Transmission dynamics and control of multidrug-resistant Klebsiella pneumoniae in neonates in a developing country
Figures
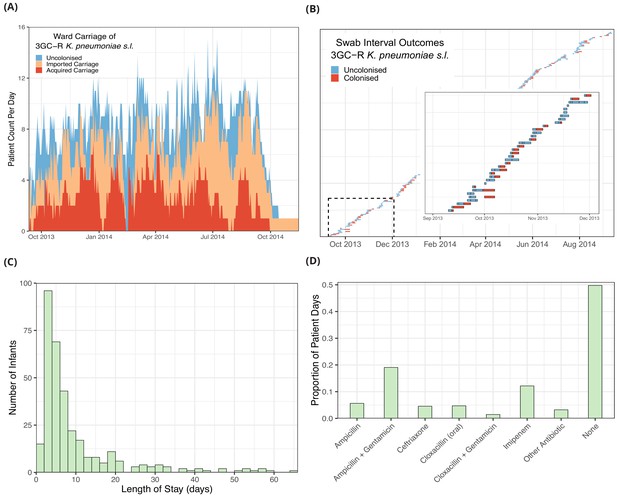
Descriptive epidemiological data from a cohort of 333 infants admitted to a neonatal unit in a Children’s Hospital in Cambodia from September 2013 to September 2014.
Daily counts of neonates colonised with third generation cephalosporin-resistant (3GC-R) Klebsiella pneumoniae sensu lato over the study period are shown in panel A, where colour reflects uncolonised, imported or acquired cases, according to case definitions. The total height of the peaks shows the ward occupancy on that day. The results from rectal swabs among the 191 infants uncolonised at entry for 3GC-R K. pneumoniae s.l. are shown in panel B, with the window highlighting the swab outcomes from the first thirty five infants uncolonised at entry. Each row represents a patient and each coloured block represents a swab interval, where the width is the number of days in the interval (i.e. time between swabs). Outcomes are shown up to the first swab positive for 3GC-R K. pneumoniae s.l., after which time the patient is assumed to be colonised until discharge. The length of stay distribution for infants in the neonatal unit is shown as a histogram in panel C, where the bin width is two days. An infant’s length of stay is the total time in the neonatal unit during the study period, including re-admissions. The 333 infants were present in the neonatal unit for a total of 3417 study days. The proportion of study days when infants took the six most common antibiotic combinations, or other antibiotics, or none are shown in panel D.
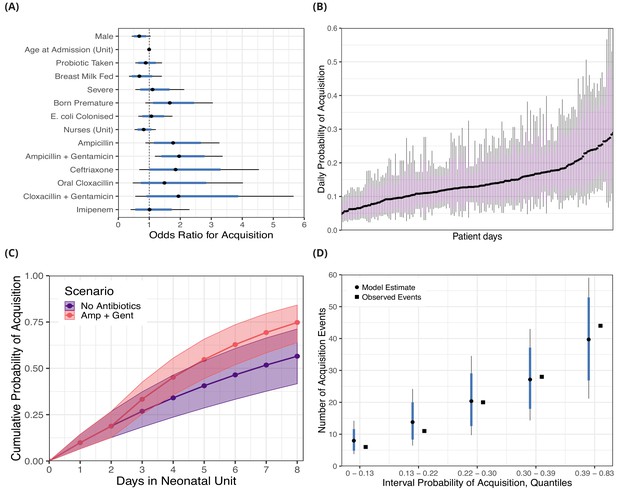
Posterior distributions for risk factors for the daily probability of acquiring third-generation cephalosporin-resistant (3GC-R) Klebsiella pneumoniae sensu lato among 191 susceptible neonates.
Odds ratios for the daily risk of colonisation are shown in panel A. The daily risk of colonisation per patient day is shown in panel B. Note that the 864 patient days have been thinned by a factor of five for visualisation. The cumulative risk for different patient scenarios is explored in panel C; a four day old girl, born full term, without severe conditions, breast milk fed and not taking antibiotics or probiotics over eight days in the neonatal unit is shown in blue. The red line shows the same infant, however ampicillin + gentamicin is taken from day three onwards. The lines and points in both cases show the cumulative probability posterior median, and the shaded area shows the 80% credible interval (CrI). In panel D, we took the probability of colonisation for each of the 400 swab interval and binned them into five quantiles. We then compared the expected number of colonisation events predicted by the model with the observed number of colonisation events (squares) in the swab intervals by quantile. In panels A, B and D points represent posterior medians, thick blue/purple lines represents the 80% CrI and thinner black lines represent the 95% CrI.
Posterior chains from Hamiltonian Markov chain Monte Carlo fitting using Stan for risk factor model A (see Table 2).
The parameters are the intercept [alpha] and 14 slopes for covariates [betas]. The Gelman-Rubin diagnostic () is <1.01 for all parameters. The tail effective sample size (ESStail) ranges from 1948 to 2499 and the bulk effective sample size (ESSbulk) ranges from 1808 to 2350.
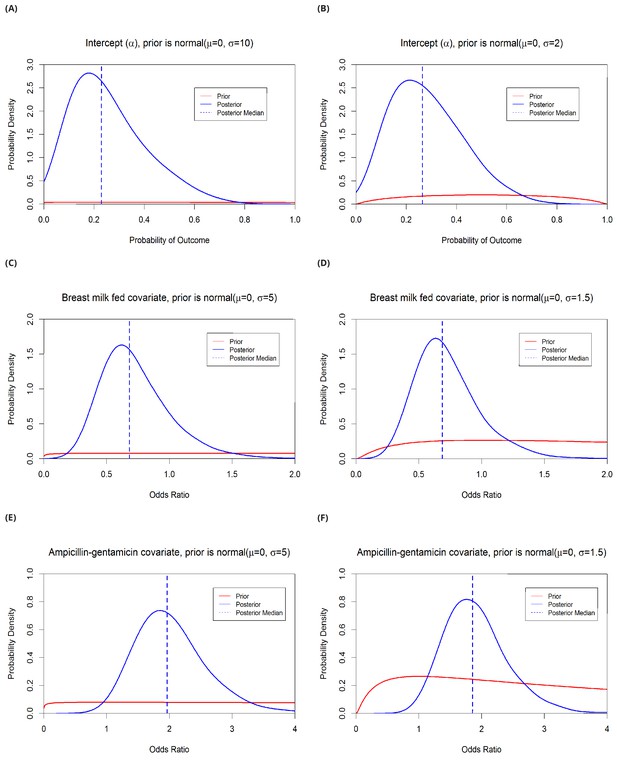
Estimates from risk factor model A with variable priors.
The intercept (α) prior is varied from normal (μ=0, σ=10) to normal (μ=0, σ=2). This changes the median posterior probability from 0.23 to 0.26 (panels A and B). Covariate prior distributions were varied from normal (μ=0, σ=5) to normal (μ=0, σ=1.5). The median odds ratio for the effect of breast feeding on the risk of acquisition/detection of resistant Klebsiella pneumoniae changes from 0.68 to 0.69 (C–D), and the effect of taking ampicillin + gentamicin within the past 48 hours changes from 2.0 to 1.9 (E–F). Note that priors are fitted on the log-odds scale and that the prior and posterior distributions shown in this figure have been logit-transformed (A–B) or exponentiated (C–F).
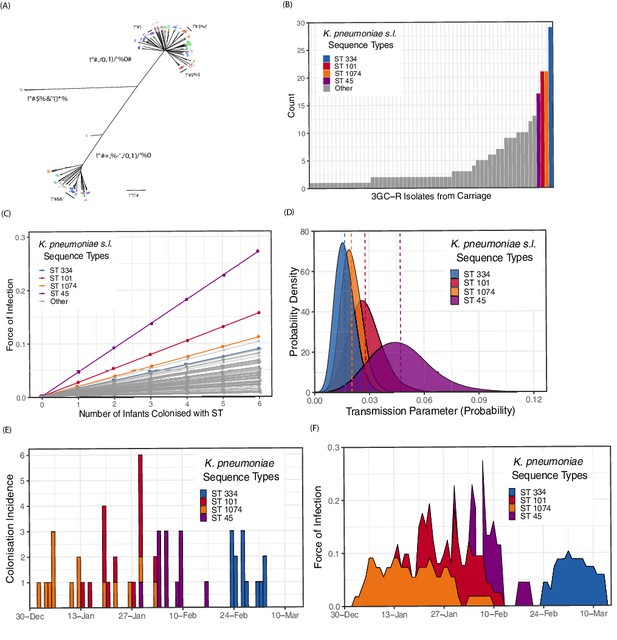
Population structure of third-generation cephalosporin-resistant (3GC-R) Klebsiella and force of infection by sequence type (ST).
An unrooted phylogeny of 317 3GC-R Klebsiella isolates cultured from rectal and environmental swabs over a four month period in a neonatal unit in a children’s hospital in Cambodia is shown in panel A, where the branch lengths correspond to the mash distance (a measure of k-mer similarity) between whole-genome assemblies. The four largest STs are labelled as well as the population subdivisions by Klebsiella species. The frequency distribution of STs is shown in panel B, with the four largest STs shown in colour. Results from a transmission model estimating the force of infection by ST are shown in panel C, where the force of infection scales linearly with the number of colonised infants with that ST. The largest four STs have again been highlighted. Horizontal jitter has been applied to prevent overplotting of points. The uncertainty around the transmission parameter estimates are shown in panel D for the four most common STs, where the posterior mean is shown with a dotted line. The daily incidence of new colonisation events with the four most frequent STs are shown between the 1st January to the 15th March 2014 in panel E, along with the estimated force of infection over the same period in panel F using parameter estimates of β from transmission model 4 (Table 3).
Posterior chains from Hamiltonian Markov chain Monte Carlo fitting for Klebsiella transmission models (see Table 3).
(A) Transmission model 1; intercept [alpha] =1.001, ESSbulk = 1359, ESStail = 1171. (B) Transmission model 2; alpha and beta; ranges from 1.001 to 1.002, ESSbulk ranges from 1144 to 1298, ESStail ranges from 1247 to 1282. (C) Transmission model 3; alpha, beta, gamma and lambda. R̂ ranges from 1.000 to 1.003, ESSbulk ranges from 1274 to 1396, ESStail ranges from 1180 to 1285. (D) Transmission model 4. alpha and beta hyper-parameters scale/shape 1, location/shape two and transmission parameters for each ST (62, not shown), ranges from 0.999 to 1.003, ESSbulk ranges from 1687 to 2887 and ESStail ranges from 2105 to 2796.
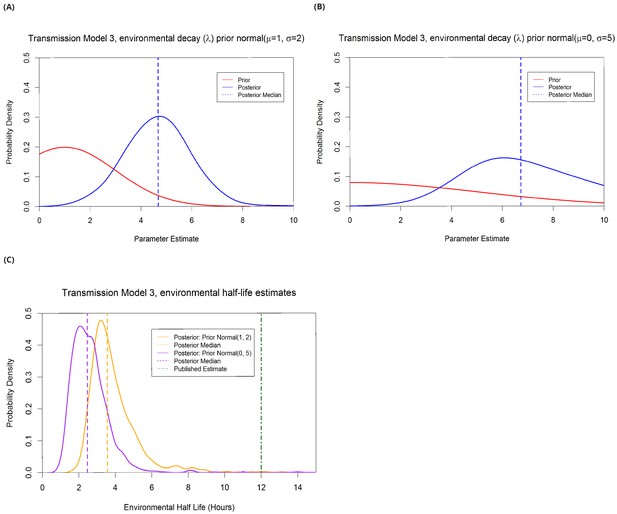
Estimation of λ from transmission model 3 under different prior assumptions.
The more informative prior, half-normal(µ=1, σ=2), gave a median parameter estimate of 4.7 (panel A) corresponding to an environmental half life of 3.6 hours (panel C). The less informative prior, half-normal(µ=0, σ=5), gave a median parameter estimate of 6.7 (panel B) corresponding to an environmental half life of 2.5 hours (panel C). The only published estimate of resistant Klebsiella pneumoniae half life on surfaces is 12 hours (λ=1.39, see Methods), therefore the informative prior gives more weight to biologically plausible values of λ. The qualitative interpretation of the model (that environmental contamination with K. pneumoniae decays quickly) is unchanged with either prior, as is WAIC.
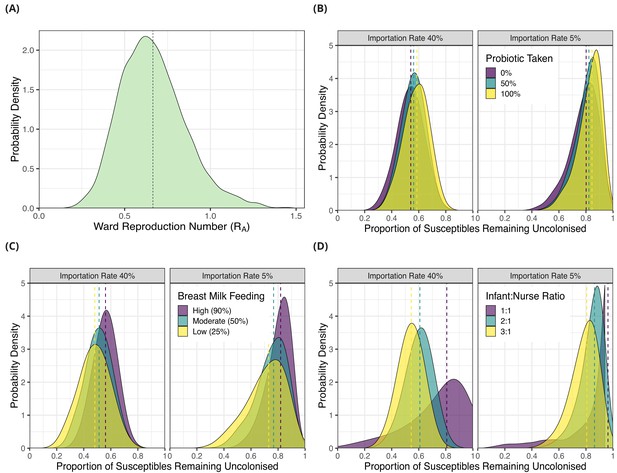
Simulation results from dynamic agent-based models using parameter estimates on acquisition of third generation cephalosporin-resistant (3GC-R) Klebsiella pneumoniae sensu lato among neonates in a Children’s Hospital in Cambodia.
The distribution of ward reproduction number (RA) values shown in panel A was obtained by taking 2000 samples from the force of infection posterior distribution, and for each sample running the agent-based simulation 100 times and taking the mean value. The results from simulating counterfactual scenarios with a dynamic agent-based model are shown in panels B, C and D. In B, the proportion of infants taking a probiotic (Lactobacillus acidophilus) on entry to the ward was varied between 0, -.5 and 1 in setting with a high proportion of imported cases (0.4) and a lower proportion of imported cases (0.05). In panel C, the proportion of infants that were breast milk fed was varied was varied between 0.25, 0.5 and 0.9in settings with a high proportion of imported cases (0.4) and a lower proportion of imported cases (0.05). In panel D, the infant nurse ratio was varied between 3:1, 2:1 and 1:1 in settings with a high proportion of imported cases (0.4) and a lower proportion of imported cases (0.05). The outcome measure in all simulations is the proportion of infants susceptible on entry that remained uncolonised with 3GC-R K. pneumoniae s.l. on discharge. The simulated outcomes are displayed as density plots, with dashed lines showing the median value.
Tables
Summary of characteristics of infants admitted to the neonatal intensive care unit at a children’s hospital in Cambodia from September 2013 to September 2014.
Colonisation status with third generation cephalosporin-resistant Klebsiella pneumoniae sensu lato was recorded through prospectively taken rectal swabs.
| Variable | Males | Females | Total |
|---|---|---|---|
| Number of Patients | 177 (53.1%) | 156 (47.8%) | 333 (100%) |
| Length of Stay in Days Median, (IQR)* | **6 (4, 11) | 6 (4, 11) | 6 (4, 11) |
| Colonised with K. pneumoniae at Entry (or Unknown Time)† | 66 (10) | 55 (11) | 121 (21) |
| Colonised with K. pneumoniae During Admission | 54/101 (54.5%) | 55/90 (61.1%) | 109/191 (57.1%) |
| Co-colonised with K. pneumoniae and E. coli at Entry (or Unknown Time)† | 26 (5) | 19 (2) | 45 (7) |
| Co-colonised with K. pneumoniae and E. coli During Admission | 49/146 (33.6%) | 53/135 (39.3%) | 102/281 (36.3%) |
| Age at Entry in Days (IQR)* | *8 (2, 15) | 9 (1, 17) | 8 (1, 16) |
| Probiotic Taken‡ | 76/177 (42.9%) | 62/156 (39.7%) | 138/333 (41.4) |
| Breast Milk Fed | 163/177 (92.1%) | 139/156 (89.1%) | 302/333 (90.1%) |
| Severe§ | 35/177 (19.8%) | 32/156 (20.5%) | 67/333 (20.1%) |
| Born Premature | 30/177 (16.9%) | 24/156 (15.4%) | 54/333 (16.2%) |
-
* Interquartile range. † Colonised at entry is defined as an initial positive swab within the first 48 hours of admission; if the first swab is positive and it was taken later than 48 hours from admission then the infant is considered to be colonised at an unknown time. ‡ Assigned by clinician to receive oral Lactobacillus acidophilus. § Severe symptoms are requiring ventilation, continuous airway pressure or inotopes.
Comparison of models for the risk of acquiring third generation cephalosporin-resistant Klebsiella pneumoniae sensu lato over 864 patient days in a neonatal intensive care unit in Cambodia.
Models vary by explanatory variables (A-C) or by permitting the intercept to vary between study months in a hierarchical model (D). Models were fitted on the log-odds scale with a logit link function, hence prior distributions are shown as log-odds. Posterior parameter distributions have been transformed using the logistic function and are shown as probabilities. Prior distributions are normal distributions, shown in brackets are the mean and standard deviation respectively.
| Risk Factor Model | Parameters | Priors | Posterior Median (95% CrI)* | WAIC† |
|---|---|---|---|---|
| (A) Single intercept Standard covariates‡ 96 hour antibiotic exposure | α (intercept) β (slopes) | normal(0, 10) normal(0, 5) | 0.23 (0.055, 0.60) ORs§ in results | 438 |
| (B) Single intercept Standard covariates‡ 48 hour antibiotic exposure | α (intercept) β (slopes) | normal(0, 10) normal(0, 5) | 0.26 (0.068, 0.63) Not shown | 441 |
| (C) Single intercept Standard covariates‡ + colonisation pressure term¶ 96 hour antibiotic exposure | α (intercept) β (slopes) | normal(0, 10) normal(0, 5) | 0.26 (0.059, 0.64) Not shown | 440 |
| (D) Intercept varies by month Standard covariates‡ 96 hour antibiotic exposure | α[month] (intercept) μ (normal mean) σ (normal standard de- viation) β (slopes) | normal(μ, σ) normal(0, 3) half-normal(0, 1) normal(0, 3) | Varies by month†† 0.21 (0.044, 0.57) 0.54 (0.51, 0.63) Not shown | 440 |
-
* 95% Credible interval. † Widely applicable information criterion (a model comparison statistic where lower values indicate better fitting models). ‡ Standard covariates: use of ampicillin, ampicillin + gentamicin, cloxacillin (oral), ceftriaxone, cloxacillin + gentamicin, and imipenem within the previous 48 or 96 hours; whether breast fed; receipt of an oral probiotic on entry (Lactobacillus acidophilus), sex, premature (born before the 37th week of pregnancy), severity (defined as severe if requiring ventilation, continuous positive airway pressure or inotopes), already colonised with 3GC-R E. coli, age in days on first admission to the NU, and the daily number of nurses on the ward. These explanatory variables were treated as binary and, where appropriate, time-varying. Covariates were recorded for every day the infant was present in the neonatal unit (see Methods for full details). § Odds ratios. ¶ Colonisation pressure is the number of known colonised patients on the ward on a given day. †† Median posterior probability ranges by month 0.20–0.23.
Transmission models fitted to prospectively collected, genotyped swab data on the acquisition of third-generation cephalosporin-resistant (3GC-R) Klebsiella pneumoniae sensu lato.
The table shows the parameters, prior and posterior distributions along with the WAIC (model comparison measure where lower values indicate a better fit to data). See methods for equations. Normal prior distributions show the mean and standard deviation respectively within brackets, beta prior distributions show the two shape parameters within brackets.
| Transmission Model (Equations in Methods) | Parameters | Priors | Posterior Median (95% CrI)* | WAIC † |
|---|---|---|---|---|
| (1) Constant risk of transmission | α (intercept) | beta(2, 8) | 0.0038 (0.0032, 0.0044) | 1919 |
| (2) Pseudo mass action (PMA) principal | α (intercept) β (transmission)‡ | beta(2, 8) beta(2, 8) | 0.0019 (0.0015, 0.0024) 0.0096 (0.0075, 0.012) | 1739 |
| (3) PMA plus environmen- tal contamination, from colonised patients, which decays over time | α (intercept) β (transmission)‡ γ (environment) λ (decay term) | beta(2, 8) beta(2, 8) beta(2, 8) half-normal(1, 2) | 0.0019 (0.0014, 0.0023) 0.0088 (0.0063, 0.011) 0.097 (0.0086, 0.37) 4.7 (2.2, 7.1) | 1740 |
| (4) Hierarchical PMA varying transmission coefficient by ST4 | α (intercept) β (transmission)‡ αs (beta shape 1) βs (beta shape 2) | beta(2, 8) beta(αs , βs ) half-normal(2, 5) half-normal(8, 5) | 0.0021 (0.0016, 0.0026) Varies by ST§ 0.23 (0.14, 0.38) 17 (9.7, 25) | 1733 |
| (5) Hierarchical PMA varying intercept by ST§ | α (intercept) β (transmission)‡ αs (beta shape 1) βs (beta shape 2) | beta(αs , βs ) beta(2, 8) half-normal(2, 5) half-normal(8, 5) | Varies by ST§ 0.0094 (0.0073, 0.012) 0.23 (0.16, 0.31) 21 (13, 29) | 1793 |
-
* 95% Credible interval. † Widely applicable information criterion (WAIC). ‡ Transmission parameter that is multiplied by the number of infants colonised with the sequence type on the same day to give the force of infection (colonisation pressure). § 3GC-R K. pneumoniae s.l. sequence type (ST).
Key epidemiological parameters estimated in this study from longitudinal swab data on third generation cephalosporin-resistant Klebsiella pneumoniae sensu lato from a neotatal intensive care unit from a Children’s Hospital in Cambodia.
| Parameter | Method | Estimate | Uncertainty interval | Key Assumptions |
|---|---|---|---|---|
| Daily risk of acquisition for neonates | Bayesian regression model | 0.15 | 0.091, 0.19 (IQR*) | Culture diagnostic 100% sensitive |
| Force of infection from one colonised infant | Bayesian transmission model | 0.016 | 0.0093, 0.027 (95% CrI†) | Expected values from transmission model 4 |
| Swab sensitivity (1) | Negatives following a positive swab Beta conjugate prior | 0.90 | 0.88, 0.92 (95% CrI†) | All positives are false negatives Beta(1,7) prior |
| Swab sensitivity (2) | Negatives following a positive swab Beta conjugate prior | 0.93 | 0.91, 0.94 (95% CrI†) | Three consecutive negatives are a true decolonisation Beta(1,7) prior |
| Environmental half life‡ | Bayesian transmission model | 3.6 hours | 2.4, 7.6 hours (95% CrI*) | Exponential decay Normal(1, 2) prior |
| Ward reproduction number RA) | Agent-based simulation | 0.65 | 0.36, 1.1 (95% interval§) | Ward size of 8 susceptible neonates |
-
* Interquartile range (IQR) taken from distribution of daily risk of acquisition (Figure 2B). † Credible interval (CrI). ‡ Inverse rate of decay of environmental contamination, as estimated in transmission model 3, multiplied by ln(2). § 95% of simulated values fell within this interval.





