Common activation mechanism of class A GPCRs
Figures
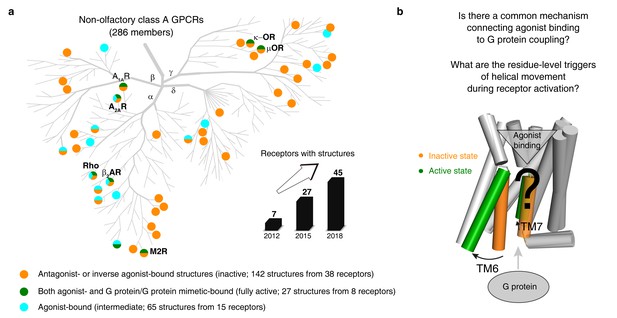
An increasing number of reported class A GPCR structures facilitates studies on common activation mechanism.
(a) Distribution of structures in different states in the non-olfactory class A GPCR tree as of October 1, 2018. (b) Common GPCR activation mechanism and the residue-level triggers are not well understood.
-
Figure 1—source data 1
The released class A GPCR structures (as of October 1, 2018).
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Disease mutations occurred in class A GPCRs.
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig1-data2-v3.xlsx
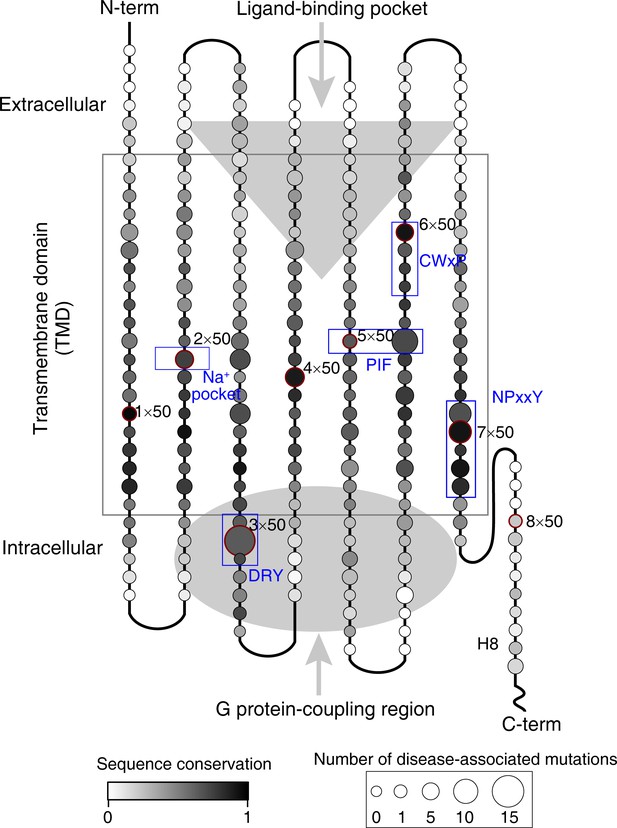
The pattern of conservation of residues and the map of number of disease-associated mutations in human class A GPCRs.
The alignment of 286 non-olfactory, class A human GPCRs were obtained from the GPCRdb (Hauser et al., 2018; Isberg et al., 2016; Pándy-Szekeres et al., 2018; Isberg et al., 2015) and sent for the sequence conservation score calculation for all residue positions by the Protein Residue Conservation Prediction (Capra and Singh, 2007) tool with scoring method ‘Property Entropy” (Mirny and Shakhnovich, 1999). To obtain disease-associated mutations, we performed database integration and literature investigation for all 286 non-olfactory class A GPCRs. Four commonly used databases (UniProt [The UniProt Consortium, 2017], OMIM [Amberger et al., 2011], Ensembl [Zerbino et al., 2018] and GPCRdb) were first filtered by disease mutations and then merged. Finally, we collected 435 disease mutations from 61 class A GPCRs (Figure 1—source data 2).
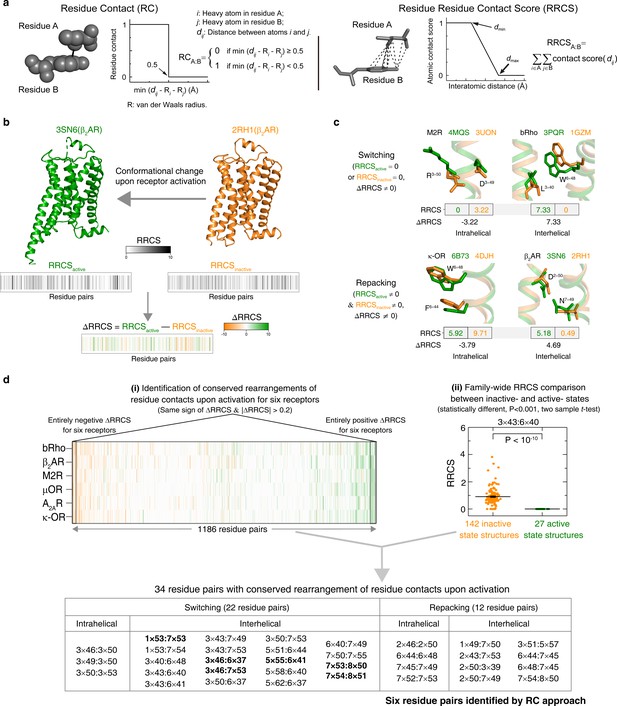
Understanding GPCR activation mechanism by RRCS and ∆RRCS.
(a) Comparison of residue contact (RC) (Venkatakrishnan et al., 2016) and residue residue contact score (RRCS) calculations. RRCS can describe the strength of residue-residue contact quantitatively in a much more accurate manner than the Boolean descriptor RC. (b) RRCS and ΔRRCS calculation for a pair of active and inactive structures can capture receptor conformational change upon activation. (c) Two types of conformational changes (i.e. switching and repacking contacts) can be defined by RRCS to quantify the global, local, major and subtle conformational changes in a systematic way. (d) Two criteria of identifying conserved residue rearrangements upon receptor activation by RRCS and ΔRRCS. Thirty-four residues pairs were identified based on the criteria (see Materials and methods, Figure 2—source datas 1 and 2 for details), only six of them were discovered before (Venkatakrishnan et al., 2016).
-
Figure 2—source data 1
Calculated RRCS of 34 residue pairs constituting the common activation pathway for released class A GPCR structures.
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig2-data1-v3.xlsx
-
Figure 2—source data 2
Thirty-four residue pairs show conserved rearrangements of residue contacts upon activation.
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig2-data2-v3.pdf
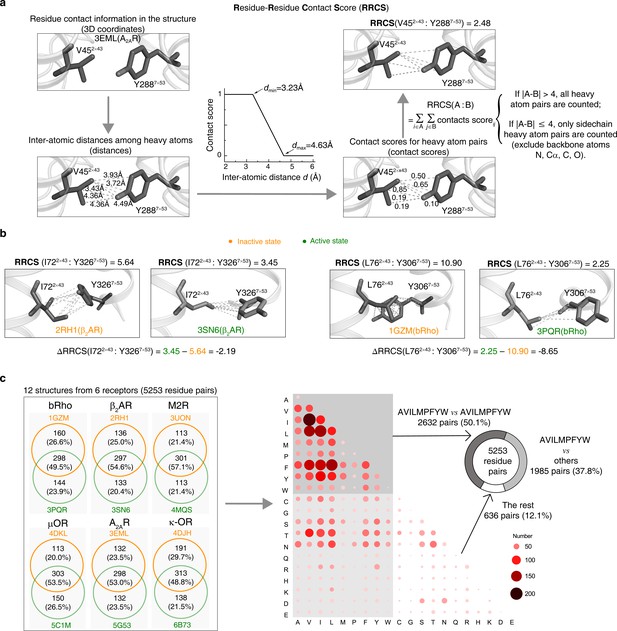
Calculation of RRCS and ΔRRCS.
(a) Workflow of RRCS calculation. (b) Examples of RRCS and ΔRRCS calculation for two residues pairs. (c) Statistics of residue contacts and contact types for six receptors (bRho, β2AR, M2R, µOR, A2AR and κ-OR) in their inactive and active states. Contact type describes physicochemical properties of two interacted amino acids that form a pair. The amino acids with hydrophobic side chains (one-letter code: A, V, I, L, M, P, F, Y, W) contribute to the majority of residue contacts, either within themselves (50.1%) or with other amino acids (37.8%).
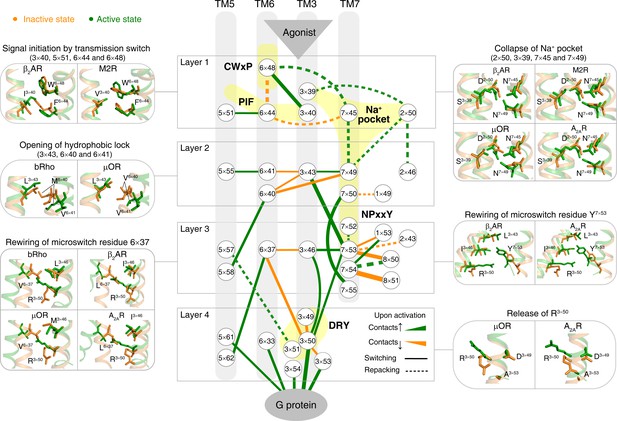
Common activation pathway of class A GPCRs.
Node represents structurally equivalent residue with the GPCRdb numbering (Isberg et al., 2016) while the width of edge is proportional to the average ∆RRCS among six receptors (bRho, β2AR, M2R, µOR, A2AR and κ-OR). Four layers were qualitatively defined based on the topology of the pathway and their roles in activation: signal initiation (layer 1), signal propagation (layer 2), microswitches rewiring (layer 3) and G-protein coupling (layer 4).
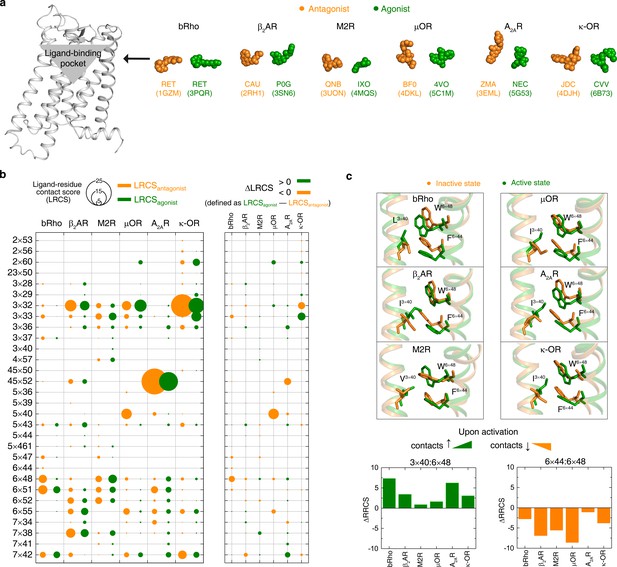
Rearrangements of ligand-residue contacts in ligand-binding pocket are not conserved, reflecting diverse ligand recognition modes.
(a) Sphere representation of antagonist- and agonist-bound receptor crystal structures. (b) Diverse LRCS and ∆LRCS reveal the repertoire of ligand recognition across class A GPCRs. The agonist or antagonist was treated as a single residue when calculating LRCS and ∆LRCS. As shown by the calculated ∆RRCS, no ligand-residue pair exhibits conserved rearrangements upon activation. (c) Conserved conformational changes were only observed at the very bottom of ligand-binding pocket (6×48, 3×40 and 6×44).
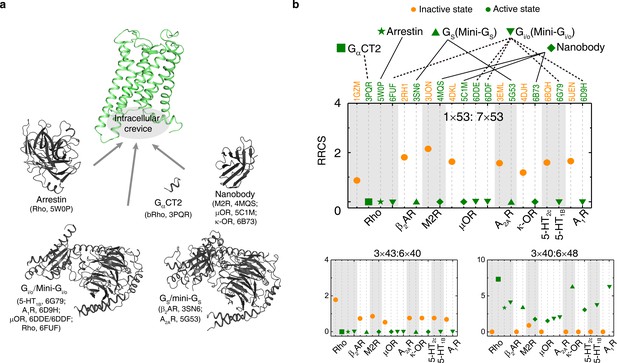
The common activation mechanism is the shared portion of various downstream pathways of different class A GPCRs.
(a) Intracellular binding partners used in the active state structures. (b) Comparison of RRCS for active (green) and inactive (orange) states of eight receptors with different intracellular binding partners, including four recently solved cryo-EM structures of Gi/o-bound receptors (5-HT1B receptor, rhodopsin, A1R and µOR) (Tsai et al., 2018; García-Nafría et al., 2018; Kang et al., 2018; Koehl et al., 2018; Draper-Joyce et al., 2018) whose resolutions were low (usually ≥3.8 Å for the GPCR part). Nevertheless, almost all conserved residue rearrangements in the pathway can be observed from them. Three of 34 residues pairs were shown here, see Figure 4—figure supplements 1 and 2 for the remaining 31 residue pairs.
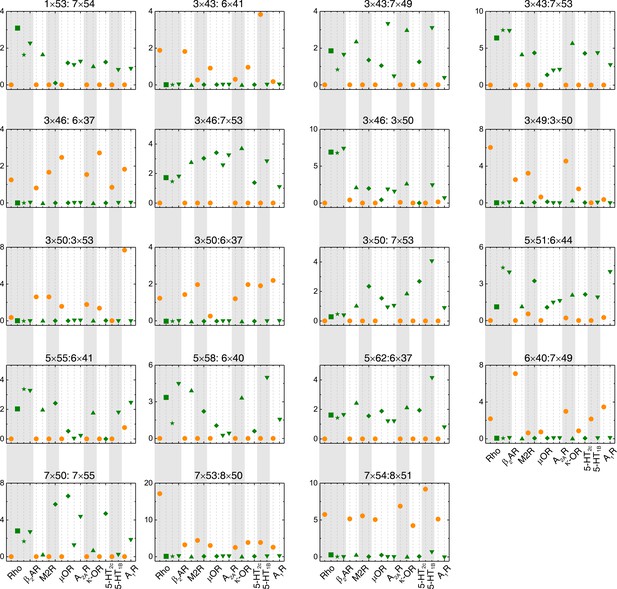
The switching conformation change is conserved upon receptor activation.
Comparison of RRCS for active (green) and inactive (orange) states of eight receptors with different intracellular binding partners, including four recently solved cryo-EM structures of Gi/o-bound receptors (5-HT1B receptor, rhodopsin, A1R and µOR) whose resolutions were low (usually ≥3.8 Å for the GPCR part). Nevertheless, almost all conserved residue rearrangements in the pathway can be observed from them. Nineteen of 34 residues pairs were shown here, see Figure 4 and Figure 4—figure supplement 2 for the remaining residue pairs.
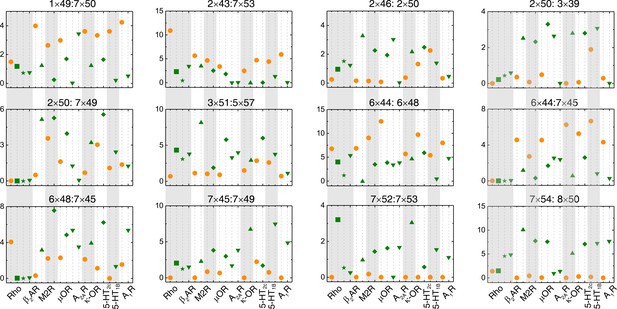
The repacking conformation change is conserved upon receptor activation.
Comparison of RRCS for active (green) and inactive (orange) states of eight receptors with different intracellular binding partners, including four recently solved cryo-EM structures of Gi/o-bound receptors (5-HT1B receptor, rhodopsin, A1R and µOR) whose resolutions were low (usually ≥3.8 Å for the GPCR part). Nevertheless, almost all conserved residue rearrangements in the pathway can be observed from them. Twelve of 34 residues pairs were shown here, see Figure 4 and Figure 4—figure supplement 1 for the remaining residue pairs.
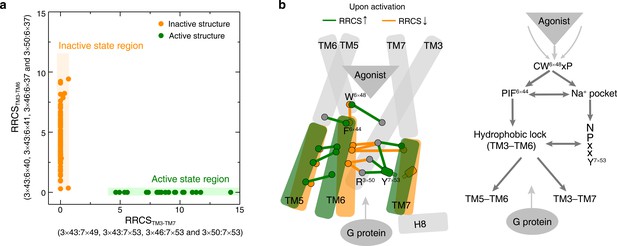
Common activation model of class A GPCRs reveals major changes upon GPCR activation.
(a) Active and inactive state structures form compact clusters in the 2D inter-helical contact space: RRCSTM3-TM7 (X-axis) and RRCSTM3-TM6 (Y-axis). GPCR activation is best described by the outward movement of TM6 and inward movement of TM7, resulting in switch in the contacts of TM3 from TM6 to TM7. (b) Common activation model for class A GPCRs. Residues are shown in circles, conserved contact rearrangements of residue pairs upon activation are denoted by lines.
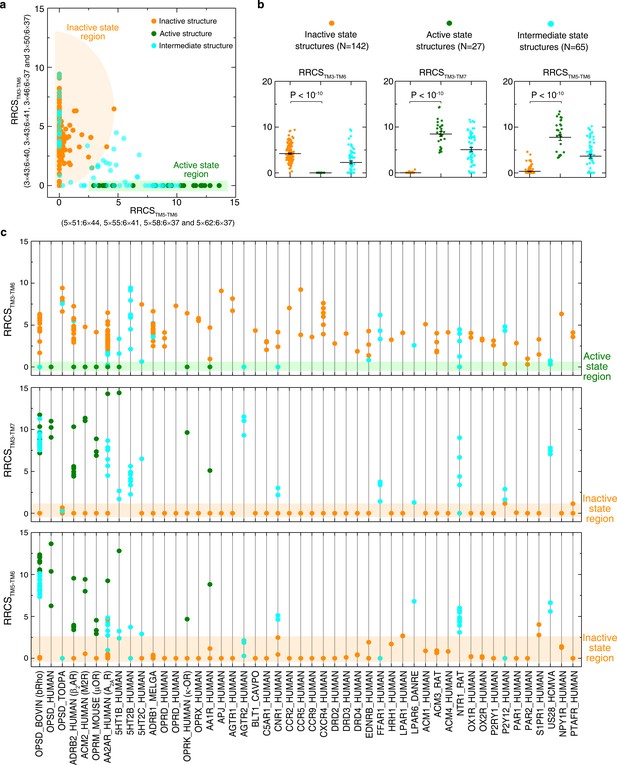
Global conformational change upon activation.
(a) Distinct clustering of inactive- and active-state structures in two-dimentional inter-helical contact space RRCSTM5-TM6 vs. RRCSTM3-TM6. (b) The inter-helical contacts comparison between inactive- and active-state structures. (c) Receptor-specific inter-helical contacts for all class A GPCR structures (inactive, intermediate and active states are colored in orange, cyan and green, respectively). These results demonstrate that receptor activation involves the elimination of TM3-TM6 contacts, formation of TM3-TM7 and TM5-TM6 contacts, reflecting the outward movement of the cytoplasmic end of TM6 away from TM3, the inward movement of TM7 towards TM3 and the repacking of TM5 and TM6.
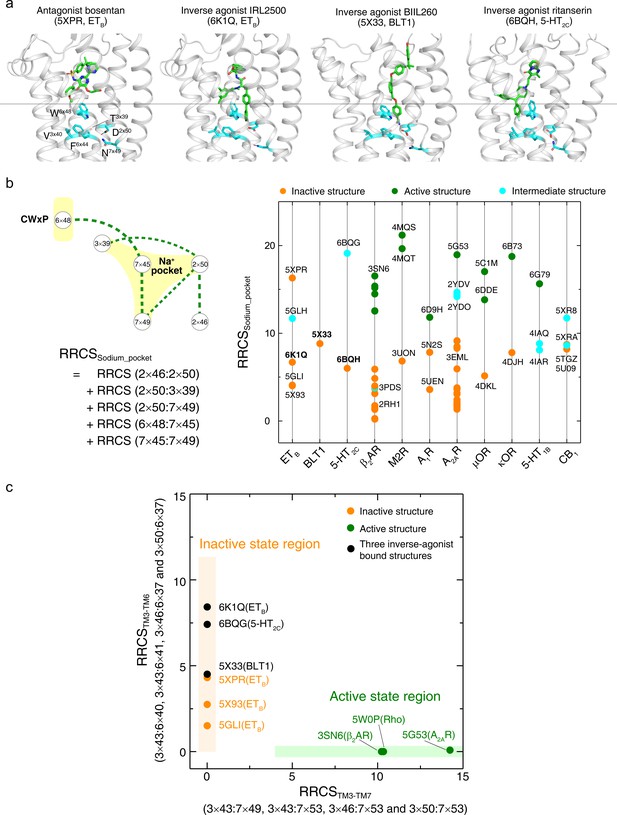
An inverse-agonism of class A GPCRs by preventing the collapse of Na+ pocket.
(a) Comparison of binding poses of antagonist (bosentan [Shihoya et al., 2017]) and inverse agonists (IRL2500 [Nagiri et al., 2019], BIIL260 [Hori et al., 2018] and ritanserin [Peng et al., 2018]). Inverse agonists diffuse deeper in the ligand-binding pocket to touch the Na+ pocket. These key residues around the Na+ pocket are shown. (b) Comparison of the sum of contact scores of the conserved residue pairs around the Na+ pocket (RRCSsodium_pocket) between inactive- and active-state structures. The collapse of the Na+ pocket leads to a denser repacking of six residues (five residue pairs), reflected by higher RRCSsodium_pocket scores compared to that of the inactive state structures. (c) Distribution of three inverse agonist bound structures in the 2D inter-helical contact space: RRCSTM3-TM7 (X-axis) and RRCSTM3-TM6 (Y-axis). These inverse agonists bound structures (5×33, 6BQG and 6K1Q) located in the inactive state region with zero RRCSTM3-TM7 but high RRCSTM3-TM6 scores, despite deeper binding modes.
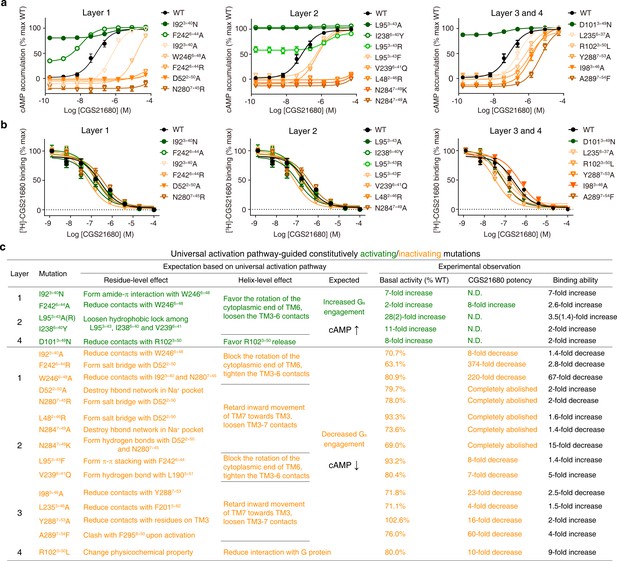
Experimental validation of the common activation mechanism.
(a) cAMP accumulation assay and (b) radioligand binding assay: both validated the common activation pathway-guided design of CAMs/CIMs for A2AR. Wildtype (WT), CAMs and CIMs are shown in black, green and orange, respectively. (c) Mechanistic interpretation of common activation pathway-guided CAMs/CIMs design. N.D.: basal activity was too high to determine an accurate EC50 value.
-
Figure 6—source data 1
Functional and ligand binding properties of A2AR mutations.
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig6-data1-v3.docx
-
Figure 6—source data 2
Analysis of the 14 unsuccessful predictions of A2AR CAMs/CIMs.
ΔStability (>0 means destabilized;<0 means stabilized) is the change of receptor stability when a mutation was introduced, calculated by Residue Scanning module in BioLuminate (Beard et al., 2013). WT, wild-type.
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig6-data2-v3.docx
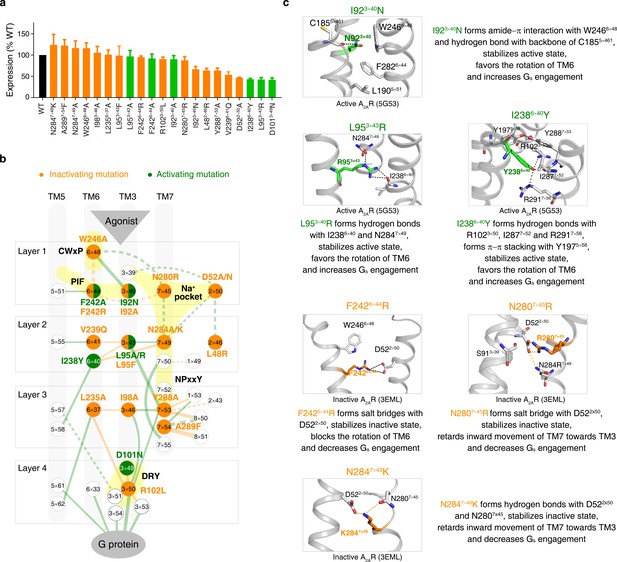
Experimental validation of common activation pathway-guided CAM/CIM design for A 2A R.
(a) Cell surface expression of the WT A 2AR and its mutants. WT, CAMs and CIMs are colored by black, orange and green, respectively. (b) Mapping of validated CAMs/CIMs to the common activation pathway. (c) The mechanisms of CAM/CIM design. CAMs and CIMs are in green and orange, respectively.
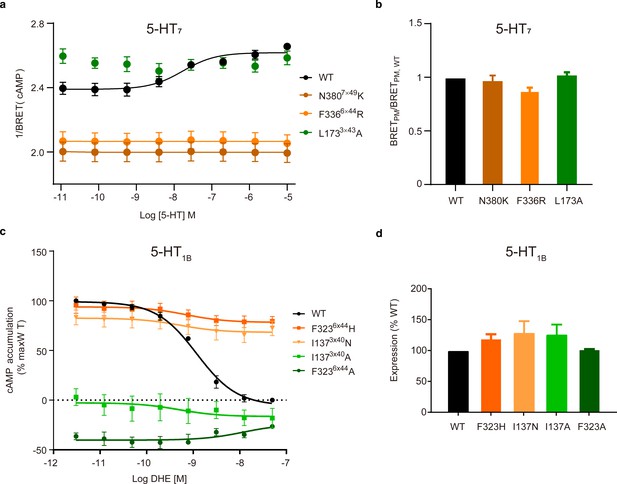
Experimental validation of common activation pathway-guided CAM/CIM design for Gs-coupled 5-HT7 and Gi-coupled 5-HT1B receptors.
(a) cAMP assay of Gs-coupled 5-HT7 receptor used an EPAC-based BRET cAMP sensor (n = 6). WT, CAMs and CIMs are colored by black, orange and green, respectively. (b) Trafficking assay measured WT and mutant 5-HT7 receptors in the plasma membrane (PM). (c) cAMP measurement of 5-HT1B receptor. CAMs and CIMs are shown in green and orange, respectively. (d) Cell surface expression of mutant 5-HT1B receptors relative to that of WT. DHE, dihydroergotamine.
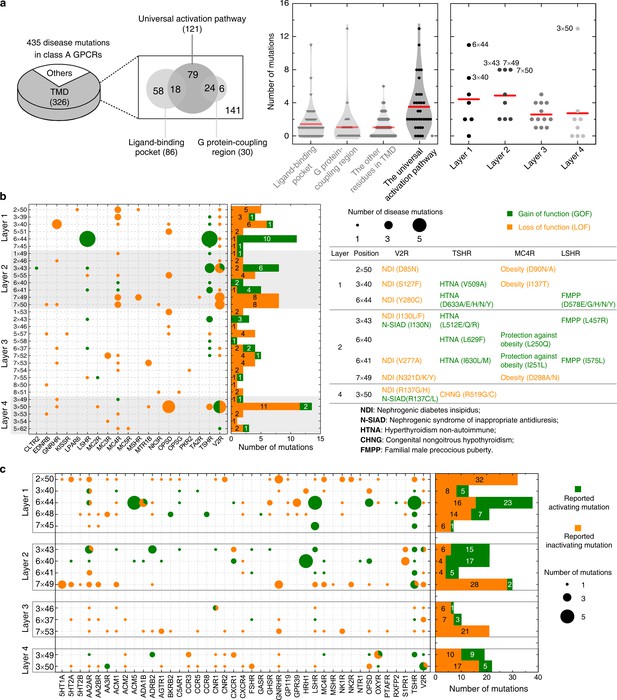
Importance of the common activation pathway in pathophysiological and biological contexts.
(a) Comparison of disease-associated mutations in the common activation pathway (further decomposed into layers 1–4), ligand-binding pocket, G-protein-coupling region and other regions. Red line denotes the mean value. (b) Mapping of disease-associated mutations in class A GPCRs to the common activation pathway. (c) Key roles of the residues constituting the common activation pathway have been reported in numerous experimental studies on class A GPCRs. Two hundred seventy two (272) CAMs/CIMs from 41 receptors were mined from the literature for the 14 hub residues (i.e. residues that have more than one edges in the pathway).
-
Figure 7—source data 1
Constitutively activating/inactivating mutations for the 14 hub residues in the common activation pathway.
- https://cdn.elifesciences.org/articles/50279/elife-50279-fig7-data1-v3.xlsx
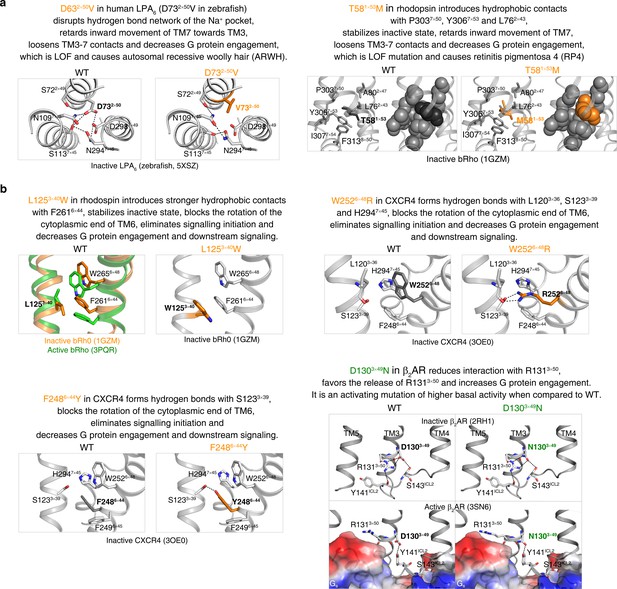
The common activation pathway can be used to mechanistically interpret disease-associated mutations and CAMs/CIMs.
(a) Pathway-guided mechanistic interpretations of two disease mutations. (b) Pathway-guided mechanistic interpretations of four CAMs/CIMs.
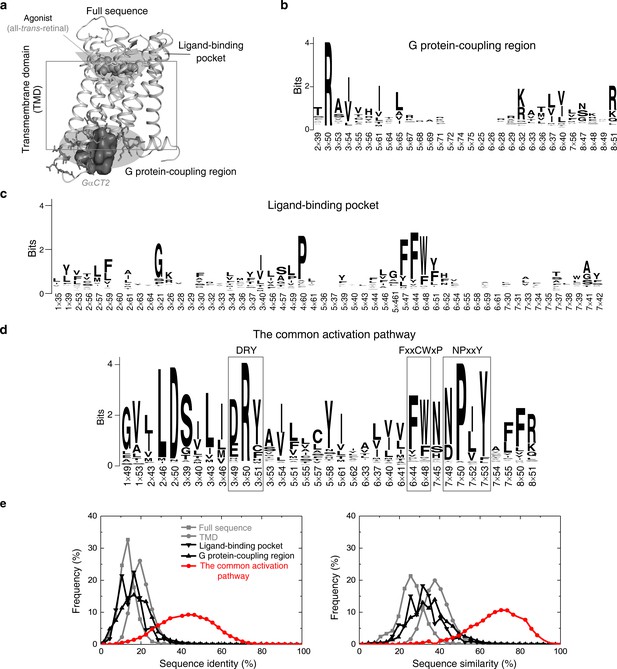
Residues in the common activation pathway are more conserved than other functional regions of GPCR.
(a) Illustration of different functional regions of GPCR. (b-d) Sequence pattern of the G protein-coupling region (b), ligand-binding pocket (c) and the common activation pathway (d). (e) Distribution of sequence identity (left) and similarity (right) for functional regions across 286 non-olfactory class A receptors.
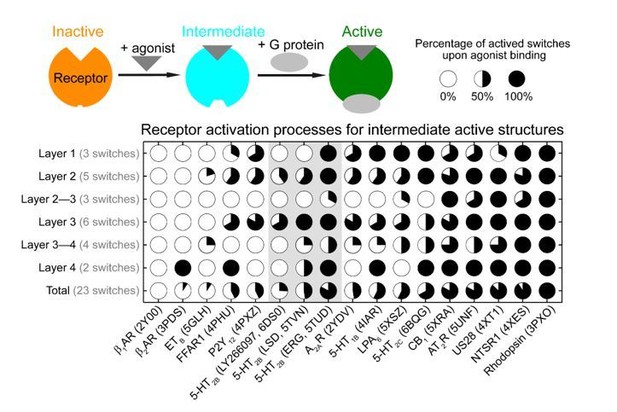
Receptor activation process via intermediate structures (i.e., only agonist but not G protein bound structures) are in receptor- and ligand-specific manner, thereby highlighting the modular nature and complexity of the activation pathways.
Tables
List of twelve structures of six receptors.
| GPCR | PDB code | Resolution (Å) | Ligand G protein/G protein memetic | State | Mutation in the TM region |
|---|---|---|---|---|---|
| bRho | 1GZM | 2.7 | 11-cis-retinal (inverse agonist, covalently bound) | Inactive | No mutation |
| 3PQR | 2.9 | All-trans-retinal (agonist, covalently bound) GαCT2 (G protein peptide) | Active | No mutation | |
| β2AR | 2RH1 | 2.4 | Carazolol (inverse agonist) | Inactive | No mutation |
| 3SN6 | 3.2 | BI-167107 (agonist) Gs (G protein) | Active | No mutation | |
| M2R | 3UON | 3.0 | 3-quinuclidinyl-benzilate (antagonist) | Inactive | No mutation |
| 4MQS | 3.5 | Iperoxo (agonist) Nb9-8 (nanobody) | Active | No mutation | |
| μOR | 4DKL | 2.8 | β-funaltrexamine (antagonist) | Inactive | No mutation |
| 5C1M | 2.1 | BU72 (agonist) Nb39 (nanobody) | Active | No mutation | |
| A2AR | 3EML | 2.6 | ZM241385 (antagonist) | Inactive | No mutation |
| 5G53 | 3.4 | NECA (agonist) Mini-Gs (engineered G protein) | Active | No mutation | |
| κOR | 4DJH | 2.9 | JDTic (antagonist) | Inactive | I135L (3×29) |
| 6B73 | 3.1 | MP1104 (agonist) Nb39 (nanobody) | Active | I135L (3×29) |
List of GPCR-G protein complex structures.
| Receptor | PDB code | Method | Resolution (Å) | Ligand + G protein/G protein memetic |
|---|---|---|---|---|
| A2AR | 5G53 | X-ray | 3.4 | NECA + Mini-Gs |
| A2AR | 6GDG | cryo-EM | 4.1 | NECA + Mini-Gs |
| β2AR | 3SN6 | X-ray | 3.2 | BI-167107 + Gs |
| β2AR | 6E67 | X-ray | 3.7 | BI-167107 + Fused Gs C-terminal |
| Rho | 6CMO | cryo-EM | 4.5 | Gi |
| Rho | 6FUF | X-ray | 3.1 | all-trans-retinal +Mini-Go |
| μOR | 6DDE/6DDF | cryo-EM | 3.5 | DAMGO + Gi |
| A1AR | 6D9H | cryo-EM | 3.6 | Adenosine + Gi2 |
| 5-HT1B | 6G79 | cryo-EM | 3.8 | Donitriptan + Go |
| CB1 | 6N4B | cryo-EM | 3.0 | MDMB-Fubinaca + Gi |
| NTSR1 | 6OSA | cryo-EM | 3.0 | JMV449 + Gi1 (NC state) |
| NTSR1 | 6OS9 | cryo-EM | 3.0 | JMV449 + Gi1 (C state) |
| M2R | 6OIK | cryo-EM | 3.6 | Iperoxo + LY2119620 +GoA |
| M1R | 6OIJ | cryo-EM | 3.3 | Iperoxo + G11 |
List of three inverse agonist-bound structures.
| GPCR | PDB code | Ligand | Ligand type | Na+ mimic group in ligand | Occupying Na+ pocket |
|---|---|---|---|---|---|
| BLT1 | 5X33 | BIIL260 | Inverse agonist | Yes | Yes |
| ETB | 6K1Q | IRL2500 | Inverse agonist | No | Yes |
| 5-HT2C | 6BQH | Ritanserin | Inverse agonist | No | No |
List of allosteric modulators among determined structures of class A GPCRs.
| Location of binding site | Receptor | Ligand | Ligand type | PDB code |
|---|---|---|---|---|
| Intracellular pocket | β2AR | CMPD-15PA | Negative allosteric modulator | 5X7D |
| CCR9 | Vercirnon | Antagonist | 5LWE | |
| CCR2 | CCR2-RA-[R] | Antagonist | 5T1A | |
| Extra-helical binding site | C5aR | NDT9513727 | Inverse agonist | 5O9H |
| C5aR | avacopan | Allosteric antagonist | 6C1R | |
| C5aR | NDT9513727 | Allosteric antagonist | 6C1Q | |
| β2AR | compound-6FA | Positive allosteric modulator | 6N48 | |
| P2Y1 | BPTU | antagonist | 4XNV | |
| GPR40 | AP8 | Full allosteric agonists (agoPAM) | 5TZY | |
| GPR40 | Compound 1 | Full agonist | 5KW2 | |
| PAR2 | AZ3451 | Antagonist | 5NDZ | |
| Non-canonical binding pocket | GPR40 | TAK-875 | Ago-allosteric modulator | 4PHU |
| GPR40 | MK-8666 | Ago-allosteric modulator | 5TZR | |
| PAR2 | AZ8838 | antagonist | 5NDD | |
| Extracellular vestibule above the orthosteric site | M2 | LY2119620 | Positive allosteric modulator | 4MQT |
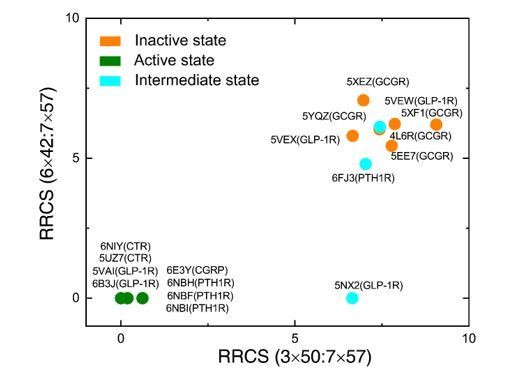
List of available class B GPCRs structures.
| Receptor | PDB code | Method | Resolution (Å) | Ligand + G protein/G protein memetic | States |
|---|---|---|---|---|---|
| CRF1R | 4K5Y | X-ray | 2.9 | Antagonist CP-376395 | Inactive |
| CRF1R | 4Z9G | X-ray | 3.2 | Antagonist CP-376395 | Inactive |
| CTR | 6NIY | cryo-EM | 3.3 | Peptide ligand + Gs | Active |
| CTR | 5UZ7 | cryo-EM | 4.1 | Peptide ligand + Gs | Active |
| CGRP | 6E3Y | cryo-EM | 3.3 | CGRP + Gs | Active |
| GCGR | 4L6R | X-ray | 3.3 | No ligand was seen | Inactive |
| GCGR | 5EE7 | X-ray | 2.5 | Antagonist MK-0893 | Inactive |
| GCGR | 5YQZ | X-ray | 3.0 | Partial agonist NNC1702 | Intermediate |
| GCGR | 5XEZ | X-ray | 3.0 | Negative allosteric modulator NNC0640 | Inactive |
| GCGR | 5XF1 | X-ray | 3.2 | Negative allosteric modulator NNC0640 | Inactive |
| GLP-1R | 5VAI | cryo-EM | 4.1 | human GLP-1 + Gs | Active |
| GLP-1R | 6B3J | cryo-EM | 3.3 | Exendin-P5 + Gs | Active |
| GLP-1R | 5NX2 | X-ray | 3.7 | Truncated peptide agonist | Intermediate |
| GLP-1R | 5VEW | X-ray | 2.7 | PF-06372222 | Inactive |
| GLP-1R | 5VEX | X-ray | 3.0 | NNC0640 | Inactive |
| PTH1R | 6FJ3 | X-ray | 2.5 | Peptide agonist ePTH | Intermediate |
| PTH1R | 6NBH | cryo-EM | 3.5 | A long-acting PTH analog + Gs | Active |
| PTH1R | 6NBI | cryo-EM | 4.0 | A long-acting PTH analog + Gs | Active |
| PTH1R | 6NBF | cryo-EM | 3.0 | A long-acting PTH analog + Gs | Active |
Additional files
-
Supplementary file 1
Key resource table.
- https://cdn.elifesciences.org/articles/50279/elife-50279-supp1-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50279/elife-50279-transrepform-v3.docx