Release of cholesterol-rich particles from the macrophage plasma membrane during movement of filopodia and lamellipodia
Figures
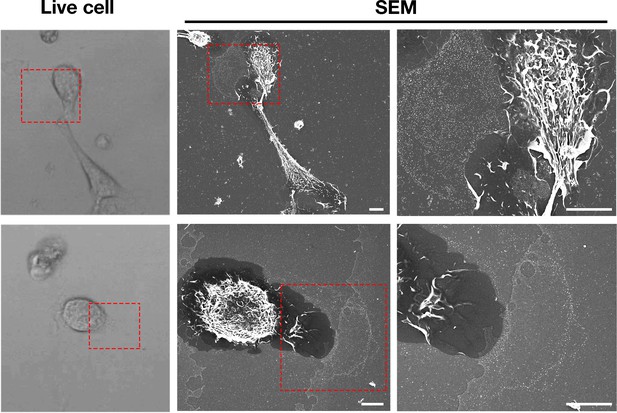
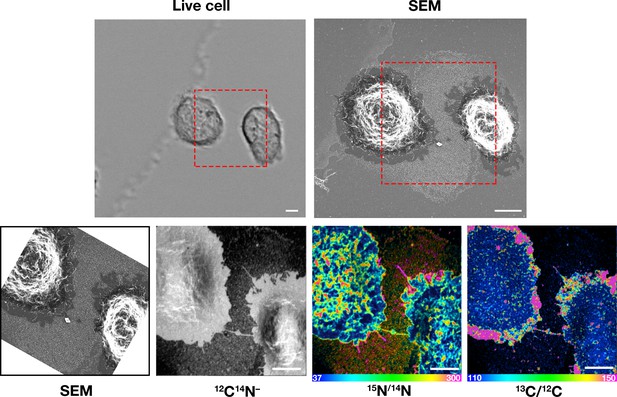
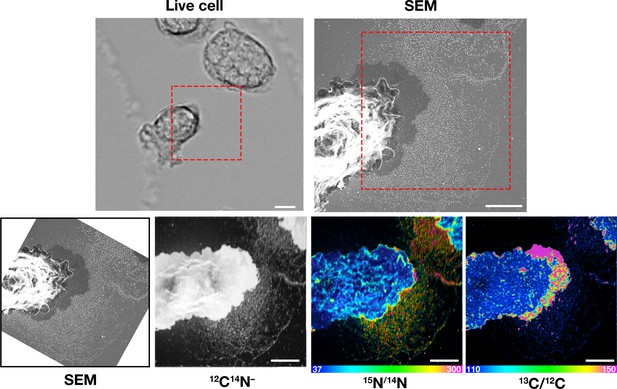
Macrophages release plasma membrane–derived particles onto the substrate during extension and retraction of filopodia and lamellipodia, as judged by correlative live-cell imaging and SEM.
Cells were plated onto poly-D-lysine–coated gridded glass-bottom Petri dishes, and videos were recorded for 24 hr at 5 min intervals (see Figure 1—videos 1–2). The ‘Live cell’ images show the final frame of the videos. The imaging of cells by SEM made it possible to visualize a lawn of particles that had been released onto the substrate during the projection and retraction of filopodia/lamellipodia. The red boxed region in the live-cell image and in the low-magnification SEM image is shown in the SEM image on the far right. Three independent experiments were performed; representative images are shown. Scale bar, 5 μm.
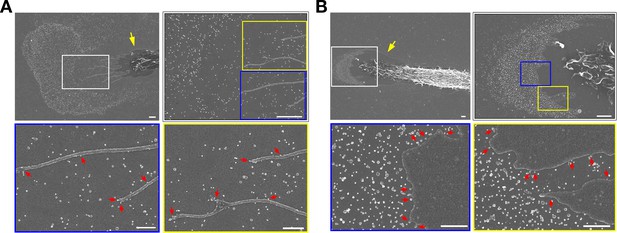
Macrophages release particles from the plasma membrane of filopodia and lamellipodia by a process that resembles budding.
(A) Upper left, scanning electron micrograph (SEM) of a mouse peritoneal macrophage (yellow arrow), revealing a lawn of ~30-nm particles on the surrounding substrate. A higher magnification image of the region in the white box is shown in the upper right image, centered on macrophage filopodia. Higher magnification images of the regions in the yellow and blue boxes are shown in the lower two images. (B) Upper left, SEM of a mouse peritoneal macrophage (yellow arrow), revealing a lawn of ~30-nm particles on the surrounding substrate. A higher magnification image of the region in the white box is shown in the image on the upper right, centered on the lamellipodium of the macrophage. Higher magnification images of the regions in the blue and yellow boxes are shown in the images below. For both panels, red arrows show the formation and release of particles from macrophage filopodia and lamellipodia. Three independent experiments were performed; representative images are shown. Scale bar for the top two images, 2 μm. Scale bar for the bottom two images, 500 nm.
Mouse peritoneal macrophages release vesicular particles onto the surrounding substrate during the extension and retraction of filopodia and lamellipodia.
Video shows a macrophage that was imaged by SEM in the top row of Figure 1. Macrophages were plated onto poly-D-lysine–coated gridded glass-bottom Petri dishes, and videos were recorded for 24 h at 5-min intervals. White arrow in videos point to the cell that was visualized by SEM; the red arrow in the videos points to a region of lamellipodia projection/retraction; the white box depicts the region of the cell that was subsequently visualized by scanning electron microscopy (SEM) (see Figure 1).
Video shows a macrophage that was imaged by SEM in the top row of Figure 1. Video shows a 24-h period of live-cell imaging.
Shows a macrophage imaged by SEM in the bottom row of Figure 1.
Video shows the final 15 h of a 24-h period of live-cell imaging.
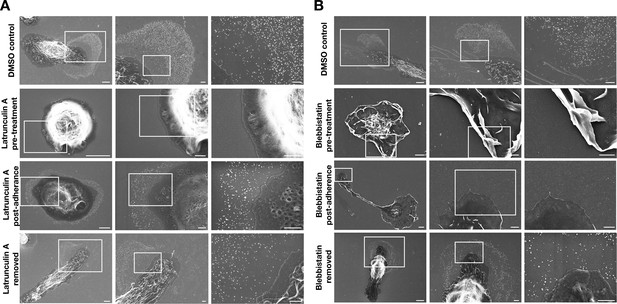
Inhibiting macrophage movement with latrunculin A or blebbistatin abolishes particle release onto the surrounding substrate.
Scanning electron micrographs (SEMs) of mouse peritoneal macrophages that had been treated with latrunculin A (A) or blebbistatin (B). Macrophages were treated with latrunculin A, blebbistatin, or vehicle alone (DMSO) overnight; the drug treatments were initiated when the cells were in suspension 1 h before plating ('pre-treatment') or 1 h after adherence to the substrate ('post-adherence'). Macrophages treated with vehicle alone (DMSO) released particles onto the substrate. No particles were visible on the substrate in cells that had been pre-treated with latrunculin A or blebbistatin—or in the blebbistatin post-adherence cells. In the latrunculin A post-adherence cells, a circumferential ring of particles was observed around cells. Latrunculin A and blebbistatin were removed from some dishes after the overnight incubation, and the cells were incubated for an additional 18 h without drugs. After removing the drugs, the release of particles onto the substrate resumed. Four independent experiments were performed; representative images are shown. Scale bars for images on the left in each panel are 4 µm; scale bars for the images in the middle and right are 1 µm.
Treatment of mouse peritoneal macrophages with latrunculin A or blebbistatin prevent projection and retraction of filopodia/lamellipodia.
Video shows macrophages treated with vehicle (DMSO) alone. Macrophages were plated on glass-bottom Petri dishes and incubated in medium containing latrunculin A (5 μM), blebbistatin (30 μM), or vehicle (DMSO) alone. Cells were imaged by live-cell microscopy for 18 h at 5-min intervals. Movement of macrophage lamellipodia was observed in the 'DMSO Control' macrophages but not in macrophages treated with latrunculin A or blebbistatin. Videos show an 18-h period of live-cell imaging.
Video shows macrophages treated with vehicle (DMSO) alone.
Shows macrophages treated with vehicle (DMSO) alone.
https://doi.org/10.7554/eLife.50231.008Shows macrophages treated with latrunculin A.
https://doi.org/10.7554/eLife.50231.009Shows macrophages treated with latrunculin A.
https://doi.org/10.7554/eLife.50231.010Shows a macrophage treated with blebbistatin.
https://doi.org/10.7554/eLife.50231.011Shows macrophages treated with blebbistatin.
https://doi.org/10.7554/eLife.50231.012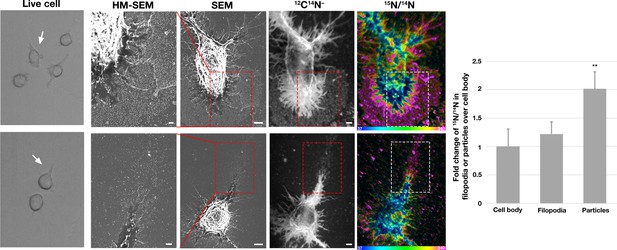
Correlative live-cell, scanning electron microscopy (SEM), and NanoSIMS imaging, revealing that particles released onto the substrate during movement of filopodia and lamellipodia are enriched in accessible cholesterol.
RAW 264.7 macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes. Videos were recorded for 24 hr at 5 min intervals (see Figure 3—videos 1–2). The ‘Live cell’ images in this figure show the final frame of the videos, with the white arrows pointing to the cells that were subsequently visualized by SEM and NanoSIMS. After live-cell imaging, cells were incubated with [15N]ALO-D4 (a modified cytolysin that binds to ‘accessible cholesterol’). The same cells that were imaged by live-cell imaging were subsequently imaged by SEM (to visualize particles) and NanoSIMS (to visualize [15N]ALO-D4 binding). The particles left behind on the substrate during movement of lamellipodia and filopodia bound [15N]ALO-D4 avidly. 12C14N– NanoSIMS images were used to visualize cell morphology; the 15N/14N images show 15N enrichment (i.e., binding of [15N]ALO-D4). The boxed region in the SEM and NanoSIMS images is shown at higher magnification in the ‘HM-SEM’ image, providing definition of individual particles. 15N enrichment was ~twofold higher in the macrophage particles than on the plasma membrane over the cell body and the filopodia. Two independent experiments were performed; representative images are shown. Quantification of 15N/14N ratios were performed on the cell body and macrophage particles [25 distinct regions of the cell body, filopodia, and particles (20 pixels in diameter) were circled on the 12C14N– image, followed by calculation of 15N/14N ratios in each region]. Images from six macrophages were used for the quantification. Graph shows the mean and standard deviation of the fold change of 15N enrichment in particles and filopodia, normalized to the macrophage cell body. **p<0.001. Scale bar, 2 μm.
RAW 264.7 macrophages release particles during projection and retraction of lamellipodia.
Video shows a macrophage imaged by SEM and NanoSIMS in the top row of Figure 3. RAW 264.7 macrophages were plated onto iridium- and poly-D-lysine–coated glass-bottom Petri dishes and incubated in media containing 10% FBS. Cells were imaged by live-cell microscopy for 24 h at 5-min intervals. White arrows point to the projection and retraction of the lamellipodia. White box indicates the region that was subsequently imaged by scanning electron microscopy (see Figure 3).
Video shows a macrophage imaged by SEM and NanoSIMS in the top row of Figure 3. Video shows the final 12 h of a 24-h period of live-cell imaging.
Shows a macrophage imaged by SEM and NanoSIMS in the bottom row of Figure 3.
Video shows the final 15 h of a 24-h period of live-cell imaging.
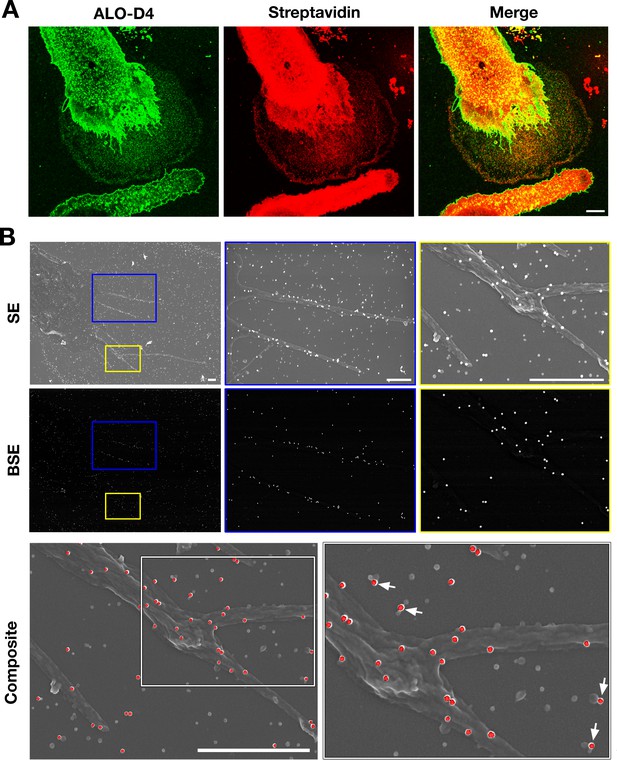
Particles released from the plasma membrane of biotinylated mouse peritoneal macrophages can be detected with streptavidin and ALO-D4 (a cytolysin that binds to the accessible pool of cholesterol).
(A) Biotinylated macrophage particles can be detected with fluorescently labeled streptavidin as judged by confocal microscopy. Macrophages in suspension were biotinylated with Sulfo-NHS-SS-biotin. After plating the cells onto glass coverslips and incubating the cells in macrophage growth medium containing 10% FBS, the cells were washed and then incubated with Atto 647N–labeled streptavidin (red) and Alexa Fluor 488–labeled [15N]ALO-D4 (green). Cells were then fixed with 3% PFA and imaged by super-resolution stimulated emission depletion (STED) microscopy. Streptavidin and ALO-D4 bound to the macrophages as well as the lawn of particles on the surrounding substrate. Scale bar, 5 µm. (B) Biotinylated macrophage particles can be detected with streptavidin-conjugated gold nanoparticles, as judged by scanning electron microscopy (SEM). After biotinylating the plasma membrane of mouse peritoneal macrophages with Sulfo-NHS-SS-biotin, cells were plated onto glass-bottom Petri dishes. On the following day, the cells were incubated with streptavidin-conjugated 40-nm gold nanoparticles. Cells were then fixed with 1% glutaraldehyde and processed for SEM. Secondary electron (SE) and backscattered electron (BSE) images revealed gold nanoparticles on the macrophage cell body, filopodia, and on macrophage particles that had been released onto the substrate. Higher magnification images of the blue and yellow boxes are shown on the right. Composite images (gray, SE; red, BSE) images show colocalization of gold nanoparticles with the particles on the substrate. Higher magnification image of the white box is shown on the right. White arrows point to gold nanoparticles binding to macrophage particles. Three independent experiments were performed; representative images are shown. Scale bar, 1 µm.
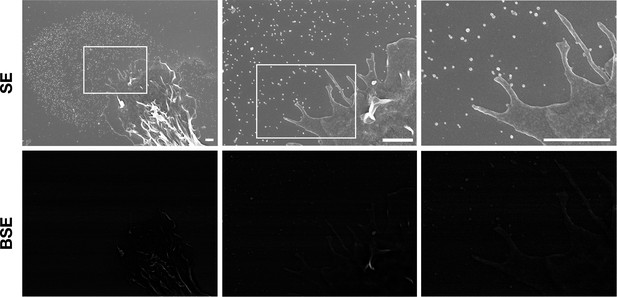
Particles released from non-biotinylated macrophages do not bind streptavidin-conjugated gold nanoparticles as judged by scanning electron microscopy (SEM).
Non-biotinylated macrophages were plated onto glass-bottom Petri dishes. On the following day, the cells were incubated with streptavidin-conjugated 40-nm gold nanoparticles. Cells were then fixed with 1% glutaraldehyde and processed for SEM. Secondary electron (SE) and backscattered electron (BSE) images revealed no binding of gold nanoparticles to the macrophage cell body, filopodia, or on particles that had been released onto the substrate. Higher magnification images of the boxed region are shown on the right. Three independent experiments were performed; representative images are shown. Scale bar, 1 μm.
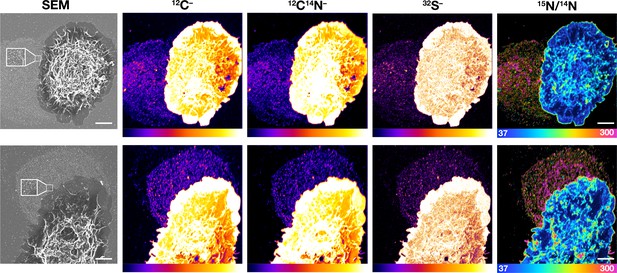
Correlative SEM and NanoSIMS imaging of macrophages and plasma membrane–derived particles on the surrounding substrate.
Mouse peritoneal macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes and incubated in medium containing 10% FBS for 24 hr. Cells were then incubated with [15N]ALO-D4 for 2 hr, washed, and prepared for NanoSIMS imaging. The lawn of particles on the surrounding substrates was detectable on 12C–, 12C14N–, and 32S– NanoSIMS images. There was strong binding of the [15N]ALO-D4 to the lawn of particles surrounding cells, evident in the 15N/14N ratio image. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
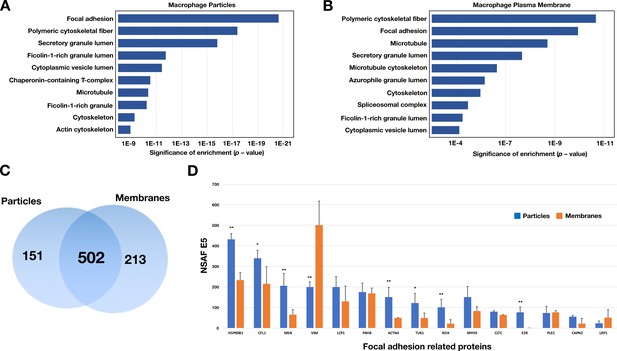
Enrichment in focal adhesion proteins in the particle preparations from RAW 264.7 macrophages.
The most abundant proteins (the top 75th percentile by spectral counts) were analyzed by Enrichr and categorized by GO Cellular Components 2018. (A–B) Analysis of proteins in macrophage particles (n = 653) and macrophage plasma membranes (n = 715) by GO categories. The top 10 cellular component categories were ordered by level of statistical significance. (C) Venn diagram depicting numbers of proteins present in the particle preparation only, the plasma membrane preparation only, or both. (D) Bar graph showing the top 15 focal adhesion–related proteins by the normalized spectral abundance factor (NSAF), multiplied by 10,000. The particle fraction is shown in blue; the plasma membrane fraction is shown in orange. The bar graph shows the mean ± SD for three independent experiments. *p<0.05; **p<0.001.
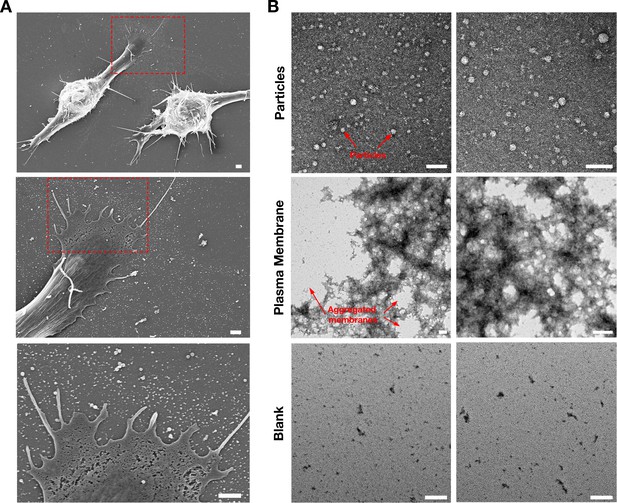
Isolation of particles released onto the substrate by RAW 264.7 macrophages.
(A) SEM images of RAW 264.7 macrophages plated onto poly-D-lysine–coated silicon wafers, revealing large numbers of particles on the substrate surrounding the macrophages. Higher magnification images of the red boxed regions are shown in the image below. Scale bar, 1 μm. (B) Negative stained transmission electron micrographs (TEMs) of the particle and plasma membrane preparations from RAW macrophages. Macrophages were plated in tissue culture flasks, and both macrophages and macrophage-derived particles were biotinylated with Sulfo-NHS-SS-biotin. After releasing the cells and the particles from the substrate with 5 mM EDTA, preparations of macrophage particles and plasma membranes were isolated as described in the Materials and methods. Particle and plasma membrane preparations were placed on carbon/formar TEM grids, negatively stained with 2% uranyl acetate, and visualized by TEM. Particles were 20–80-nm in diameter (red arrows). Images of the plasma membrane fractions revealed aggregated membranous material (red arrows). A blank grid, which was also subjected to negative staining, contained no particles or membranes. Three independent experiments were performed; representative images are shown. Scale bar, 100 nm.
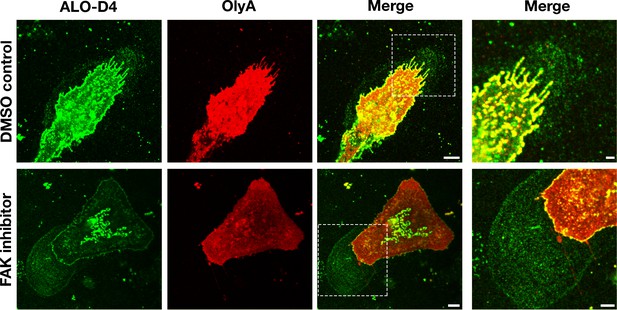
Particles released by mouse peritoneal macrophages onto the surrounding substrate are enriched in accessible cholesterol but not sphingomyelin-sequestered cholesterol.
Mouse peritoneal macrophages were plated onto poly-D-lysine–coated glass coverslips and incubated overnight in medium containing 10% FBS and either an FAK inhibitor (CAS 4506-66-5, 2 μM) or vehicle (DMSO) alone. On the next day, the cells were incubated with Alexa Fluor 488–labeled [15N]ALO-D4 (green), which binds to accessible cholesterol, and Atto 647N–labeled [13C]OlyA (red), which binds to sphingomyelin-bound cholesterol (both at 20 μg/ml). Cells were then washed, fixed with 3% PFA, and imaged by STED microscopy. STED images were obtained from the bottom of the macrophage (optical section of ~200 nm). The lawn of particles surrounding macrophages was readily detectable with ALO-D4, but the binding of OlyA to particles was negligible. Two independent experiments were performed; representative images are shown. Scale bar, 5 µm. Higher magnification images of the boxed regions are shown on the right. Scale bar, 2 µm.
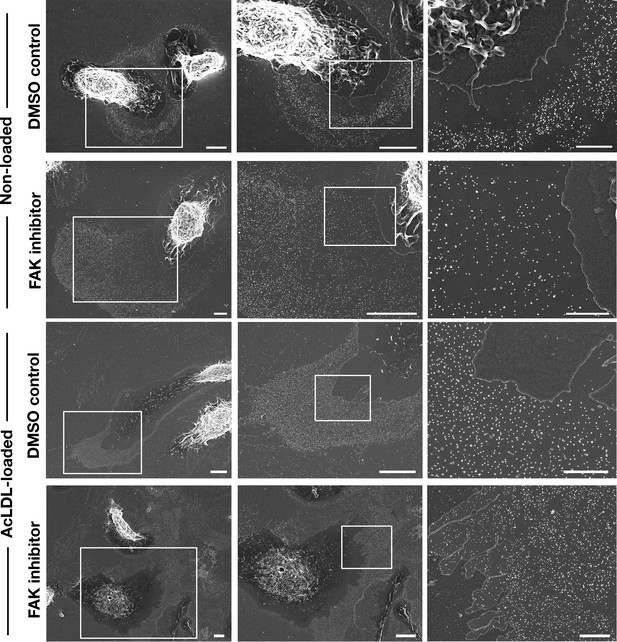
Inhibiting focal adhesion kinase in mouse peritoneal macrophages is accompanied by large lawns of particles on the surrounding substrate.
Macrophages were plated onto poly-D-lysine–coated glass-bottom Petri dishes and incubated with a focal adhesion kinase (FAK) inhibitor, 2 μM)] or vehicle (DMSO) alone. In some experiments, macrophages were loaded with acetyl-LDL (AcLDL, 50 μg/ml). The lawn of particles surrounding macrophages was imaged by SEM. Higher magnification images of the white boxed areas are shown on the right. Four independent experiments were performed, with a minimum of 20 cells visualized per experiment. The lawns of particles outside of FAK inhibitor–treated macrophages were ~twice the size of the macrophage cell body, whereas the lawns were only ~40% of the area of the cell body in DMSO-treated macrophages. Scale bars for the images in the left and middle columns, 5 μm. Scale bars for images in the right column, 2 μm.
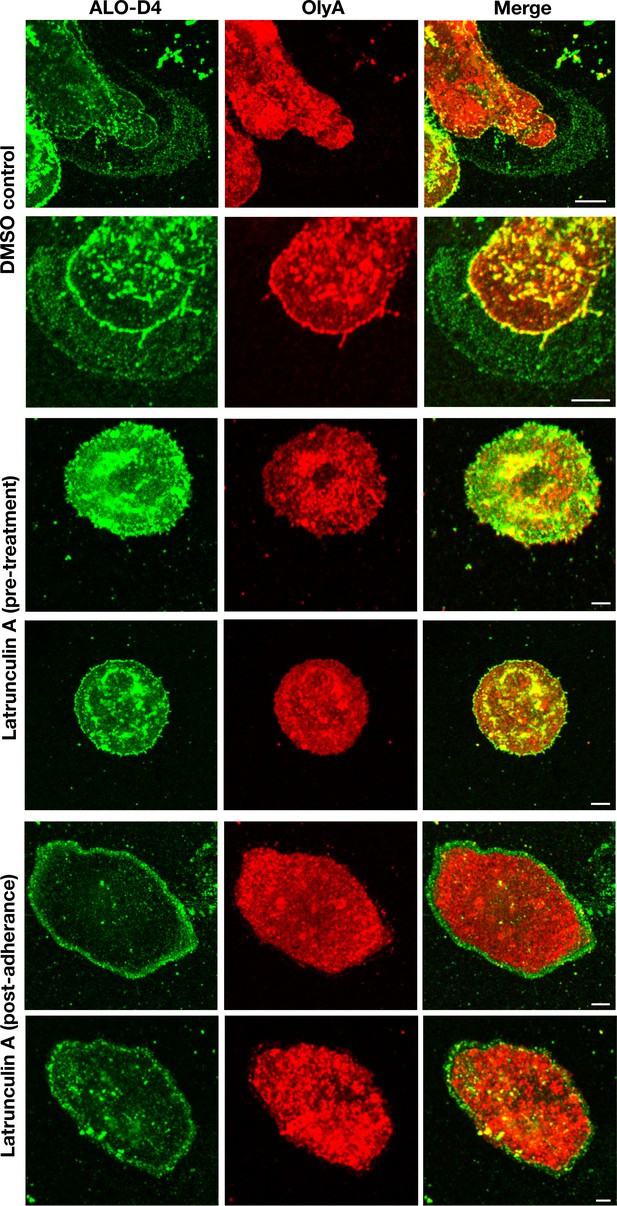
Incubating mouse peritoneal macrophages with latrunculin A alters the distribution of ALO-D4 binding.
Macrophages were plated onto poly-D-lysine–coated glass coverslips and incubated with latrunculin A (5 μM) or vehicle alone (DMSO control). The incubation of latrunculin A was initiated either 1 hr prior to plating the cells (‘pre-treatment’) or after the cells had been plated and were adherent to the substrate (‘post-adherence’). On the next day, cells were incubated with Alexa Fluor 488–labeled ALO-D4 and Atto 647N–labeled OlyA (20 μg/ml each). Cells were then fixed with 3% PFA and imaged by STED microscopy. The lawn of particles surrounding macrophages was readily detectable with ALO-D4 but not with OlyA. In post-adherence cells, a circumferential ring of ALO-D4 binding was detected, reflecting ALO-D4 binding to particles released onto the substrate during retraction of the macrophage cell body. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
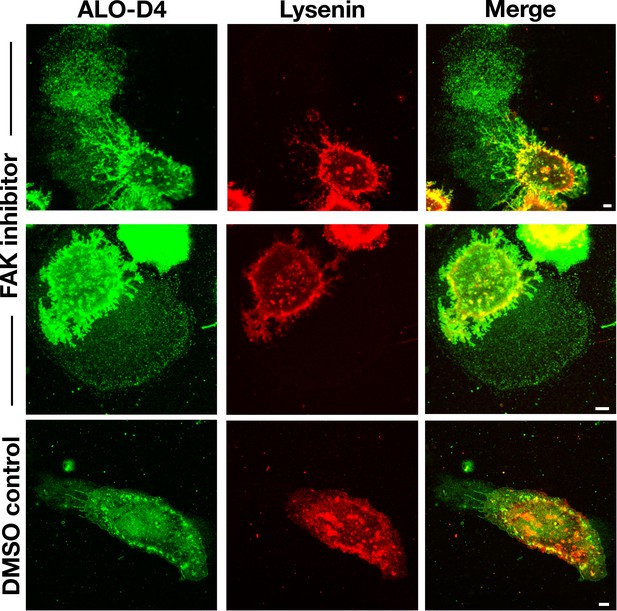
Particles released by mouse peritoneal macrophages are enriched in accessible cholesterol but not in sphingomyelin.
Mouse peritoneal macrophages were plated onto poly-D-lysine–coated glass coverslips and incubated overnight in medium containing 10% FBS and either a focal adhesion kinase (FAK) inhibitor (2 μM) or vehicle (DMSO) alone. On the next day, macrophages were incubated with Alexa Fluor 488–labeled [15N]ALO-D4, a marker of accessible cholesterol (green, 20 μg/ml), and a mCherry–lysenin fusion protein, which binds to sphingomyelin (red, 10 μg/ml). Cells were then washed, fixed with 3% PFA, and imaged by STED microscopy. STED images were obtained from the bottom of the macrophage (optical section of ~200 nm). The lawn of particles around macrophages could be detected by ALO-D4 but not by the mCherry–lysenin fusion protein. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
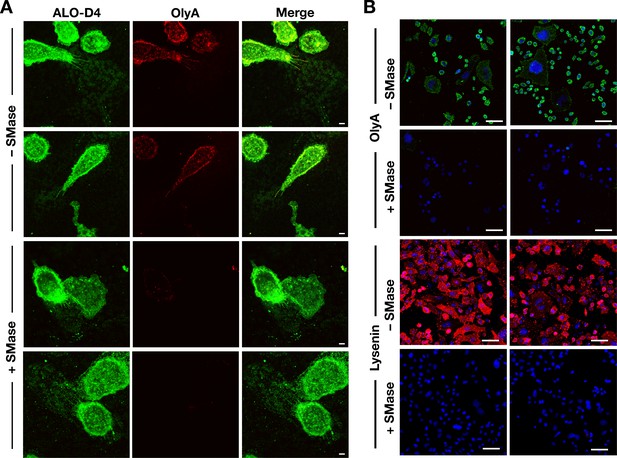
Sphingomyelinase treatment reduces OlyA and lysenin binding to the plasma membrane.
(A) Sphingomyelinase markedly reduces OlyA binding to the macrophage plasma membrane. Mouse peritoneal macrophages were plated onto poly-D-lysine–coated glass coverslips and incubated overnight in medium containing 10% FBS. On the next day, cells were incubated for 30 min at 37°C in the presence or absence of sphingomyelinase (SMase) from Staphylococcus aureus (100 milliunits/ml). After washing, cells were incubated with Alexa Fluor 488–labeled ALO-D4, a marker of accessible cholesterol (green, 20 μg/ml), and Atto 647N–labeled OlyA, which detects sphingomyelin-bound cholesterol (red, 20 μg/ml). Scale bar, 5 μm. (B) Sphingomyelinase treatment abolishes OlyA and lysenin binding to CHO-K1 cells. CHO-K1 cells were plated onto poly-D-lysine–coated glass coverslips and incubated overnight in Ham’s F-12 medium containing 10% FBS. On the next day, cells were incubated for 30 min at 37°C in the presence or absence of sphingomyelinase (SMase) (100 milliunits/ml). After washing, the cells were incubated for with Alexa Fluor 488–labeled OlyA, a marker of sphingomyelin-bound cholesterol (green, 20 μg/ml), or with the mCherry–lysenin fusion protein, which binds to sphingomyelin (red, 10 μg/ml). Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
Movement of mouse peritoneal macrophages under different experimental conditions.
Video shows macrophages treated with vehicle (DMSO) alone. Macrophages were plated onto glass-bottom Petri dishes and incubated in medium containing an FAK inhibitor (2 μM), acetylated LDL (acLDL; 50 μg/ml), acLDL and the FAK inhibitor, or vehicle alone (DMSO). Cells were imaged by live-cell microscopy for 30 h at 5-min intervals. Videos show a 30-h period of live-cell imaging.
Video shows macrophages treated with vehicle (DMSO) alone.
Shows macrophages treated with an FAK inhibitor.
https://doi.org/10.7554/eLife.50231.027Shows acLDL-loaded macrophages treated with vehicle (DMSO) alone.
https://doi.org/10.7554/eLife.50231.028Shows acLDL-loaded macrophages treated with an FAK inhibitor.
https://doi.org/10.7554/eLife.50231.029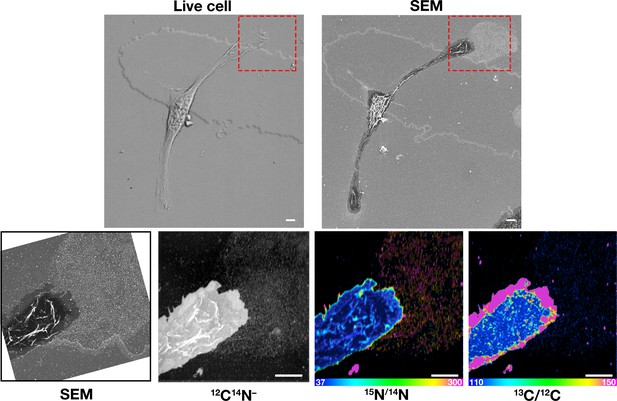
Correlative live-cell, SEM, and NanoSIMS imaging of mouse peritoneal macrophages, demonstrating that particles released onto the substrate during movement of filopodia and lamellipodia are enriched in accessible cholesterol but not sphingolipid-sequestered cholesterol.
Macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes, and videos of cell movement were recorded for 24 h at 5-min intervals (see Figure 7—video 1). The red arrow in the video points to an area of lamellipodia projection/retraction; the white box depicts the area that was later visualized by SEM and NanoSIMS. The 'Live cell' image in this figure shows the final frame of the video. After live-cell imaging, the cells were then incubated with [15N]ALO-D4 (which binds to accessible cholesterol) and [13C]OlyA (which binds to sphingomyelin-sequestered cholesterol). The cells were then imaged by SEM (to visualize particles) and then by NanoSIMS to visualize [15N]ALO-D4 and [13C]OlyA binding. The NanoSIMS images were created from secondary ions released from the top of the macrophage plasma membrane and the surrounding macrophage particles. Thus, the image differs from confocal fluorescence images in Figure 6, where we focused on the bottom surface of the macrophage (optical section of ~200 nm). The particles released onto the substrate were enriched in accessible cholesterol but not sphingomyelin-sequestered cholesterol. 12C14N– images were useful for cell morphology; 15N/14N images depict binding of [15N]ALO-D4; 13C/12C images depict binding of [13C]OlyA. Here, the 13C/12C and 15N/14N scales were adjusted for visualization of macrophage-derived particles. Additional NanoSIMS images of this cell, with different scales, are shown in Figure 8A. Two independent experiments were performed; representative images are shown. Scale bar, 5 µm.
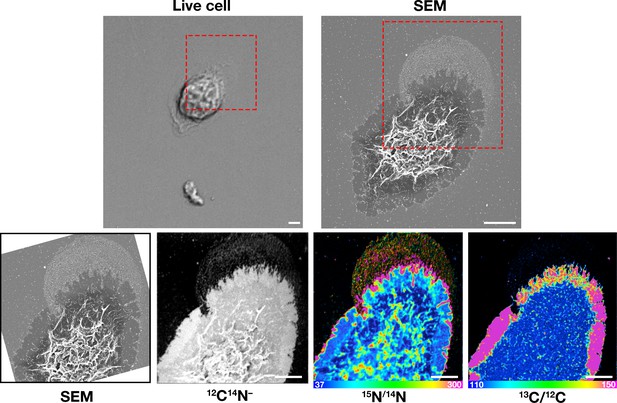
Correlative live-cell, SEM, and NanoSIMS imaging of mouse peritoneal macrophages, demonstrating that particles released onto the substrate during movement of filopodia and lamellipodia are enriched in accessible cholesterol but not sphingolipid-bound cholesterol.
Macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes, and videos of cell movement were recorded for 24 hr at 5 min intervals (see Figure 7—video 2). The red arrow in the video points to an area of lamellipodia projection/retraction; the white box depicts the area that was later visualized by SEM and NanoSIMS. The ‘Live cell’ image in this figure shows the final frame of the video. After live-cell imaging, the cells were then incubated with [15N]ALO-D4 (which binds to accessible cholesterol) and [13C]OlyA (which binds to sphingomyelin-bound cholesterol). The cells were then imaged by SEM (to visualize particles) and then by NanoSIMS to visualize [15N]ALO-D4 and [13C]OlyA binding. The NanoSIMS images were created from secondary ions released from the top surface of the macrophage plasma membrane and the surrounding macrophage particles. Thus, the image differs from confocal fluorescence images in Figure 6, where we focused on the bottom surface of the macrophage (optical section of ~ 200 nm). 12C14N– images were useful for cell morphology; 15N/14N images depict binding of [15N]ALO-D4; 13C/12C images depict binding of [13C]OlyA. In this figure, the scales for the 13C/12C and 15N/14N NanoSIMS images were adjusted for optimal visualization of macrophage-derived particles. Other NanoSIMS images of this cell (with different scales) are shown in Figure 8A. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
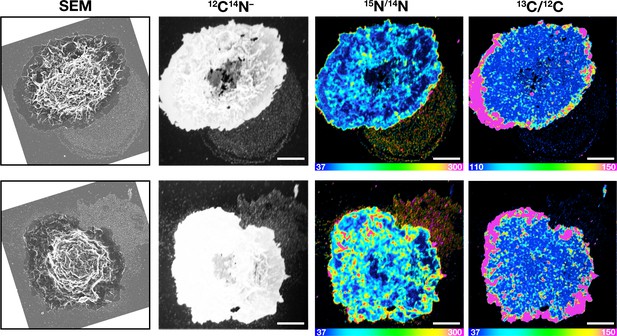
Correlative SEM and NanoSIMS imaging, demonstrating that particles released onto the substrate by macrophages are enriched in accessible cholesterol but not sphingolipid-bound cholesterol.
Mouse peritoneal macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes. Cells were then incubated at 4°C with [15N]ALO-D4 (which binds accessible cholesterol) and [13C]OlyA (which binds sphingomyelin-bound cholesterol). The cells were then imaged by SEM (to visualize particles) and then by NanoSIMS to quantify [15N]ALO-D4 and [13C]OlyA binding. The particles released onto the substrate were enriched in accessible cholesterol but not sphingomyelin-bound cholesterol. 12C14N– images were useful for cell morphology; 15N/14N images show binding of [15N]ALO-D4; 13C/12C images show binding of [13C]OlyA. In this figure, the scales for the 13C/12C and 15N/14N NanoSIMS images were adjusted for optimal visualization of macrophage-derived particles. Other NanoSIMS images of this cell (with different scales) are shown in Figure 8B. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
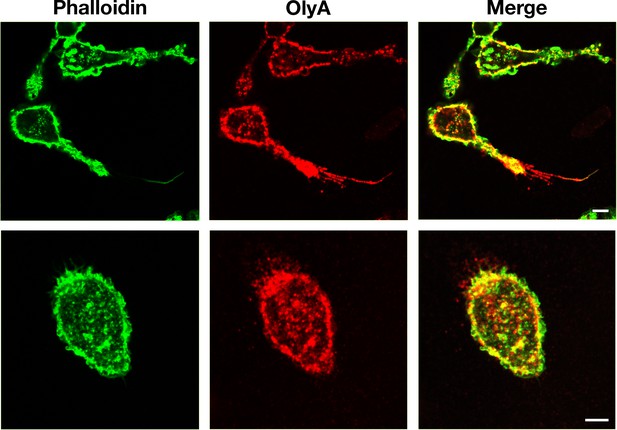
The binding of fluorescently labeled OlyA to the plasma membrane overlaps the staining of the actin cytoskeleton with phalloidin.
Mouse peritoneal macrophages were plated onto poly-D-lysine–coated glass coverslips and incubated in macrophage growth medium containing 10% FBS. On the next day, cells were incubated at 4°C with Atto 647N–labeled OlyA (20 μg/ml). Cells were then fixed with 3% PFA, permeabilized with 0.2% Triton X-100, and stained with Alexa Fluor 546–labeled phalloidin. Images were recorded using STED microscopy. OlyA binding (red) to cells largely colocalized with phalloidin binding (green) to the actin cytoskeleton. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
Correlative live-cell, SEM, and NanoSIMS imaging of mouse peritoneal macrophages treated with an FAK inhibitor, demonstrating that particles released onto the substrate during movement of filopodia and lamellipodia are enriched in accessible cholesterol but not sphingolipid-bound cholesterol.
Mouse peritoneal macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes and incubated in medium containing an FAK inhibitor (2 μM). Videos of cell movement were recorded for 24 hr at 5 min intervals (see Figure 7—video 3). The red arrow in the video points to an area of lamellipodia projection/retraction; the white box depicts the area that was later visualized by SEM and NanoSIMS. The ‘Live cell’ image in this figure shows the final frame of the video. After live-cell imaging, the cells were then incubated with [15N]ALO-D4 (which binds to accessible cholesterol) and [13C]OlyA (which binds to sphingomyelin-bound cholesterol). The cells were then imaged by SEM (to visualize particles) and then by NanoSIMS to visualize [15N]ALO-D4 and [13C]OlyA binding. The NanoSIMS images were created from secondary ions released from the top surface of the macrophage plasma membrane and the surrounding macrophage particles. Thus, the image differs from confocal fluorescence images in Figure 6, where we focused on the bottom surface of the macrophage (optical section of ~ 200 nm). 12C14N– images were useful for cell morphology; 15N/14N images depict binding of [15N]ALO-D4; 13C/12C images depict binding of [13C]OlyA. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
Correlative live-cell, SEM, and NanoSIMS imaging of mouse peritoneal macrophages treated with an FAK inhibitor, demonstrating that particles released onto the substrate during movement of filopodia and lamellipodia are enriched in accessible cholesterol but not sphingolipid-bound cholesterol.
Mouse peritoneal macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes and incubated in medium containing an FAK inhibitor (2 μM). Videos of cell movement were recorded for 24 hr at 5 min intervals (see Figure 7—video 4). The red arrow in the video points to an area of lamellipodia projection/retraction; the white box depicts the area that was later visualized by SEM and NanoSIMS. The ‘Live cell’ image in this figure shows the final frame of the video. After live-cell imaging, the cells were then incubated with [15N]ALO-D4 (which binds to accessible cholesterol) and [13C]OlyA (which binds to sphingomyelin-bound cholesterol). The cells were then imaged by SEM (to visualize particles) and then by NanoSIMS to visualize [15N]ALO-D4 and [13C]OlyA binding. The particles released onto the substrate were enriched in accessible cholesterol but not sphingomyelin-bound cholesterol. 12C14N– images were useful for cell morphology; 15N/14N images depict binding of [15N]ALO-D4; 13C/12C images depict binding of [13C]OlyA. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm.
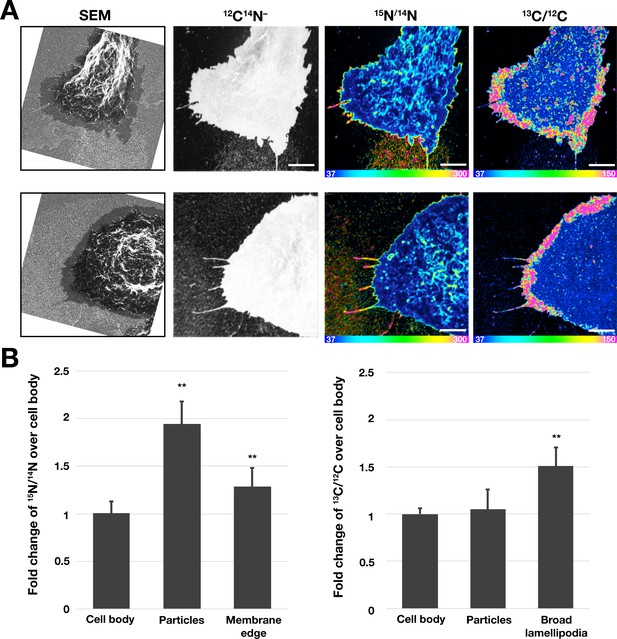
Correlative SEM and NanoSIMS imaging, demonstrating that particles released onto the substrate by macrophages treated with an FAK inhibitor are enriched in accessible cholesterol but not sphingolipid-bound cholesterol.
(A) Mouse peritoneal macrophages were plated onto iridium- and poly-D-lysine–coated gridded glass-bottom Petri dishes and incubated in medium containing an FAK inhibitor (2 μM). Cells were then incubated at 4°C with [15N]ALO-D4 (which binds accessible cholesterol) and [13C]OlyA (which binds sphingomyelin-bound cholesterol). The cells were then imaged by SEM (to visualize particles) and then by NanoSIMS to quantify [15N]ALO-D4 and [13C]OlyA binding. The particles released onto the substrate were enriched in accessible cholesterol but not sphingomyelin-bound cholesterol. 12C14N– images were useful for cell morphology; 15N/14N images show binding of [15N]ALO-D4; 13C/12C images show binding of [13C]OlyA. Two independent experiments were performed; representative images are shown. Scale bar, 5 μm. (B) Quantification, by NanoSIMS, of [15N]ALO-D4 and [13C]OlyA binding to macrophages that had been incubated with the FAK inhibitor and to the surrounding particles on the substrate. 15N/14N ratios were quantified for the cell body, macrophage-derived particles, and the thin line of 15N enrichment at the edge of the cell. 13C/12C ratios were quantified for the cell body, macrophage particles, and the broad lamellipodia near the edge of the plasma membrane. For each category, twenty-five regions, 20 pixels in diameter areas were circled, and the 15N/14N and 13C/12C ratios in each circle were calculated. A minimum of 6 macrophage images were used. Graph shows the mean and standard deviation of the fold change of 15N or 13C enrichment, normalized to the macrophage cell body. **p<0.001.
Macrophage release particles during movement of filopodia and lamellipodia movement.
Video shows a macrophage treated with vehicle (DMSO) imaged by SEM and NanoSIMS in Figure 7. Mouse peritoneal macrophages were plated onto iridium- and poly-D-lysine–coated glass-bottom Petri dishes and incubated in media containing 10% FBS and an FAK inhibitor (2 μM) or vehicle (DMSO) only. Cells were imaged by live-cell microscopy for 24 h at 5-min intervals. Red arrows point to the projection and retraction of the macrophage lamellipodia. White box indicates the area that is subsequently imaged by SEM.
Video shows a macrophage treated with vehicle (DMSO) imaged by SEM and NanoSIMS in Figure 7. Video shows a 24-h period of live-cell imaging.
Shows a macrophage treated with vehicle (DMSO) imaged by SEM and NanoSIMS in Figure 7—figure supplement 2.
Video shows a 24-h period of live-cell imaging.
Shows a macrophage treated with an FAK inhibitor imaged by SEM and NanoSIMS in Figure 7—figure supplement 4.
Video shows the final 10 h of a 24-h period of live-cell imaging.
Shows a macrophage treated with an FAK inhibitor imaged by SEM and NanoSIMS in Figure 7—figure supplement 5.
Video shows the final 10 h of a 24-h period of live-cell imaging.
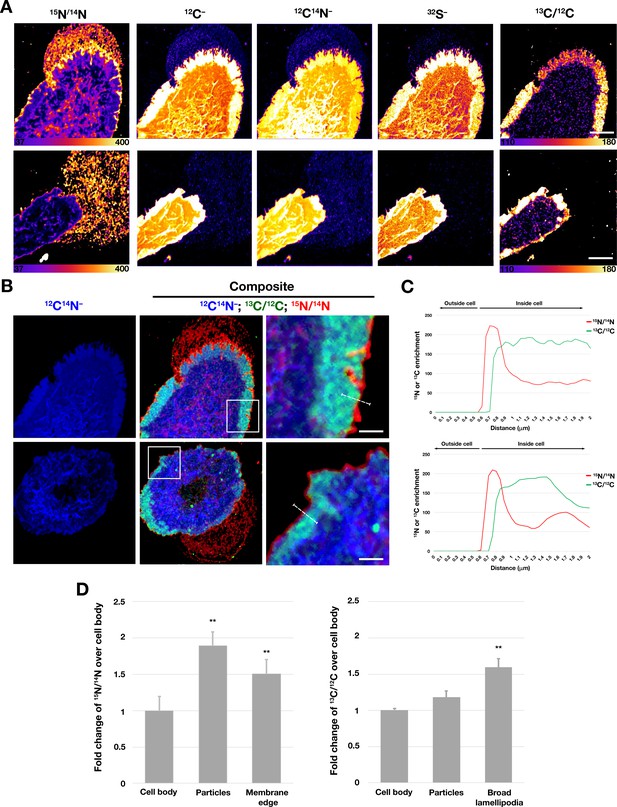
Distribution of distinct pools of cholesterol in the macrophage plasma membrane.
Mouse peritoneal macrophages were plated onto poly-D-lysine–coated MatTek dishes and incubated in medium containing 10% FBS. On the next day, cells were incubated with [15N]ALO-D4 and [13C]OlyA (20 μg/ml each) and then imaged by NanoSIMS. (A) Distributions of [15N]ALO-D4 and [13C]OlyA binding to the plasma membrane. 13C/12C NanoSIMS images revealed a broad band of 13C enrichment near the outer edge of the cell (corresponding to the flat lamellipodia of cells, apparent in the 12C–, 12C14N–, and 32S– NanoSIMS images). The 15N/14N images revealed a thin band of 15N enrichment at the far outer edge of the cells and in the lawn of particles on the surrounding substrate. Additional NanoSIMS images of these cells are shown (with different scales) in Figure 7 and Figure 7—figure supplement 1. (B) Composite 12C14N– (blue), 13C/12C (green), and 15N/14N (red) NanoSIMS images, revealing 15N enrichment at the outer edge of the plasma membrane and in the lawn of particles on the surrounding substrate. Of note, the 15N enrichment at the outer edge of the cell extended beyond the thick band of 13C enrichment. In this figure, the scales for the 13C/12C and 15N/14N NanoSIMS images were adjusted for optimal visualization of the lamellipodia of macrophages. Also, 12C– and 32S– images were included to visualize the lamellipodia. Scale bar, 5 μm. Additional NanoSIMS images of the cell in the top row of this panel are shown in Figure 8A and Figure 7—figure supplement 1; additional NanoSIMS images of the cell in the bottom row of this panel is shown in Figure 7—figure supplement 2. Two independent experiments were performed; representative images are shown. (C) Line scans comparing the 15N/14N and 13C/12C isotope ratios over the outer edge of the plasma membrane (white line in the upper and lower right images of panel B). (D) Quantification, by NanoSIMS, of [15N]ALO-D4 and [13C]OlyA binding to macrophages and to the surrounding particles on the substrate. 15N/14N ratios were quantified for the cell body, macrophage-derived particles, and the thin line of 15N enrichment at the edge of the cell. 13C/12C ratios were quantified for the cell body, macrophage particles, and the broad lamellipodia near the edge of the plasma membrane. For each category, twenty-five regions, 20 pixels in diameter areas were circled on the 12C14N– images, and the 15N/14N and 13C/12C ratios in each circle were calculated. A minimum of six macrophage images were used. Graph shows the mean and standard deviation of the fold change of 15N or 13C enrichment, normalized to the macrophage cell body. **p<0.001.
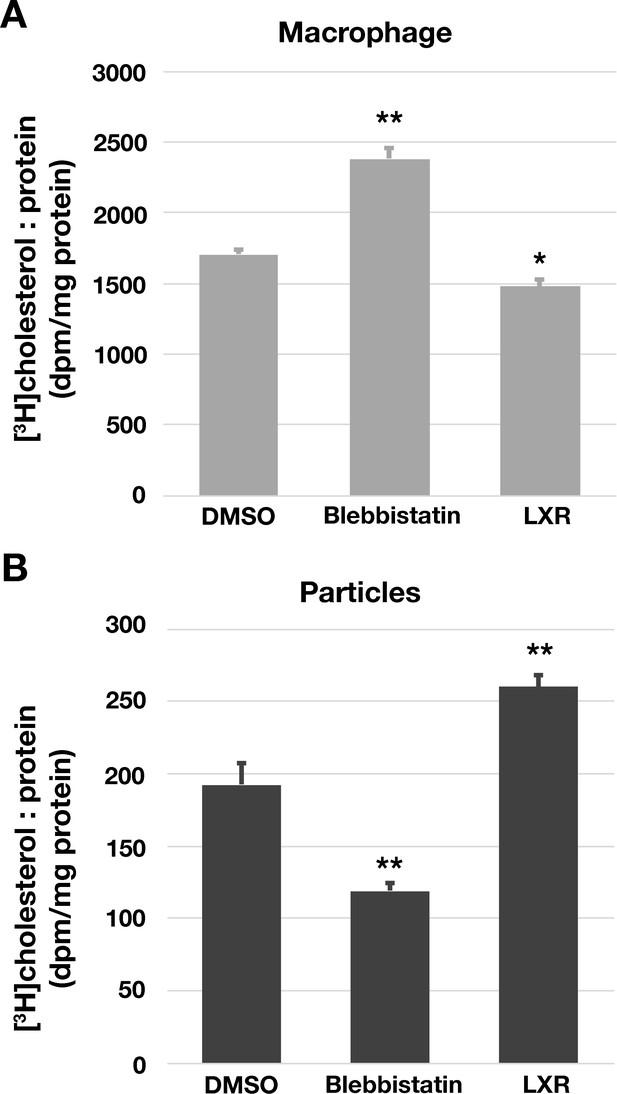
Reduced efflux of [3H]cholesterol from [3H]cholesterol-loaded macrophages when the release of particles was blocked with blebbistatin.
Mouse peritoneal macrophages were loaded with [3H]cholesterol (1 μCi/ml) and acLDL (20 μg/ml) in macrophage growth media containing 1% lipoprotein-deficient serum (LPDS) for 24 hr. On the next day, the cells were washed and the cells were released with 5 mM EDTA. The cells were then re-plated in six-well plates (1 × 106 cells/well) in medium containing blebbistatin (30 μM), an LXR agonist (1 μM), or vehicle (DMSO) alone. After 24 hr, the macrophages were washed, and both the macrophages and the particles were released from the plates by incubating the cells with 5 mM EDTA. (The complete release of particles and cells from the substrate was documented by SEM). Radioactivity in macrophages (A) and particles (B) was measured by scintillation counting. Graph shows the mean and standard deviation of radioactive counts normalized to the protein content from three independent experiments. **p<0.001 or *p<0.01 compared to cells incubated with DMSO alone.
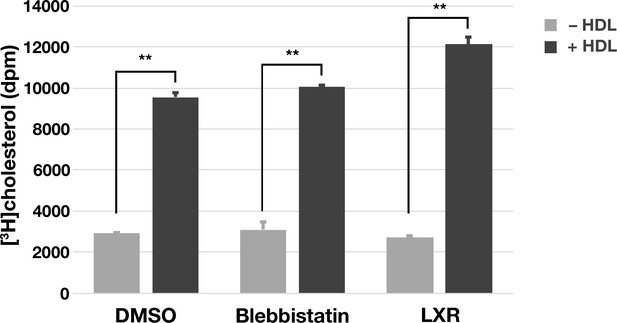
Increased efflux of [3H]cholesterol from [3H]cholesterol-loaded macrophages in the presence of HDL.
Mouse peritoneal macrophages were loaded with [3H]cholesterol (1 μCi/ml) and acLDL (20 μg/ml) in macrophage growth media containing 1% LPDS for 24 hr. On the next day, cells were washed, released from the plate with 5 mM EDTA, and then re-plated on a 6-well plate (1 × 106 cells/well) in serum-free macrophage growth medium containing blebbistatin (30 μM), an LXR agonist (1 μM), or vehicle (DMSO) alone. In some samples, HDL (20 μg/ml) was included in the medium. Cells were grown for 24 hr and then released with EDTA. The cell culture media was removed, centrifuged to remove cellular debris, and the radioactivity in the cell culture medium was measured by scintillation counting. Graph shows the mean and standard deviation of radioactive counts normalized to the protein content from two independent experiments. **p<0.001.
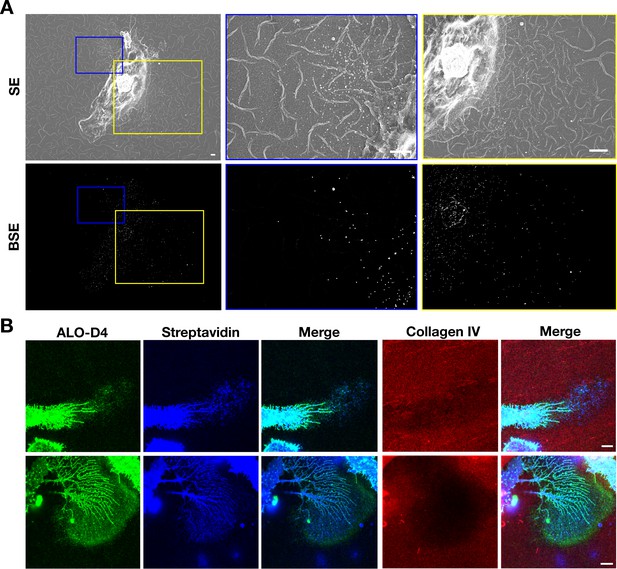
Mouse peritoneal macrophages release accessible cholesterol–enriched particles onto a polymerized collagen IV matrix.
After biotinylating the cell-surface proteins of macrophages with Sulfo-NHS-SS-biotin, the cells were plated onto glass-bottom Petri dishes that had been coated with polymerized Alexa Fluor 647–labeled collagen IV and then fixed with 0.1% glutaraldehyde (for SEM) or 1% paraformaldehyde (for fluorescence microscopy). (A) SEM images show binding of streptavidin-conjugated 40-nm gold nanoparticles to macrophages as well as particles on the collagen IV substrate. Secondary electron (SE) and backscattered electron (BSE) images show macrophage-derived particles on the collagen fibers and the binding of gold nanoparticles to the macrophage cell body, filopodia, and the plasma membrane–derived particles. BSE images were helpful to identify gold nanoparticles. Higher magnification images of the yellow and blue boxed regions are shown on the right. Scale bar, 2 μm. (B) Fluorescent microscopy studies of mouse peritoneal macrophages that had been plated for 24 hr on a polymerized Alexa Fluor 647–labeled collagen IV substrate and then incubated with Alexa Fluor 488–labeled ALO-D4 and Alexa Fluor 568–labeled streptavidin. The cells were then fixed with 3% PFA and images recorded by STED microscopy. ALO-D4 (green) and streptavidin (blue) were visualized on a lawn of particles on the collagen IV matrix (red). Four independent experiments were performed; representative images are shown. Scale bar, 5 μm.
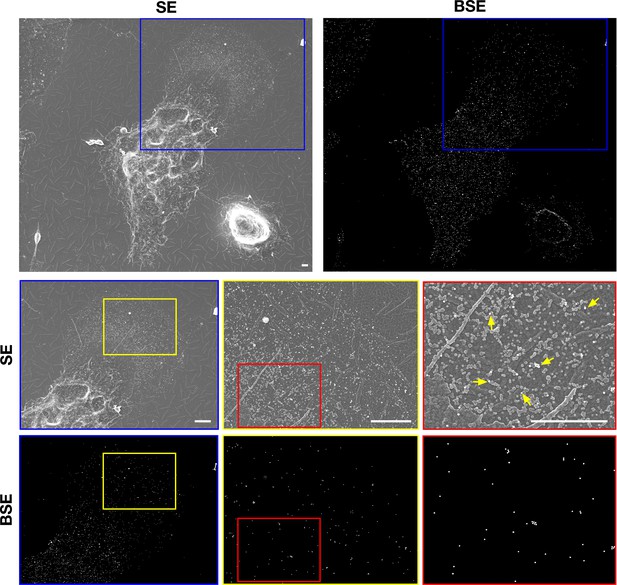
Mouse peritoneal macrophages release particles onto a polymerized collagen IV matrix.
After biotinylating the cell-surface proteins of macrophages with Sulfo-NHS-SS-biotin, the cells were plated onto glass-bottom Petri dishes coated with polymerized collagen IV. SEM images show binding of streptavidin-conjugated 40-nm gold nanoparticles to both macrophages and macrophage-derived particles. Secondary electron (SE) images show plasma membrane–derived particles on the collagen fibers as well as the binding of gold nanoparticles to macrophage cell body, the filopodia, and plasma membrane–derived particles. Backscattered electron (BSE) images were helpful for identifying gold nanoparticles. Higher magnification images of the blue boxed region are shown below. Higher magnification images of the yellow and red boxed regions are shown to the right. Yellow arrows point to gold nanoparticles binding to macrophage particles on the collagen IV substrate. Four independent experiments were performed; representative images are shown. Scale bar, 2 μm.
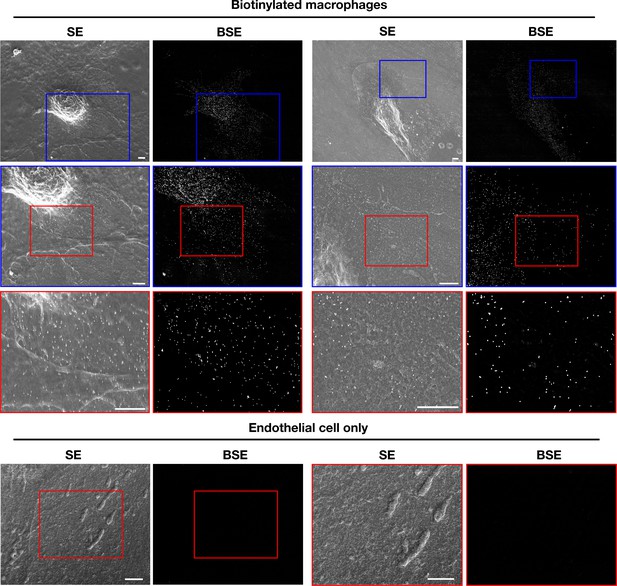
Biotinylated mouse peritoneal macrophages release plasma membrane–derived material onto the surface of dead endothelial cells.
Mouse brain endothelial cells (bEnd.3) were plated onto glass- bottom Petri dishes and allowed to grow to confluency. After fixing the endothelial cells with 0.1% glutaraldehyde in PBS, they were washed extensively with PBS. Next, biotinylated macrophages (i.e., macrophages in which the cell-surface proteins had been biotinylated with Sulfo-NHS-SS-biotin) were plated onto bEnd.3 cells. Secondary electron (SE) and backscattered electron (BSE) images were obtained with a scanning electron microscope. As expected, the SEM images revealed binding of the streptavidin- conjugated gold nanoparticles to both the cell body and filopodia of macrophages. In addition, gold nanoparticles were observed on the surface of adjacent endothelial cells. BSE images were helpful in identifying gold nanoparticles. Higher magnification images of the blue and red boxed regions in each image are shown below. As an experimental control, bEnd.3 cells without macrophages were fixed and incubated with streptavidin-conjugated gold nanoparticles. Higher magnification images of the red boxed regions are shown on the right. No binding of gold nanoparticles was detected. Three independent experiments were performed; representative images are shown. Scale bar, 2 µm.
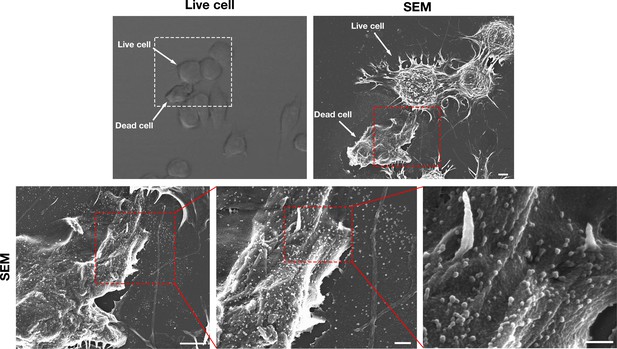
Correlative live-cell and SEM images, revealing the release of particles by a live macrophage onto the surface of a dead macrophage.
RAW 264.7 macrophages were plated onto poly-D-lysine–coated gridded glass-bottom Petri dishes, and videos of cell movement were recorded for 20 hr at 5 min intervals (see Figure 10—video 1). Live-cell microscopy revealed a live macrophage extending its lamellipodium (red arrow in the video) over both the surrounding substrate and over the dead macrophage (white arrow in the video). The white box in the video shows the region that was imaged by SEM. The SEM revealed that vesicular particles had been released onto both the surrounding substrate and onto the surface of the dead macrophage. Scale bar for the SEM on the top right, 5 μm. The region encompassed by a red box in the SEM on the top right is shown at higher magnification in the SEM on the lower left. Scale bar for the lower left SEM, 5 μm. The red boxed areas in the lower left and lower middle SEMs are shown at higher magnification on the right. Images shown are from one experiment. Scale for lower middle and lower right images, 500 nm.
Release of vesicular particles by a live RAW 264.7 macrophage onto the surrounding substrate and onto a dead RAW 264.7 macrophage.
RAW macrophages were plated onto poly-D-lysine–coated glass-bottom Petri dishes and incubated in media containing 10% FBS. Cells were imaged by live-cell microscopy for 20 hr at 5 min intervals. White arrow points to a dead macrophage; red arrow points to the projection and retraction of the live macrophage lamellipodia over the dead macrophage; white box indicates the region that was subsequently imaged by SEM (see Figure 10—figure supplement 1). Video shows a 20 hr period of live-cell imaging.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (M. musculus) | RAW 264.7 | ATCC | Catalog No. TIB-71 RRID: CVCL_0493 | |
| Cell line (M. musculus) | bEnd.3 | ATCC | Catalog No. CRL-2299 RRID: CVCL_0170 | |
| Recombinant DNA reagent | ALO-D4 plasmid | PMID: 25809258 | Dr. Arun Radhakrishnan (UT Southwestern) | |
| Recombinant DNA reagent | OlyA plasmid | PMID: 30712872 | Dr. Arun Radhakrishnan (UT Southwestern) | |
| Chemical compound, drug | N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (carbodiimide) | Millipore-Sigma | Catalog No. 03449 | |
| Chemical compound, drug | Glutaraldehyde25% solution | Electron Microscopy Sciences | Catalog No. 16220 | |
| Chemical compound, drug | Osmium tetroxide4% solution | Electron Microscopy Sciences | Catalog No. 18459 | |
| Chemical compound, drug | Paraformaldehyde16% solution | Electron Microscopy Sciences | Catalog No. 15170 | |
| Chemical compound, drug | EMbed 812 | Electron Microscopy Sciences | Catalog No. 14120 | |
| Chemical compound, drug | Sodium cacodylate trihydrate | Electron Microscopy Sciences | Catalog No. 12300 | |
| Chemical compound, drug | Uranyl acetate | SPI-Chem | Catalog No. 02624AB | |
| Chemical compound, drug | Latrunculin A | Sigma | Catalog No. L5163 | |
| Chemical compound, drug | Blebbistatin | Abcam | Catalog No. ab120425 | |
| Chemical compound, drug | FAK inhibitor | Calbiochem | Catalog No. CAS 4506-66-5 | |
| Chemical compound, drug | LXR agonist | Sigma | Catalog No. G6295 | |
| Chemical compound, drug | HDL | Alfa Aesar | Catalog No. J64903 | |
| Chemical compound, drug | [3H]cholesterol | PerkinElmer | Catalog No. NET139250UC | |
| Chemical compound, drug | Gold–streptavidin conjugation reagent | Abcam | Catalog No. ab186864 | |
| Chemical compound, drug | EZ-Link Sulfo-NHS-SS-biotin | ThermoFisher | Catalog No. 21331 | |
| Chemical compound, drug | Sphingomyelinase from Staphylococcus aureus | Sigma | Catalog No. S8633 | |
| Other | 35 mm glass-bottom gridded MatTek dish | MatTek | Catalog No. P35G-1.5–14-CGRD |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.50231.049





