoblots of indicated proteins in cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of negatively selected hits and on-target inhibition of EGFR pathway by erlotinib treatment. β-Actin was used as a loading control. Individual knockout cell lines were generated by lentivirus-mediated expression of sgRNA targeting indicated genes in HCC827 cells with constitutive Cas9 expression. (B) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (C) Quantification of colony formation in (B), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± standard deviation (SD) is shown. (D) Immunoblots of indicated proteins in cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of positively selected hits and on-target inhibition of EGFR pathway by erlotinib treatment. β-Actin was used as a loading control. (E) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (F) Quantification of colony formation in (E), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown. Statistical significance was tested using unpaired two-tailed t test (C and F); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
-
Figure 2—source data 1
Raw data from Figure 2.
- https://cdn.elifesciences.org/articles/50223/elife-50223-fig2-data1-v2.xlsx
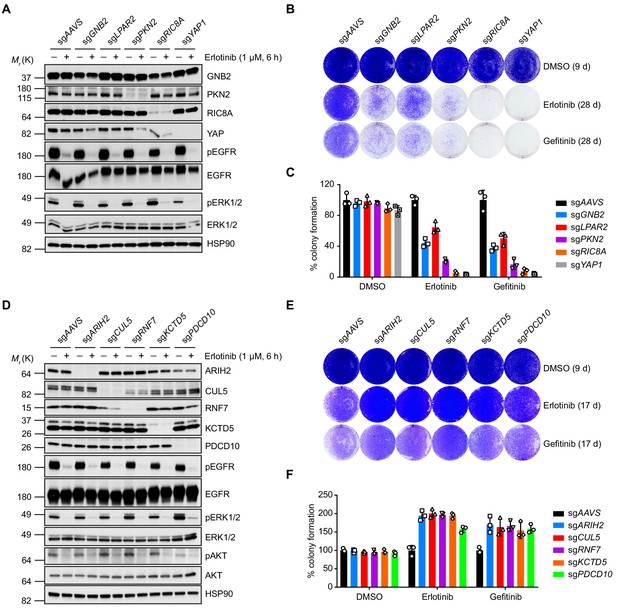
Validation of selected hits by individual knockout in NCI-H3255 cells.
(A) Immunoblots of indicated proteins in cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of negatively selected hits and erlotinib efficacy. HSP90 was used as a loading control. Individual knockout cell lines were generated by lentivirus-mediated expression of sgRNA targeting indicated genes in NCI-H3255 cells with constitutive Cas9 expression. (B) Crystal violet staining colony formation assay of indicated NCI-H3255 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (C) Quantification of colony formation in (B), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± standard deviation (SD) is shown. (D) Immunoblots of indicated proteins in cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of positively selected hits and erlotinib efficacy. HSP90 was used as a loading control. (E) Crystal violet staining colony formation assay of indicated NCI-H3255 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (F) Quantification of colony formation in (E), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown.
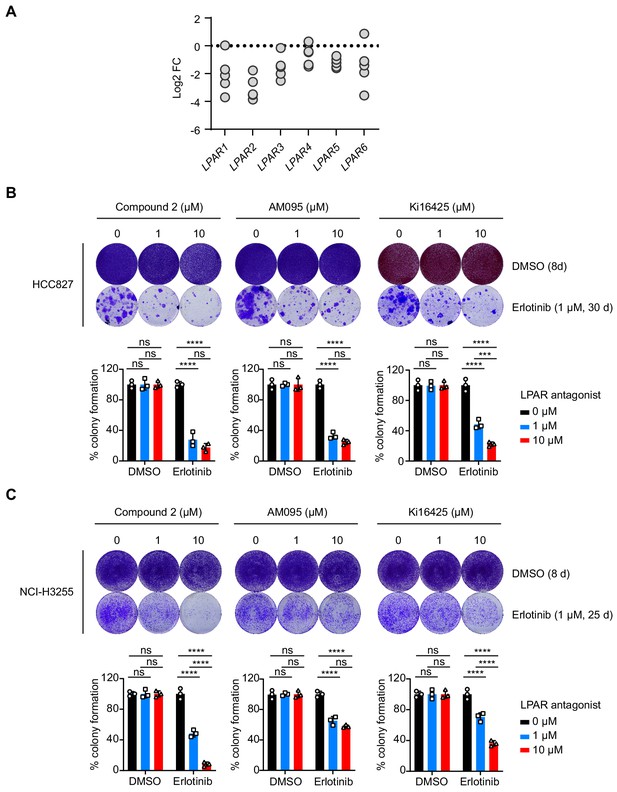
Targeting LPARs sensitizes EGFR-mutant NSCLC cells to EGFR inhibition.
(A) Dot plot showing the distribution of individual sgRNAs targeting all six members of the LPAR family in the CRISPR screen. Data are presented as log2 fold change of each sgRNA sequence based on the abundance in the erlotinib-treated versus DMSO-treated cell population. (B) Crystal violet staining colony formation assay of HCC827 cells treated with indicated compounds (upper panel). Bottom: quantification of colony formation in the upper panel. Error bars represent mean ± SD; n = 3. (C) Crystal violet staining colony formation assay of NCI-H3255 cell treated with indicated compounds (upper panel). Bottom: quantification of colony formation in the upper panel. Error bars represent mean ± SD; n = 3.
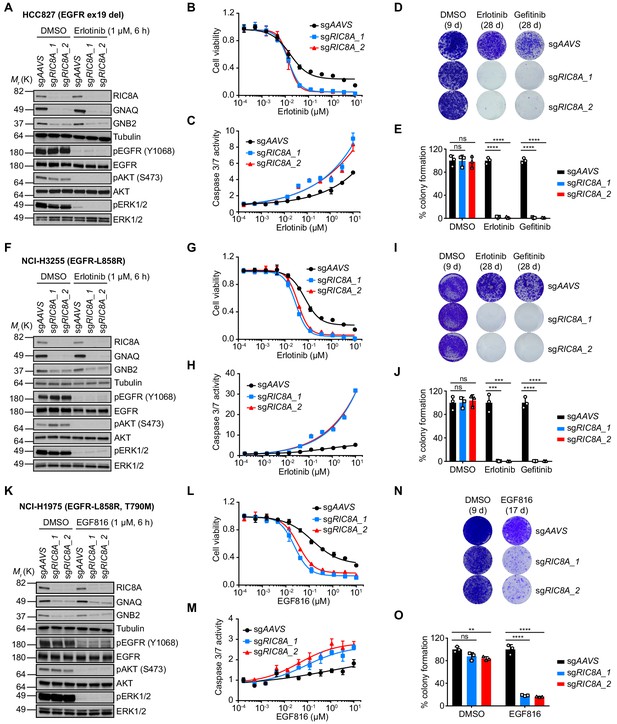
RIC8A depletion causes synthetic lethality with EGFR-TKI in EGFR-mutant NSCLC.
(A) Immunoblots of indicated proteins in control (sgAAVS) or RIC8A knockout (sgRIC8A) HCC827 cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of RIC8A and on-target inhibition of EGFR pathway by erlotinib treatment. Tubulin was used as a loading control. (B) Cell viability assessment by CellTiter-Glo assay of HCC827 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± standard deviation (SD); n = 4. (C) Activated caspase 3/7 measurement of HCC827 cells treated with serial dilutions of erlotinib for 24 hr. Error bars represent mean ± SD; n = 4. (D) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (E) Quantification of colony formation in (D), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown. (F) Immunoblots of indicated proteins in control or RIC8A knockout NCI-H3255 cells treated with DMSO or erlotinib (1 µM) for 6 hr to confirm specific knockout of RIC8A and on-target inhibition of EGFR pathway by erlotinib treatment. Tubulin was used as a loading control. (G) Cell viability assessment by CellTiter-Glo assay of NCI-H3255 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (H) Activated caspase 3/7 measurement of NCI-H3255 cells treated with serial dilutions of erlotinib for 24 hr. Error bars represent mean ± SD; n = 4. (I) Crystal violet staining colony formation assay of indicated NCI-H3255 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (J) Quantification of colony formation in (I), shown as percentage of the sgAAVS sample. Mean (three biological replicates)± SD is shown. (K) Immunoblots of indicated proteins in control or RIC8A knockout NCI-H1975 cells treated with DMSO or EGF816 (1 µM) for 6 hr to confirm specific knockout of RIC8A and on-target inhibition of EGFR pathway by EGF816 treatment. Tubulin was used as a loading control. (L) Cell viability assessment by CellTiter-Glo assay of NCI-H1975 cells treated with serial dilutions of EGF816 for 72 hr. Error bars represent mean ± SD; n = 4. (M) Activated caspase 3/7 measurement of NCI-H1975 cells treated with serial dilutions of EGF816 for 24 hr. Error bars represent mean ± SD; n = 4. (N) Crystal violet staining colony formation assay of indicated NCI-H1975 cell lines treated with DMSO or EGF816 (1 µM). (O) Quantification of colony formation in (N), shown as percentage of the sgAAVS sample. Mean (three biological replicates) ± SD is shown. Statistical significance was tested using unpaired two-tailed t test (E, J and O); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
-
Figure 3—source data 1
Raw data from Figure 3.
- https://cdn.elifesciences.org/articles/50223/elife-50223-fig3-data1-v2.xlsx
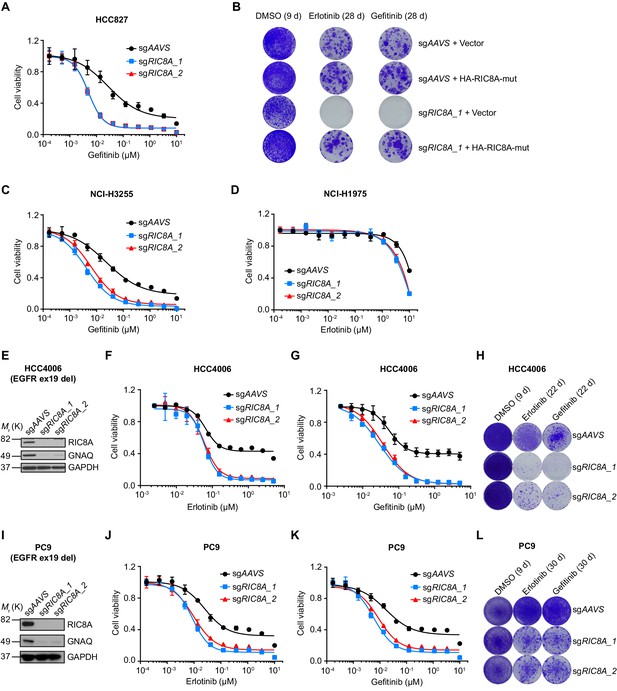
RIC8A loss is synthetic lethal with EGFR-TKI in EGFR-mutant NSCLC.
(A) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout HCC827 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (B) Crystal violet staining colony formation assay showing the rescued phenotype by overexpression of CRISPR/Cas9-resistant RIC8A. (C) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout NCI-H3255 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (D) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout NCI-H1975 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (E) Immunoblots of RIC8A and GNAQ confirm the RIC8A knockout efficiency and functional outcome of RIC8A knockout in HCC4006 cells. GAPDH was used as a loading control. (F) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout HCC4006 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (G) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout HCC4006 cells treated with serial dilutions of gefitnib for 72 hr. Error bars represent mean ± SD; n = 4. (H) Crystal violet staining colony formation assay of control or RIC8A knockout HCC4006 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (I) Immunoblots of RIC8A and GNAQ confirm the RIC8A knockout efficiency and functional outcome of RIC8A knockout in PC9 cells. GAPDH was used as a loading control. (J) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout PC9 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (K) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout PC9 cells treated with serial dilutions of gefitnib for 72 hr. Error bars represent mean ± SD; n = 4. (L) Crystal violet staining colony formation assay of control or RIC8A knockout PC9 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM).
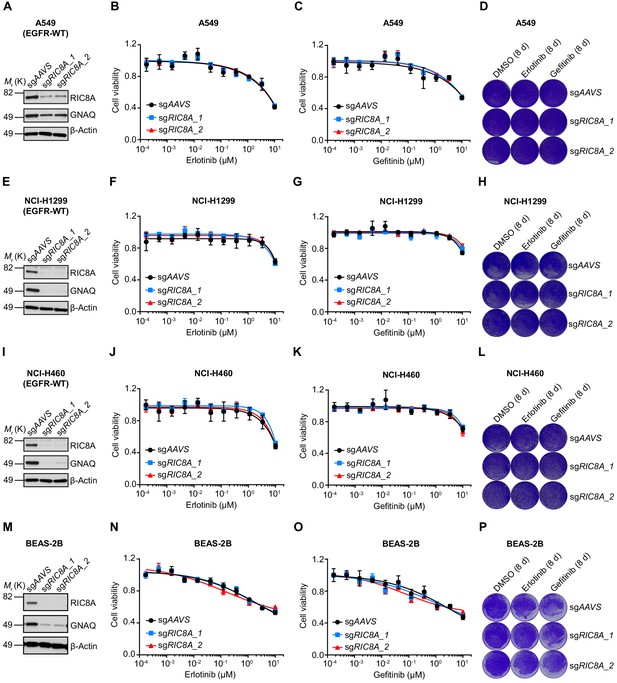
RIC8A loss exhibits no effect on EGFR TKI sensitivity in EGFR-WT NSCLC cells or normal human bronchial epithelial cells.
(A) Immunoblots of RIC8A and GNAQ confirm the RIC8A knockout efficiency and functional outcome of RIC8A knockout in A549 cells. β-Actin was used as a loading control. (B) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout A549 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (C) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout A549 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (D) Crystal violet staining colony formation assay of control or RIC8A knockout A549 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (E) Immunoblots of RIC8A and GNAQ confirm the RIC8A knockout efficiency and functional outcome of RIC8A knockout in NCI-1299 cells. β-Actin was used as a loading control. (F) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout NCI-H1299 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (G) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout NCI-H1299 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (H) Crystal violet staining colony formation assay of control or RIC8A knockout NCI-H1299 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (I) Immunoblots of RIC8A and GNAQ confirm the RIC8A knockout efficiency and functional outcome of RIC8A knockout in NCI-H460 cells. β-Actin was used as a loading control. (J) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout NCI-H460 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (K) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout NCI-H460 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (L) Crystal violet staining colony formation assay of control or RIC8A knockout NCI-H460 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (M) Immunoblots of RIC8A and GNAQ confirm the RIC8A knockout efficiency and functional outcome of RIC8A knockout in BEAS-2B cells. β-Actin was used as a loading control. (N) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout BEAS-2B cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (O) Cell viability assessment by CellTiter-Glo assay of control or RIC8A knockout BEAS-2B cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (P) Crystal violet staining colony formation assay of control or RIC8A knockout BEAS-2B cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM).
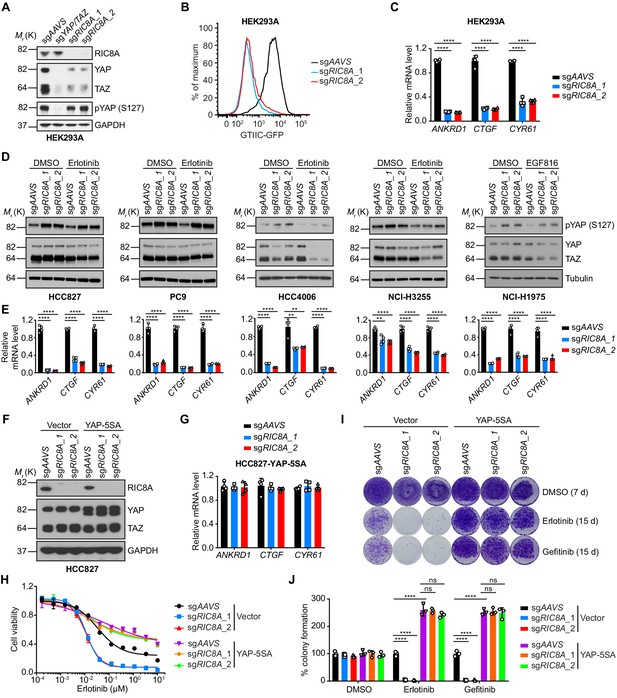
RIC8A loss attenuates YAP signaling to synergize with EGFR-TKI in EGFR-mutant NSCLC.
(A) Immunoblots of indicated proteins in HEK293A cells upon knockout of indicated genes. GAPDH was used as a loading control. (B) RIC8A knockout decreases the YAP reporter activity in HEK293A cells, assessed by flow cytometry analysis of GFP. (C) Quantitative RT-PCR analysis of relative mRNA levels of YAP target genes in control or RIC8A knockout HEK293A cells. Error bars represent mean ± SD; n = 4. (D) Immunoblots of pYAP (S127) and YAP/TAZ in indicated EGFR-mutant NSCLC cell lines treated with DMSO or 1 µM EGFR-TKI (Erlotinib, except EGF816 for NCI-H1975) for 24 hr. Tubulin was used as a loading control. (E) Quantitative RT-PCR analysis of relative mRNA levels of YAP target genes in control or RIC8A knockout EGFR-mutant NSCLC cell lines. Error bars represent mean ± SD; n = 4. (F) Immunoblots of RIC8A and YAP/TAZ in indicated HCC827 cells to confirm RIC8A knockout and YAP-5SA overexpression. GAPDH was used as loading control. (G) Quantitative RT-PCR analysis of relative mRNA levels of YAP target genes in control or RIC8A knockout HCC827 cells with ectopic expression of constitutively active YAP (YAP-5SA). Error bars represent mean ± SD; n = 4. (H) Cell viability assessment by CellTiter-Glo assay of indicated HCC827 cell lines treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (I) Crystal violet staining colony formation assay of indicated HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (J) Quantification of colony formation in (I), shown as percentage of the Vector-sgAAVS sample. Error bars represent mean ± SD; n = 3. Statistical significance was tested using unpaired two-tailed t test (C and E) or ordinary two-way ANOVA (J); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
-
Figure 4—source data 1
Raw data from Figure 4.
- https://cdn.elifesciences.org/articles/50223/elife-50223-fig4-data1-v2.xlsx
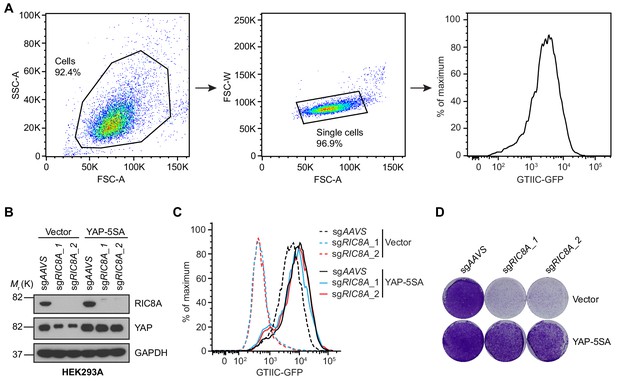
Overexpression of constitutively active YAP (YAP-5SA) blocks RIC8A loss induced reduction in YAP signaling and growth defect in HEK293A cells.
(A) Representative flow cytometry gating strategy to analyze GTIIC-GFP YAP reporter activity in HEK293A cells. Cells were gated using FSC-A/SSC-A characteristics, singlets were gated using FSC-A/FSC-W characteristics, and GFP signal was plotted as a histogram. (B) Immunoblots of RIC8A and YAP in indicated HEK293A cells to confirm RIC8A knockout and YAP-5SA overexpression. GAPDH was used as loading control. (C) Histogram showing that RIC8A knockout decreases the YAP reporter activity in HEK293A cells, which can be rescued by ectopic expression of YAP-5SA, assessed by flow cytometry analysis of GFP. (D) Crystal violet staining colony formation assay of the indicated HEK293A cell lines showing that RIC8A knockout reduces HEK293A cell growth, which can be rescued by ectopic expression of YAP-5SA.
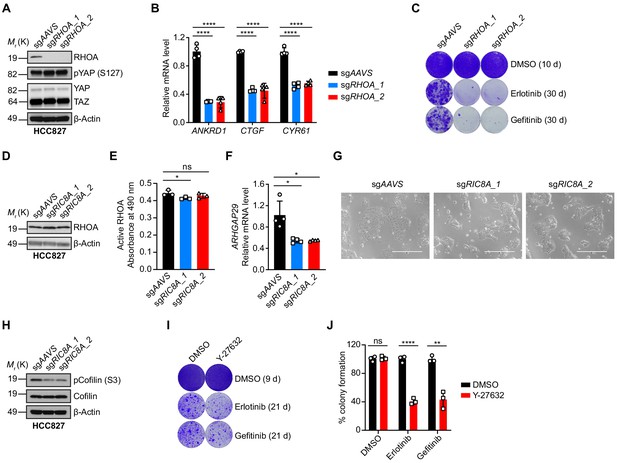
RHOA signaling is involved in RIC8A-mediated regulation of EGFR TKI sensitivity.
(A) Immunoblots of indicated proteins in control or RHOA knockout HCC827 cells. β-Actin was used as a loading control. (B) Quantitative RT-PCR analysis of relative mRNA levels of YAP target genes in control or RHOA knockout HCC827 cells. Error bars represent mean ± SD; n = 4. (C) Crystal violet staining colony formation assay showing the synthetic lethality of RHOA knockout with EGFR inhibition in HCC827 cells. (D) Immunoblot of RHOA in control or RIC8A knockout HCC827 cells. β-Actin was used as a loading control. (E) RHOA G-LISA activation assay to detect the active RHOA signal from control or RIC8A knockout HCC827 cells. Error bars represent mean ± SD; n = 3. (F) Quantitative RT-PCR analysis of relative ARHGAP29 mRNA level in control or RIC8A knockout HCC827 cells. Error bars represent mean ± SD; n = 4. (G) Photomicrographs of control or RIC8A knockout HCC827 cells showing the morphological change upon RIC8A knockout. (H) Immunoblots of indicated proteins in control or RIC8A knockout HCC827 cells. β-Actin was used as a loading control. (I) Crystal violet staining colony formation assay showing the synthetic lethality of Y-27632 treatment with EGFR inhibition in HCC827 cells. (J) Quantification of colony formation in (I), shown as percentage of the DMSO sample. Error bars represent mean ± SD; n = 3. Statistical significance was tested using unpaired two-tailed t test (B, E, F and J); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
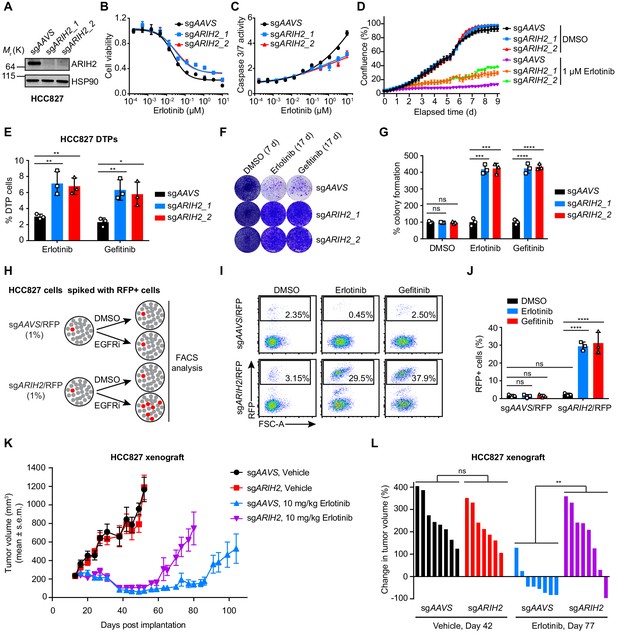
ARIH2 loss confers resistance to EGFR-TKI in vitro and in vivo.
(A) Immunoblot of ARIH2 confirms the ARIH2 knockout efficiency in HCC827 cells. HSP90 was used as a loading control. (B) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout HCC827 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (C) Activated caspase 3/7 measurement of control or ARIH2 knockout HCC827 cells treated with serial dilutions of erlotinib for 24 hr. Error bars represent mean ± SD; n = 4. (D) Kinetic cell proliferation assay monitored by IncuCyte for indicated HC827 cell lines cultured in the presence of DMSO control or 1 µM erlotinib over a 9 day period. (E) Drug-tolerant persister (DTP) cells were generated by treating control and ARIH2 knockout HCC827 cells with 1 µM of erlotnib or gefitinib for 9 d. Percentage of DTP cells is shown relative to DMSO-treated cells. Error bars represent mean ± SD; n = 3. (F) Crystal violet staining colony formation assay of control or ARIH2 knockout HCC827 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (G) Quantification of colony formation in (F), shown as percentage of the sgAAVS sample. Error bars represent mean ± SD; n = 3. (H) Schematic outline of the competitive proliferation assay to assess the selective outgrowth of ARIH2 knockout HCC827 cells upon EGFR-TKI treatment. RFP-negative HCC827 cells were spiked with approximately 1% RFP-positive sgRNA-infected HCC827 cells, control (sgAAVS) or ARIH2 knockout (sgARIH2), and grown for 3 weeks in the absence or presence of 1 µM erlotinib or gefitinib. Cells were collected and analyzed for RFP positivity by FACS. (I) Selective outgrowth of ARIH2 knockout cells in the presence of EGFR-TKI in HCC827 cell line. The percentage of RFP-positive cells is indicated. FSC, forward scatter. (J) Quantification of the selective outgrowth of ARIH2 knockout cells in the presence of EGFR-TKI as shown in (I). Mean (three biological replicates) ± SD is shown. (K) ARIH2 knockout promotes acquired resistance to erlotinib in HCC827 xenograft model. Mice bearing HCC827 xenografts, control (sgAAVS) or ARIH2 knockout (sgARIH2), were dosed once daily with 10 mg/kg erlotinib or vehicle for the indicated time frame. Data are represented as mean tumor volume (mm3) ± s.e.m., n = 8 mice for each line. (L) Percentage change in tumor volume compared to baseline (the start of dosing, day 13 post-implantation) for individual cell xenografts treated for 29 d (day 42 post-implantation) with vehicle or 64 d (day 77 post-implantation) with 10 mg/kg erlotinib. Statistical significance was tested using unpaired two-tailed t test (E, G and L) or ordinary two-way ANOVA (J); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
-
Figure 5—source data 1
Raw data from Figure 5.
- https://cdn.elifesciences.org/articles/50223/elife-50223-fig5-data1-v2.xlsx
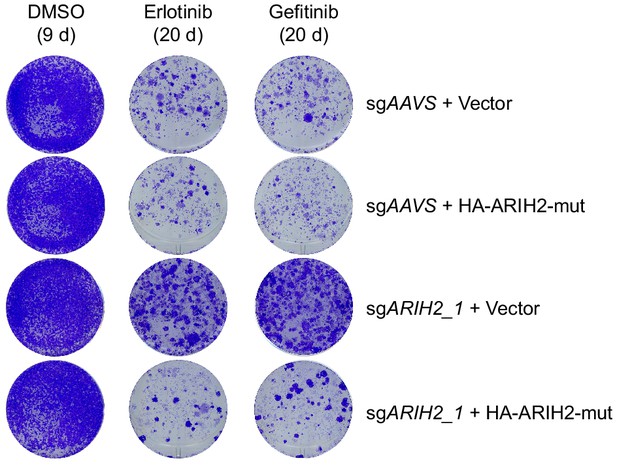
Crystal violet staining colony formation assay showing the rescued phenotype by overexpression of CRISPR/Cas9-resistant ARIH2.
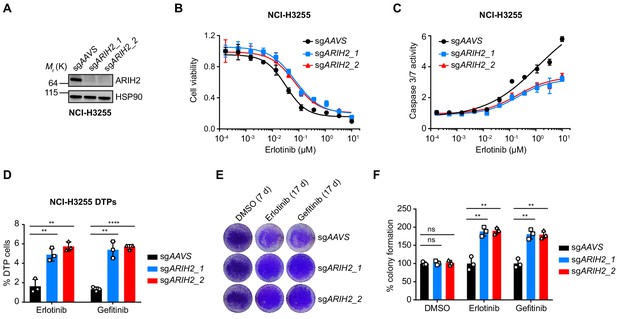
ARIH2 loss confers resistance to EGFR-TKI.
(A) Immunoblot of ARIH2 confirms the ARIH2 knockout efficiency in NCI-H3255 cells. HSP90 was used as a loading control. (B) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout NCI-H3255 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (C) Activated caspase 3/7 measurement of control or ARIH2 knockout NCI-H3255 cells treated with serial dilutions of erlotinib for 24 hr. Error bars represent mean ± SD; n = 4. (D) Drug-tolerant persister (DTP) cells were generated by treating control and ARIH2 knockout NCI-H3255 cells with 1 µM of erlotnib or gefitinib for 9 d. Percentage of DTP cells is shown relative to DMSO-treated cells. Error bars represent mean ± SD; n = 3. (E) Crystal violet staining colony formation assay of control or ARIH2 knockout NCI-H3255 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (F) Quantification of colony formation in (E), shown as percentage of the sgAAVS sample. Error bars represent mean ± SD; n = 3. Statistical significance was tested using unpaired two-tailed t test (D and F); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
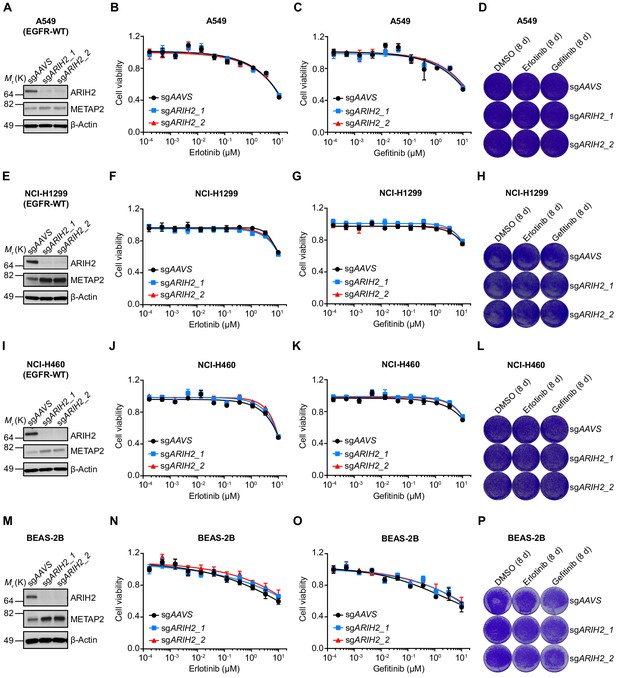
ARIH2 loss exhibits no effect on EGFR TKI sensitivity in EGFR-WT NSCLC cells or normal human bronchial epithelial cells.
(A) Immunoblots of ARIH2 and METAP2 in control or ARIH2 knockout A549 cells. β-Actin was used as a loading control. (B) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout A549 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (C) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout A549 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (D) Crystal violet staining colony formation assay of control or ARIH2 knockout A549 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (E) Immunoblots of ARIH2 and METAP2 in control or ARIH2 knockout NCI-H1299 cells. β-Actin was used as a loading control. (F) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout NCI-H1299 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (G) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout NCI-H1299 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (H) Crystal violet staining colony formation assay of control or ARIH2 knockout NCI-H1299 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (I) Immunoblots of ARIH2 and METAP2 in control or ARIH2 knockout NCI-H460 cells. β-Actin was used as a loading control. (J) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout NCI-H460 cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (K) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout NCI-H460 cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (L) Crystal violet staining colony formation assay of control or ARIH2 knockout NCI-H460 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (M) Immunoblots of ARIH2 and METAP2 in control or ARIH2 knockout BEAS-2B cells. β-Actin was used as a loading control. (N) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout BEAS-2B cells treated with serial dilutions of erlotinib for 72 hr. Error bars represent mean ± SD; n = 4. (O) Cell viability assessment by CellTiter-Glo assay of control or ARIH2 knockout BEAS-2B cells treated with serial dilutions of gefitinib for 72 hr. Error bars represent mean ± SD; n = 4. (P) Crystal violet staining colony formation assay of control or ARIH2 knockout BEAS-2B cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM).
Graph showing the gain in body weight relative to day 13 post-implantation (the start of dosing).
Error bars represent mean ± s.e.m., n = 8 mice for each line.

Mechanistic insights into ARIH2 loss-mediated EGFR-TKI resistance.
(A) Mass spectrometry analysis of global protein changes between control and ARIH2-deficient HCC827 cells. (B) Immunoblots of indicated proteins in control and ARIH2-deficient HCC827 cells. β-Actin was used as a loading control. (C) Quantitative RT-PCR analysis of relative mRNA levels of indicated genes in control or ARIH2-deficient HCC827 cells. Error bars represent mean ± SD; n = 4. (D) Immunoblots of indicated proteins showing ectopic expression of HA-METAP2 in HCC827 cells. GAPDH was used as a loading control. (E) Crystal violet staining colony formation assay of HCC827-Vector or HCC827-HA-METAP2 cell lines treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (F) Quantification of colony formation in (E), shown as percentage of the HCC827-Vector sample. Mean (three biological replicates) ± SD is shown. (G) De novo protein synthesis of HCC827-Vector or HCC827-HA-METAP2 cells after treatment with DMSO or erlotinib (1 µM, 24 hr) as determined by L-azidohomoalanine (AHA) labeling. Cells were starved of methionine for 1 hr and incubated with AHA for 1 hr. Lysates were subjected to a Click-iT chemistry reaction to switch azido-modified nascent proteins to alkyne-biotin, and visualized by Streptavidin-HRP immunoblotting. β-Actin was used as a loading control. (H) De novo protein synthesis of control or ARIH2-deficient HCC827 cells after treatment with DMSO or erlotinib (1 µM, 24 hr) as determined by AHA labeling. β-Actin was used as a loading control. (I) Immunoblot of METAP2 in HCC827 cells upon proteasome inhibitor bortezomib treatment. β-Actin was used as a loading control. (J) Immunoblots of ARIH2 and METAP2 in control or ARIH2 knockout HCC827 cells upon bortezomib treatment. β-Actin was used as a loading control. (K) De novo METAP2 protein synthesis in control or ARIH2-deficient HCC827 cells as determined by AHA labeling and streptavidin pulldown. β-Actin was used as a loading control. (L) Immunoblots of indicated proteins showing ectopic expression of HA-ALDOA and HA-PSAT1 in HCC827 cells. GAPDH was used as a loading control. (M) Crystal violet staining colony formation assay of HCC827-Vector, HCC827-HA-ALDOA or HCC827-HA-PSAT1 cells treated with DMSO, erlotinib (1 µM), or gefitinib (1 µM). (N) Quantification of colony formation in (M), shown as percentage of the HCC827-Vector sample. Mean (three biological replicates) ± SD is shown. Statistical significance was tested using unpaired two-tailed t test (C, F and N); *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
-
Figure 6—source data 1
Raw data from Figure 6.
- https://cdn.elifesciences.org/articles/50223/elife-50223-fig6-data1-v2.xlsx
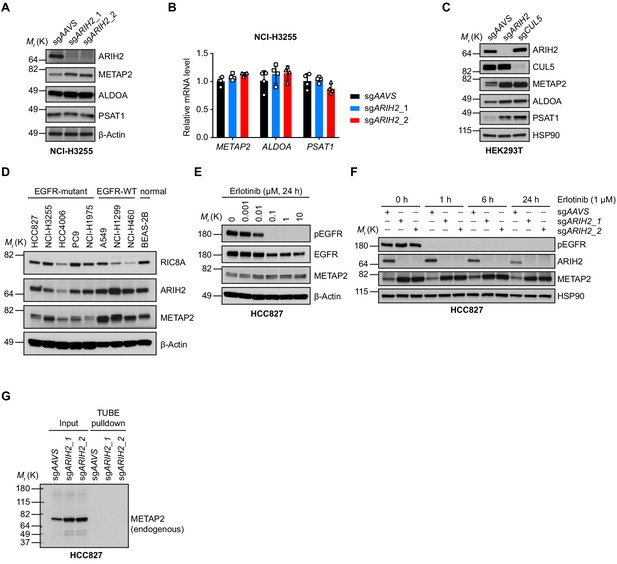
ARIH2 loss increases protein abundance of METAP2, ALDOA and PSAT1.
(A) Immunoblots of indicated proteins in control and ARIH2-deficient NCI-H3255 cells. β-Actin was used as a loading control. (B) Quantitative RT-PCR analysis of relative mRNA levels of indicated genes in control or ARIH2-deficient NCI-H3255 cells. Error bars represent mean ± SD; n = 4. (C) Immunoblots of indicated proteins in control and ARIH2- or CUL5-deficient HEK293T cells. HSP90 was used as a loading control. (D) Immunoblots of indicated proteins in a panel of EGFR-mutant and EGFR-WT NSCLC cell lines and the normal human bronchial epithelial cell line BEAS-2B. β-Actin was used as a loading control. (E) Immunoblots of indicated proteins in HCC827 cells upon erlotinib treatment. β-Actin was used as a loading control. (F) Immunoblots of indicated proteins in control or ARIH2 knockout HCC827 cells upon erlotinib treatment. β-Actin was used as a loading control. (G) Detection of endogenous METAP2 protein ubiquitination in HCC827 cells using the TUBE assay. Control or ARIH2 knockout HCC827 cells were treated with 0.1 µM bortezomib for overnight and harvested for TUBE assay.
Additional files
-
Supplementary file 1
Selected hits from the erlotinib resistance CRISPR-Cas9 screen.
A threshold of RSA ≤ −3 and Q1 z-score ≤ −1 generated a list of 122 genes whose loss sensitized HCC827 cells to erlotinib treatment. A threshold of RSA ≤ −3 and Q3 z-score ≥1 generated a list of 171 genes whose loss conferred resistance to erlotinib in HCC827 cells.
- https://cdn.elifesciences.org/articles/50223/elife-50223-supp1-v2.xlsx
-
Supplementary file 2
Individual sgRNAs and log2 fold change for selected hits.
Individual sgRNA target sequences and their respective log2 fold change based on the comparison of sgRNA abundance in the erlotinib-treated versus DMSO-treated cell population were listed in this table.
- https://cdn.elifesciences.org/articles/50223/elife-50223-supp2-v2.xlsx
-
Supplementary file 3
Key resources table.
- https://cdn.elifesciences.org/articles/50223/elife-50223-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50223/elife-50223-transrepform-v2.pdf





