Extraction of active RhoGTPases by RhoGDI regulates spatiotemporal patterning of RhoGTPases
Figures
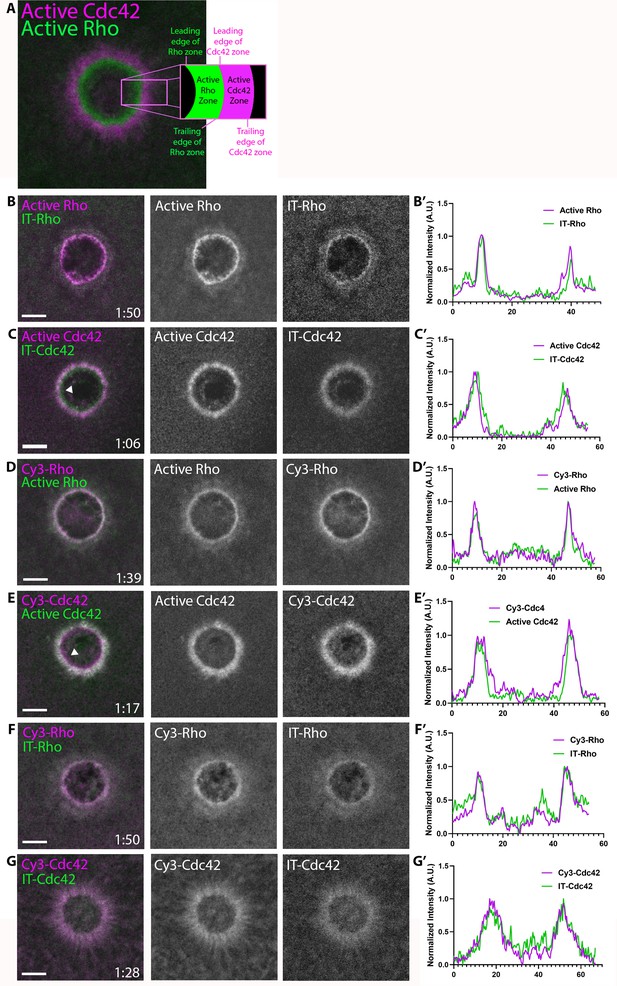
Direct visualization of Rho and Cdc42 during cell wound repair.
(A) Left: image of active Cdc42 (magenta) and active Rho (green) around single-cell wound in Xenopus laevis oocyte; right: schematic diagram indicating zone regions; (B) Wound in oocyte microinjected with rGBD (active Rho, magenta) and IT-Rho (green); (B’) Line scan of normalized fluorescence intensity from (B); (C) As in B but with wGBD (active Cdc42, magenta) and IT-Cdc42 (green); (D,D') As in B but with Cy3-Rho (magenta) and rGBD (green); (E,E') As in B but with Cy3-Cdc42 (magenta) and wGBD (green); (F,F') As in B but with Cy3-Rho (magenta) and IT-Rho (green); (G,G') As in B but with Cy3-Cdc42 (magenta) and IT-Cdc42 (green) and line scan. Scale bar 10 μm, time min:sec.
-
Figure 1—source data 1
Direct visualization of Rho and Cdc42 during cell wound repair.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig1-data1-v2.xlsx
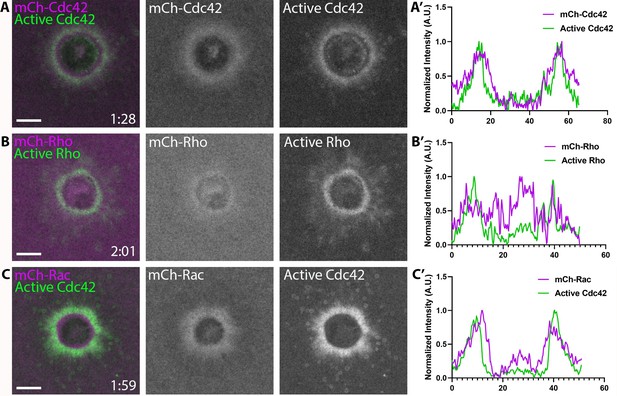
Amino-terminally tagged RhoGTPases do not localize properly to wounds.
Oocytes injected with (A) mCh-Cdc42 (magenta) and wGBD (green), (B) mCh-Rho (magenta) and rGBD (green) or C) mCh-Rac (magenta) and wGBD (green) with A’-C’) Corresponding line scans. Scale bar 10 μm, time min:sec.
-
Figure 1—figure supplement 1—source data 1
Amino-terminally tagged RhoGTPases do not localize properly to wounds.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig1-figsupp1-data1-v2.xlsx
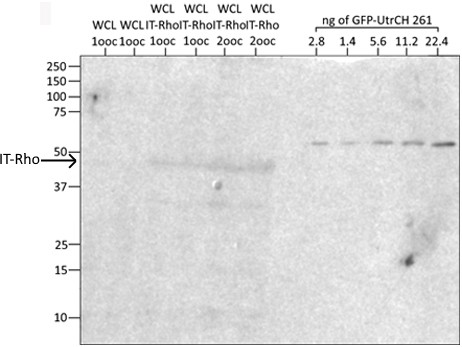
Expression level of Rho internally-tagged with GFP.
Western blot stained with αGFP antibody to determine expression of Rho internally-tagged (IT) with GFP in X. laevis oocytes; lanes 1,2: whole cell lysate (WCL) of 1 X. laevis oocyte, lanes 3,4: WCL of 1 oocyte expressing IT-Rho, lanes 5,6: WCL of 2 oocytes expressing IT-Rho; lanes 8–12: purified GFP-UtrCH 261, used to generate a standard curve.
-
Figure 1—figure supplement 2—source data 1
Expression level of Rho internally-tagged with GFP.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig1-figsupp2-data1-v2.xlsx
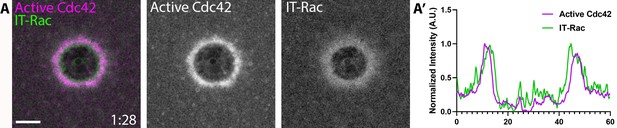
Internally-tagged Rac localizes to wounds.
(A) Oocyte injected with wGBD (magenta) and IT-Rac (green); (A’) Corresponding line scan. Scale bar 10 μm, time min:sec.
-
Figure 1—figure supplement 3—source data 1
Internally-tagged Rac localizes to wounds.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig1-figsupp3-data1-v2.xlsx
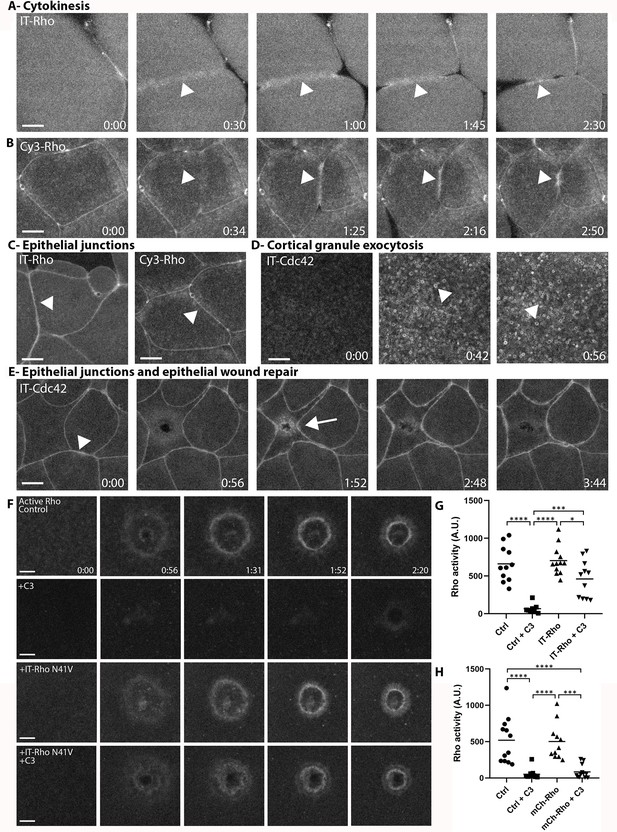
Directly-labeled Rho and Cdc42 during cytokinesis, cortical granule exocytosis, epithelial wound repair and at junctions.
(A) Cytokinesis in X. laevis embryo microinjected with IT-Rho; IT-Rho accumulates at nascent cleavage furrow (arrowhead); (B) Cytokinesis in X. laevis embryo microinjected with Cy3-Rho; Cy3-Rho accumulates at nascent cleavage furrow (arrowhead); (C) X. laevis embryo microinjected with IT-Rho (left) and Cy3-Rho (right); both are enriched at cell-cell junctions; (D) Meiotically mature Xenopus egg microinjected with IT-Cdc42; IT-Cdc42 is recruited to exocytosing cortical granules (arrowheads) following egg activation (0:42); (E) X. laevis embryo microinjected with IT-Cdc42; IT-Cdc42 concentrates at cell-cell junctions (arrowhead; 0:00) and, following damage, is recruited to the wound and becomes enriched at junctions (arrow); (F) C3-insensitive IT-Rho rescues Rho activity in presence of C3. Control: oocyte microinjected with rGBD shows normal Rho activation and wound closure; C3: cell microinjected with rGBD fails to activate Rho in presence of C3; IT-Rho-N41V: cell microinjected with rGBD and C3-insensitive IT-Rho activates Rho normally; IT-Rho-N41V+C3: cell microinjected with rGBD and C3-insensitive IT-Rho rescues Rho activity in presence of C3. Scale bar 10 μm, time min:sec; (G) Quantification of Rho activity, corrected for background (n = 8–12); (H) As in G but with amino-terminally tagged mCh-Rho (n = 8–9). One-way ANOVA with Tukey post-test statistical analysis. *p<0.05, ***p<0.001, ****p<0.0001.
-
Figure 2—source data 1
Directly-labeled Rho and Cdc42 during cytokinesis, cortical granule exocytosis, epithelial wound repair and at junctions.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig2-data1-v2.xlsx
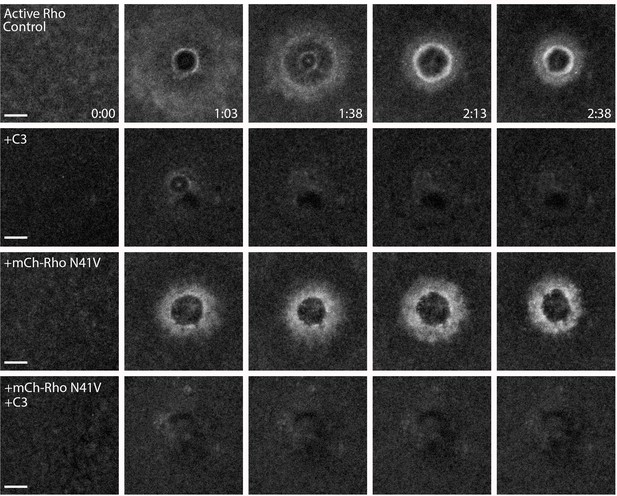
Amino-terminally tagged Rho fails to rescue Rho activity upon inhibition of endogenous Rho.
C3-insensitive amino-terminally tagged mCh-Rho fails to rescue Rho activity in presence of C3. Control: oocyte microinjected with rGBD shows normal Rho activation and wound closure; C3: cell microinjected with rGBD fails to activate Rho in presence of C3; mCh-Rho-N41V: cell microinjected with rGBD and C3-insensitive mCh-Rho activates Rho, albeit it an ill-defined zone; mCh-Rho-N41V+C3: cell microinjected with rGBD and C3-insensitive mCh-Rho fails to rescue Rho activity in presence of C3. Scale bar 10 μm, time min:sec.
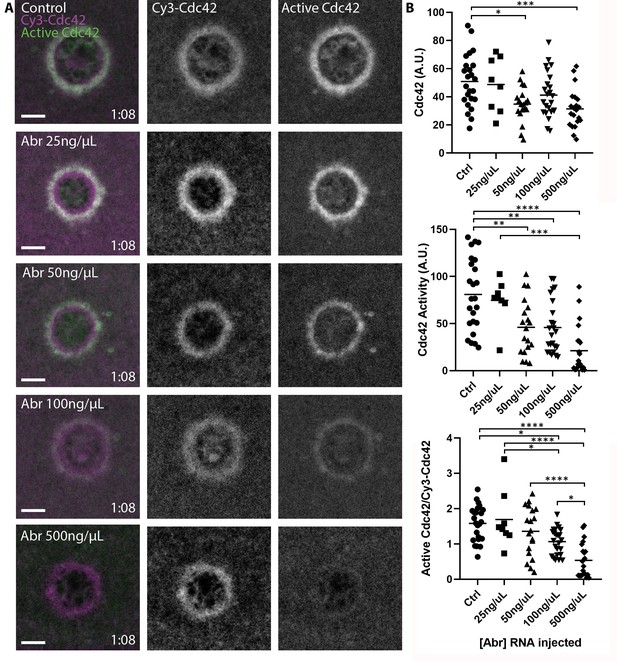
Pools of inactive and active Cdc42 at the plasma membrane.
(A) Oocytes microinjected with wGBD (green), Cy3-Cdc42 (magenta) and indicated concentrations of mRNA encoding the Cdc42-GAP Abr. Scale bar 10 μm, time min:sec.; (B) Quantification of Cy3-Cdc42, active Cdc42 and ratio of active Cdc42 to Cy3-Cdc42 for each condition. n = 8–24. One-way ANOVA with Tukey post-test statistical analysis. *p<0.05, **p<0.01, ***p<0.001, ***p<0.0001.
-
Figure 3—source data 1
Pools of inactive and active Cdc42 at the plasma membrane.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig3-data1-v2.xlsx
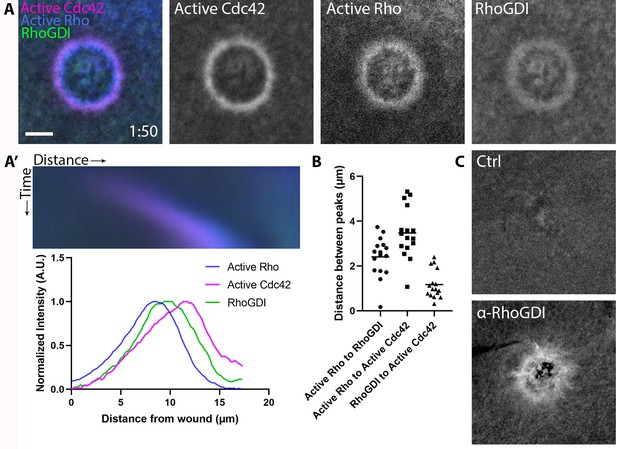
RhoGDI is recruited to single-cell wounds enriched in Rho and Cdc42 activity.
(A) Oocytes microinjected with wGBD (magenta), rGBD (blue) and GDI (green); (A’) Kymograph of wound closure and line scan of radially-averaged fluorescence intensity from (A); (B) Quantification of distance between peaks (n = 16); (C) Wounded oocytes fixed and stained with anti-X. laevis GDI. Scale bar 10 μm, time min:sec.
-
Figure 4—source data 1
RhoGDI is recruited to single-cell wounds enriched in Rho and Cdc42 activity.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig4-data1-v2.xlsx
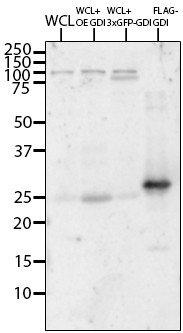
X. laevis RhoGDI antibody specificity.
Western blot stained with αGDI antibody to determine specificity; lane 1: X. laevis oocyte whole cell lysate (WCL), lane 2: WCL of oocytes overexpressing GDI, lane 3: WCL of oocytes expressing 3xGFP-GDI, lane 4: purified FLAG-GDI.
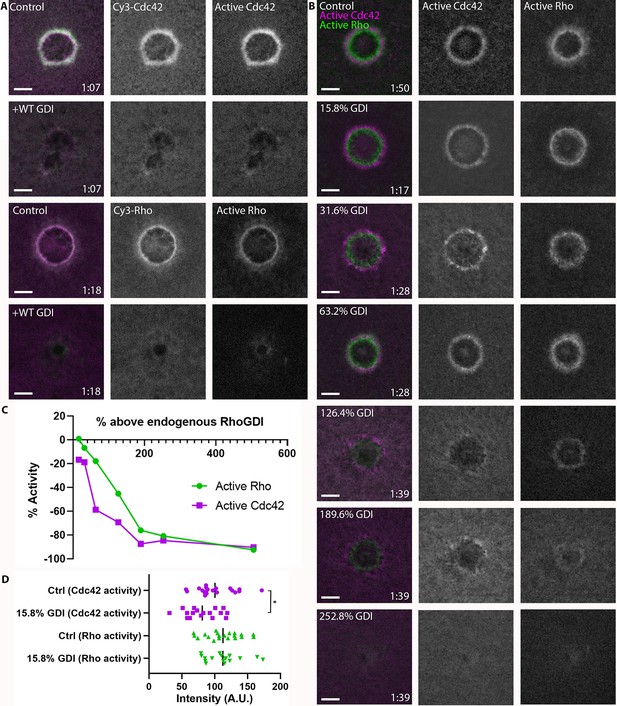
RhoGDI overexpression differentially regulates Rho and Cdc42 activity.
(A) Top two rows: oocytes microinjected with Cy3-Cdc42 (magenta) and wGBD (green) alone or with GDI. Bottom two rows: Oocytes microinjected with Cy3-Rho (magenta) and rGBD (green) alone or with GDI; (B) Oocytes microinjected with wGBD (magenta), rGBD (green) and increasing concentrations of GDI protein. Scale bar 10 μm, time min:sec; (C) Standard curve of Rho and Cdc42 activity with increasing concentrations of GDI (n = 10–23 for each concentration); (D) Quantification of Rho and Cdc42 activity at 15.8% above endogenous GDI (n = 17–20). Unpaired student’s t-test, 2-tailed distribution, equal variance statistical analysis. *p<0.05.
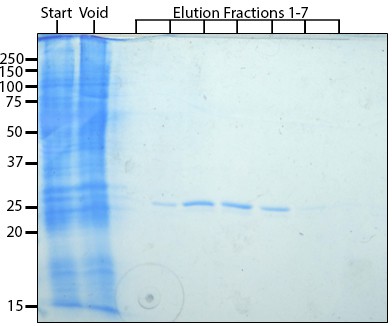
Purified X. laevis RhoGDI protein.
Coomassie stain of 12% SDS-PAGE to assess purity of FLAG-GDI; lane 1: start, lane 2: void, lanes 3–10: elution fractions.
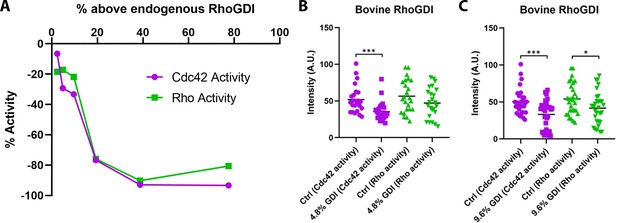
Bovine RhoGDI decreases Rho and Cdc42 activity in a dose-dependent manner in vivo.
(A) Standard curve of Rho and Cdc42 activity with increasing concentrations of bovine GDI (n = 2–30 for each concentration); (B) Quantification of Rho and Cdc42 activity at 4.8% above endogenous GDI (n = 24); (C) As in B but at 9.6% above endogenous GDI (n = 30–31). Unpaired student’s t-test, 2-tailed distribution, equal variance statistical analysis. *p<0.05, ***p<0.001.
-
Figure 5—figure supplement 2—source data 1
Bovine RhoGDI decreases Rho and Cdc42 activity in a dose-dependent mannerin vivo.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig5-figsupp2-data1-v2.xlsx
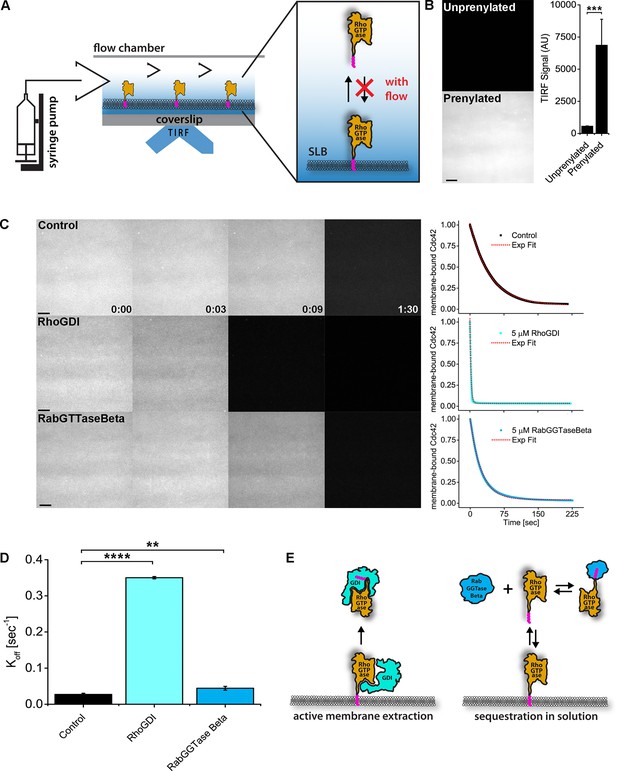
RhoGDI extracts RhoGTPases from membranes in vitro.
(A) Experimental setup of in vitro experiments: prenylated RhoGTPases were reconstituted on supported lipid bilayers (SLBs) in flow chambers and imaged by TIRF. Wash off experiments were designed to avoid RhoGTPase rebinding to membranes and performed controlling the flow rate via a syringe pump; (B) TIRF imaging allows for selective imaging of RhoGTPases at the membrane. Prenylated and unprenylated Cdc42 were imaged in the same conditions and TIRF signal at membranes was quantified (n = 4 for each condition); (C) Wash off experiments: prenylated Cdc42 reconstituted on SLBs were washed with imaging buffer only (control), in presence of 5 μM RhoGDI or RabGTTase Beta. Time lapse images at selected time points and quantification of the full experiments are shown. Decay curves were fitted with a monoexponential function; (D) Comparison of the Koff values obtained by fitting the decay curves (n = 3 for control and RabGTTase Beta, n = 2 for RhoGDI); (E) Schematic representation of the proposed mode of action of the two RhoGTPases solubilizers. RabGGTase Beta sequesters RhoGTPases in solution, whereas RhoGDI actively extracts RhoGTPases from the membranes. Scale bar 10 μm. Unpaired student’s t-test, 2-tailed distribution, equal variance statistical analysis. **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 6—source data 1
RhoGDI extracts RhoGTPases from membranesin vitro.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig6-data1-v2.xlsx
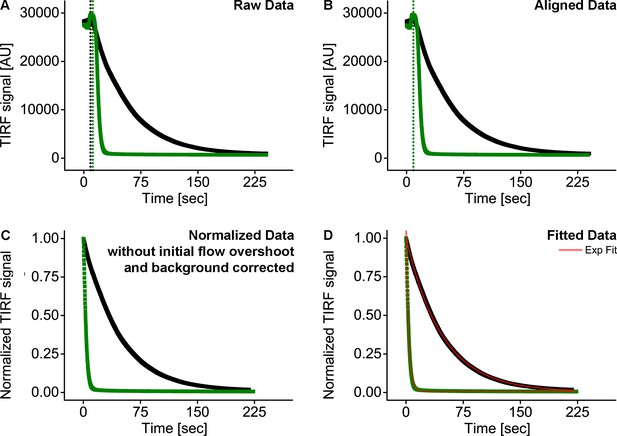
In vitro data analysis.
(A) Raw data. Wash off experiments were started after the GTPase signal at the membrane was stable. After starting the syringe pump, a signal overshoot occurred; (B) The overshoot signal was used as a reference point to align different experiments and cut off for further analysis; (C) Data were background corrected and normalized to display multiple curve on the same graph; (D) Data were fitted with a monoexponential decay curve to obtain dissociation constants (Koff) of the GTPases from the membrane.
Purified RhoGDI protein.
Coomassie stain of 12% SDS-PAGE to assess purity of RhoGDI purified from bacteria; lane 1: start, lane 2: flow through, lane 3: TEV cleavage, lane 4: tag removal, lane 5: end product after gel filtration.
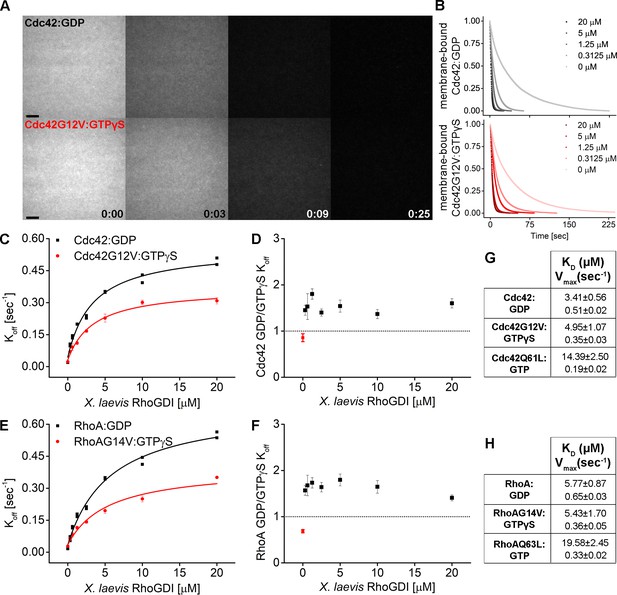
RhoGDI extracts both inactive and active RhoGTPases from membranes in vitro.
(A) Wash off experiments: prenylated Cdc42 in both inactive (Cdc42:GDP) and constitutively-active (Cdc42G12V:GTPγS) states were reconstituted on SLBs and washed in presence of 5 μM GDI. Time lapse images at selected time points are shown; (B) Quantification of wash off experiments in which the concentration of GDI was titrated between 0 and 20 μM; (C) Koff values obtained for inactive and constitutively-active Cdc42G12V fitting the decay curves with a monoexponential decay function are plotted against GDI concentration. Extraction rates were fitted with a hyperbolic function; fitting parameters Kd and Vmax are reported in table G; (D) Ratio of Koff obtained for inactive and constitutively-active Cdc42G12V at the same GDI concentration; (E–F, H) Same as in C-D and G for inactive (Rho:GDP) and constitutively-active (RhoG14V:GTPγS) Rho. Scale bar 10 μm.
-
Figure 7—source data 1
RhoGDI extracts both inactive and active RhoGTPases from membranesin vitro.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig7-data1-v2.xlsx
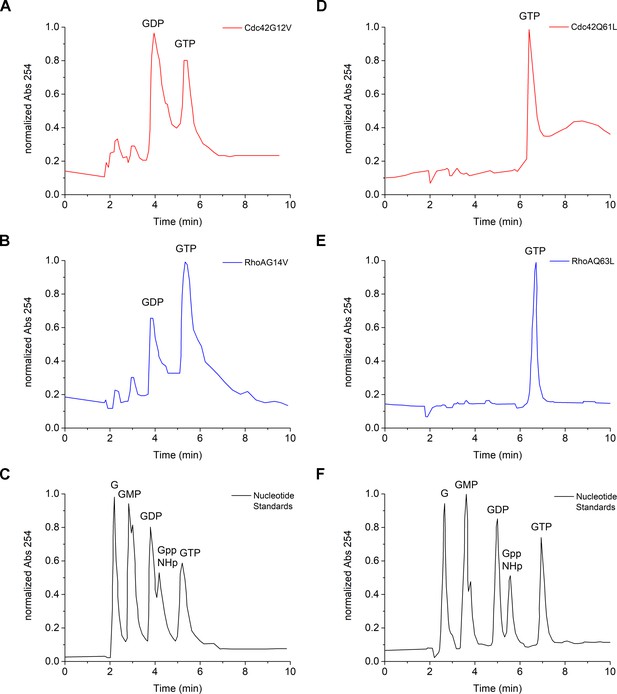
Nucleotide state of constitutively-active RhoGTPase variants after purification.
The nucleotide state of Cdc42G12V (A), RhoG14V (B), Cdc42Q61L (D) and RhoQ63L (E) was assessed by reversed phase chromatography monitoring absorbance at 254 nm. A sample containing nucleotide standards was run on the same day of the samples (C for A and B; F for D and E).
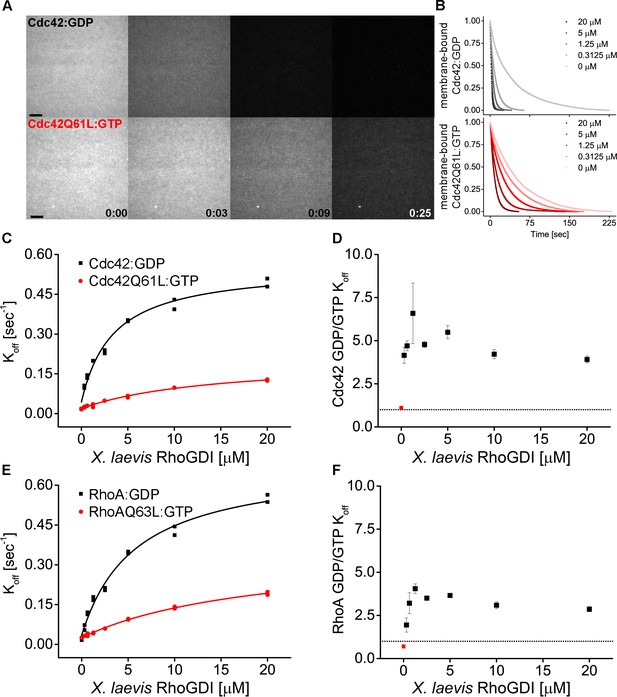
RhoGDI extracts both inactive and active RhoGTPases from membranes in vitro.
(A) Wash off experiments: prenylated Cdc42 in both inactive (Cdc42:GDP) and constitutively-active (Cdc42Q61L:GTP) states were reconstituted on SLBs and washed in presence of 5 μM GDI. Time lapse images at selected time points are shown; (B) Quantification of wash off experiments in which the concentration of GDI was titrated between 0 and 20 μM; (C) Koff values obtained for inactive and constitutively-active Cdc42Q61L fitting the decay curves with a monoexponential decay function are plotted against GDI concentration. Extraction rates were fitted with a hyperbolic function; (D) Ratio of Koff obtained for inactive and constitutively-active Cdc42Q61L at the same GDI concentration; (F) Same as in C-D for inactive (Rho:GDP) and constitutively-active (RhoQ63L:GTP) Rho. Scale bar 10 μm.
-
Figure 7—figure supplement 2—source data 1
RhoGDI extracts both inactive and active RhoGTPases from membranesin vitro.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig7-figsupp2-data1-v2.xlsx
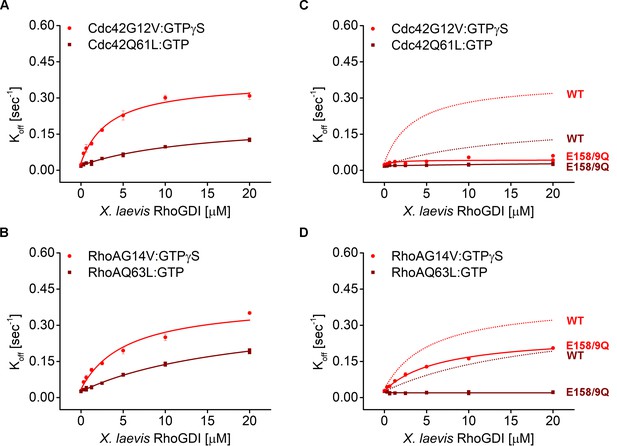
Comparison of G12V and Q61L constitutively-active RhoGTPases.
Comparison of Koff values obtained for the two constitutively-active variants of Cdc42 (Cdc42G12V and Cdc42Q61L, (A and C) and Rho (RhoG14V and RhoQ63L, (B and D) from wash off experiments in presence of either WT (A–B) or E158/9Q GDI (C–D). Extraction rates were fitted with a hyperbolic function.
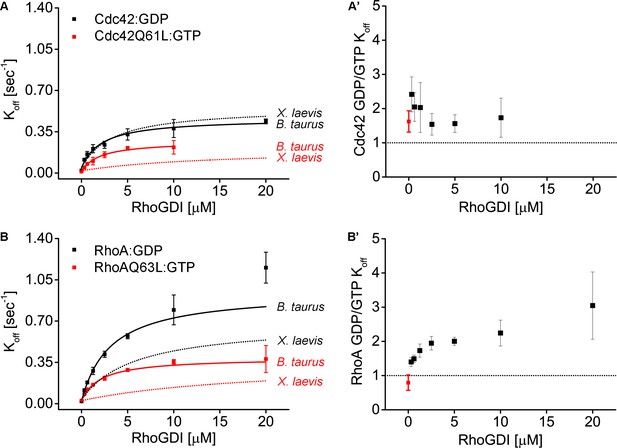
Comparison of bovine and Xenopus RhoGDI in their ability to extract both inactive and active RhoGTPases from synthetic membranes.
(A) Koff values obtained for inactive (Cdc42:GDP) and constitutively-active (Cdc42Q61L:GTP) Cdc42 at different bovine GDI concentrations. Extraction rates were fitted with a hyperbolic function; (A’) Ratio of Koff obtained for inactive and constitutively-active Cdc42 at the same bovine GDI concentration; (B–B’) same as in A-A’ for inactive (Rho:GDP) and constitutively-active (RhoQ63L:GTP) Rho.
-
Figure 7—figure supplement 4—source data 1
Comparison of bovine andXenopusRhoGDI in their ability to extract both inactive and active RhoGTPases from synthetic membranes.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig7-figsupp4-data1-v2.xlsx
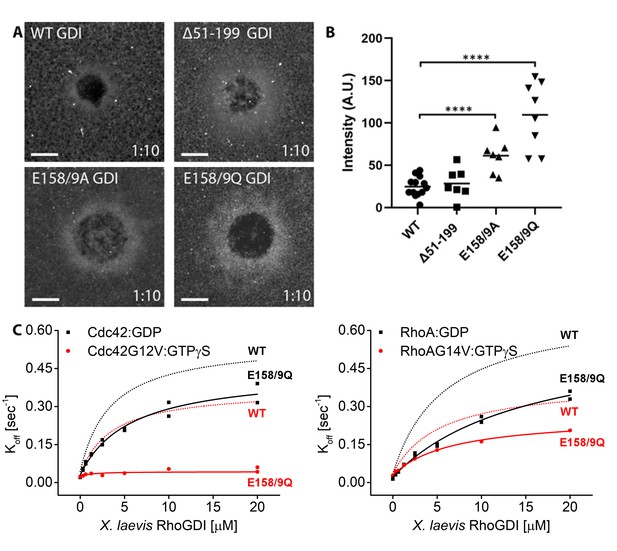
Identification of mutant RhoGDI deficient in extraction of active RhoGTPase.
(A) Oocytes microinjected with halo-tagged WT, Δ51–199, E158/9A and E158/9Q GDI mutants. Scale bar 10 μm, time min:sec; (B) Quantification of GDI intensity at wounds (n = 7–13). Unpaired student’s t-test, 2-tailed distribution, unequal variance statistical analysis to WT. ****p<0.0001. (C) Comparison of Koff values obtained for inactive (Cdc42:GDP, Rho:GDP) and constitutively-active (Cdc42G12V: GTPγS, RhoG14V: GTPγS) Cdc42 and Rho from wash off experiments in presence of either WT (black) or E158/9Q (QQ) (red) GDI. Extraction rates were fitted with a hyperbolic function.
-
Figure 8—source data 1
Identification of mutant RhoGDI deficient in extraction of active RhoGTPase.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig8-data1-v2.xlsx
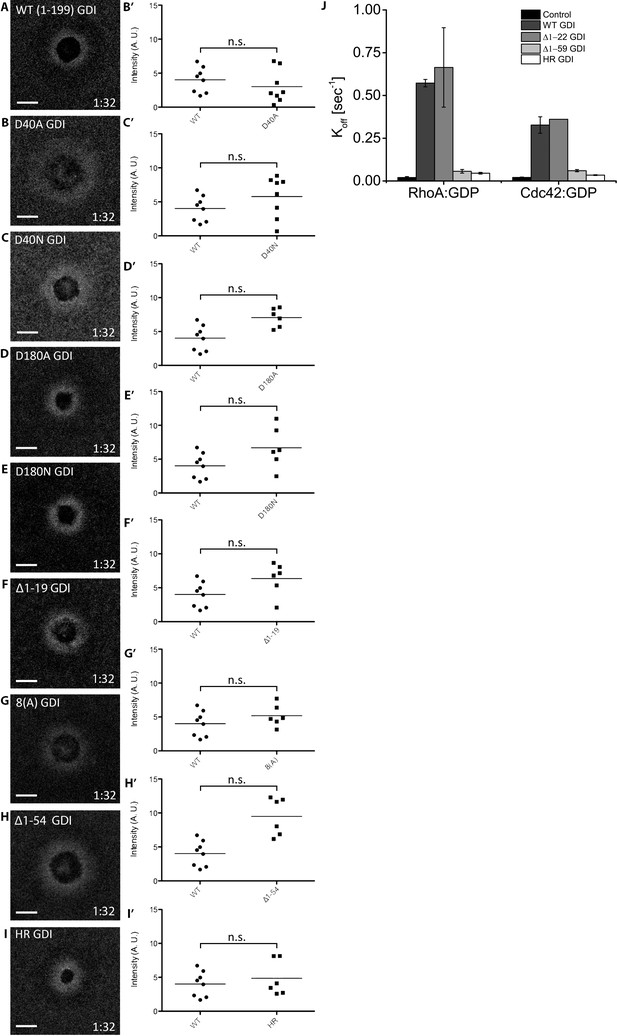
Analysis of previously-described extraction-deficient RhoGDI mutants.
(A–I) Oocytes microinjected with WT or mutant GDI and quantification of localization relative to WT GDI; Unpaired student’s t-test, 2-tailed distribution, unequal variance statistical analysis; (J) Average Koff values obtained for inactive Rho and Cdc42 in absence (control) and presence of 5 μM of either bovine GDI WT or bovine GDI mutants.
-
Figure 8—figure supplement 1—source data 1
Analysis of previously-described extraction-deficient RhoGDI mutants.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig8-figsupp1-data1-v2.xlsx
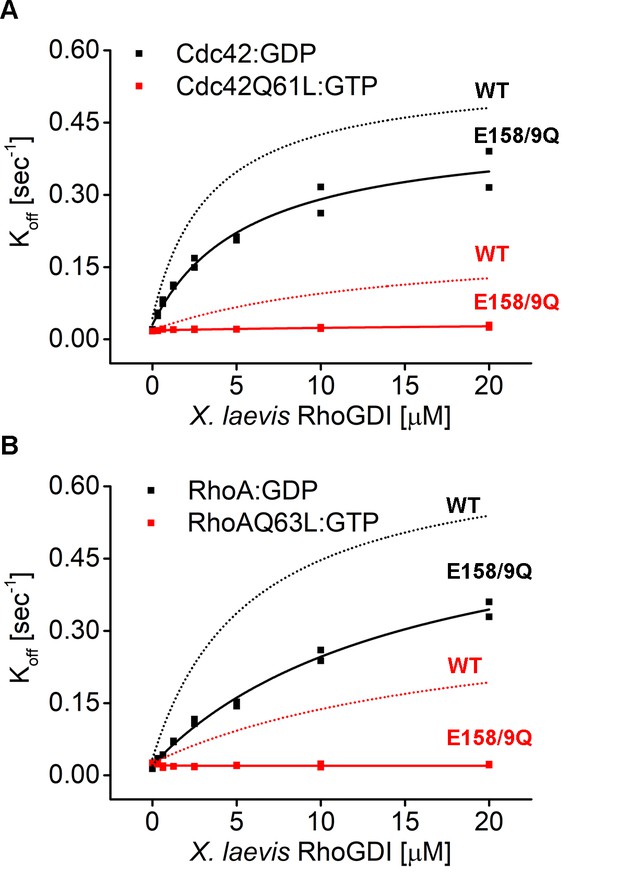
Mutant E158/9Q RhoGDI is deficient in extraction of constitutively-active Cdc42Q61L and RhoQ63L in vitro..
(A–B) Comparison of Koff values obtained for inactive (Cdc42:GDP, Rho:GDP) and constitutively-active (Cdc42Q61L:GTP, RhoQ63L:GTP) Cdc42 and Rho from wash off experiments in presence of either WT (black) or E158/9Q (QQ) (red) GDI. Extraction rates were fitted with a hyperbolic function.
-
Figure 8—figure supplement 2—source data 1
Mutant E158/9Q RhoGDI is deficient in extraction of constitutively-active Cdc42Q61L and RhoQ63Lin vitro.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig8-figsupp2-data1-v2.xlsx
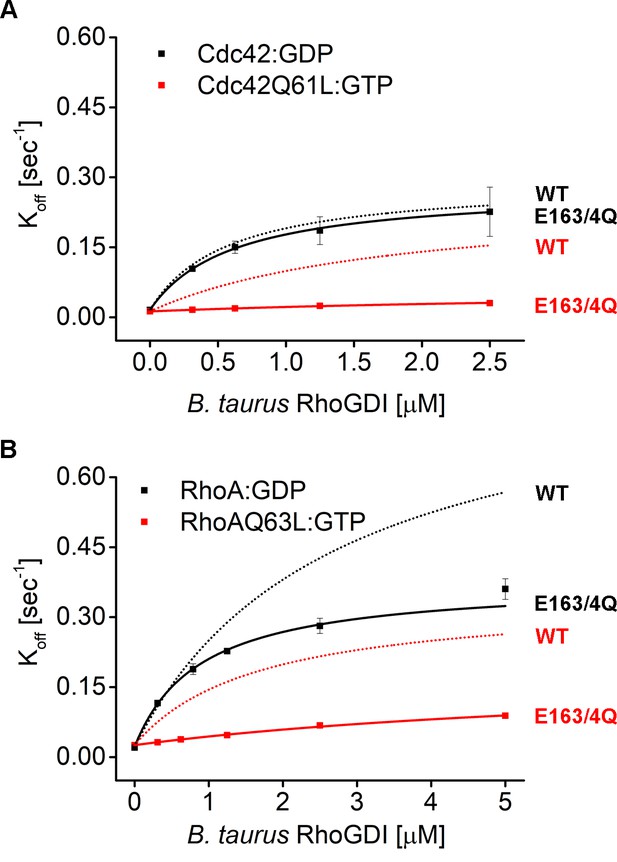
Mutant E163/4Q bovine RhoGDI is deficient in extraction of active Cdc42 and Rho in vitro.
Comparison of Koff values obtained for inactive (Cdc42:GDP, Rho:GDP) and constitutively-active (Cdc42Q61L:GTP, RhoQ63L:GTP) Cdc42 and Rho from wash off experiments in presence of either WT (black) or E163/4Q (QQ) (red) bovine GDI. Extraction rates were fitted with a hyperbolic function.
-
Figure 8—figure supplement 3—source data 1
Mutant E163/4Q bovine RhoGDI is deficient in extraction of active Cdc42 and Rhoin vitro.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig8-figsupp3-data1-v2.xlsx
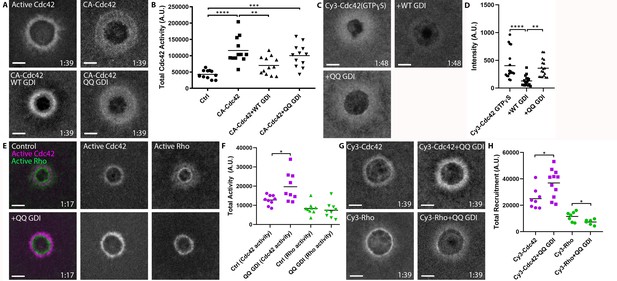
RhoGDI extracts active Cdc42 in vivo.
(A) Oocytes microinjected with wGBD alone or with constitutively-active Cdc42 (CA:G12V), WT or QQ GDI; B) Quantification of total Cdc42 activity for (A), (n = 12); C) oocytes microinjected with Cy3-Cdc42 bound to GTPɣS alone or with WT or QQ GDI; D) Quantification of intensity for (C), (n = 18). Scale bar 10 μm, time min:sec. One-way ANOVA with Tukey post-test statistical analysis; E) Oocytes microinjected with wGBD (magenta), rGBD (green) alone or with QQ GDI; F) Quantification of total Cdc42 (magenta) and Rho (green) activity from (E) (n = 9); G) Cy3-Cdc42 or Cy3-Rho alone or with QQ GDI; H) Quantification of total recruitment of Cy3-Cdc42 (magenta) and Cy3-Rho (green) (n = 6–11). Scale bar 10 μm, time min:sec. Unpaired student’s t-test, 2-tailed distribution, unequal variance statistical analysis. *p<0.05, **p<0.01, ***p<0.001, ***p<0.0001.
-
Figure 9—source data 1
RhoGDI extracts active Cdc42in vivo.
- https://cdn.elifesciences.org/articles/50471/elife-50471-fig9-data1-v2.xlsx
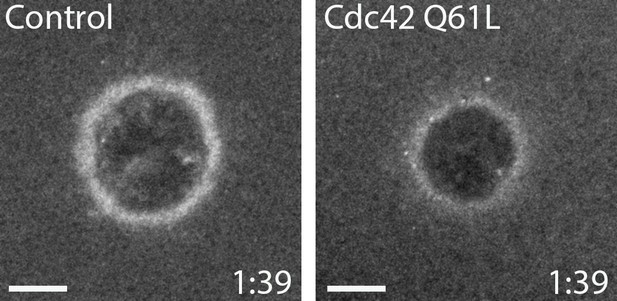
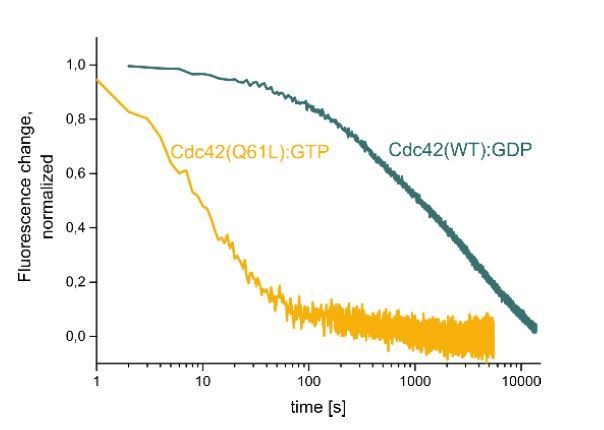
Cdc42 Q61L does not behave like constitutively-active Cdc42 in vivo.
Oocytes injected with wGBD alone or with Cdc42 Q61L. Scale bar 10 μm, time min:sec.

Schematic of proposed update to RhoGTPase cycle.
We propose that in addition to the canonical GTPase cycle, GDI can extract active GTPase from the plasma membrane. Based on evidence that GTPase:GDI binding prevents GTP hydrolysis and nucleotide exchange (Hart et al., 1992; Ueda et al., 2001), active GTPase extracted by GDI would still be active upon its release back into the plasma membrane.
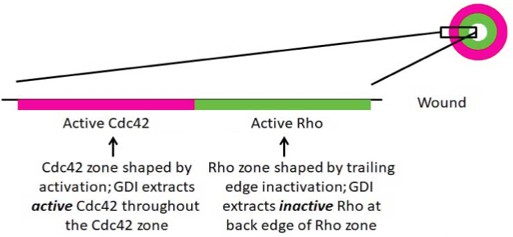
Schematic of RhoGDI’s role in RhoGTPase zone definition around wounds.
Active Rho (green) and active Cdc42 (red) are activated in discrete, concentric zones that close inward as the wound heals. Cdc42 inactivation is variable throughout its zone, thus its zone is shaped by activation (Burkel et al., 2012). Conversely, the Rho zone is shaped by inactivation as it is subject to RhoGAPs 1/8 at the trailing edge of its zone (Davenport, 2016). Thus, we hypothesize that GDI extracts active Cdc42 throughout the Cdc42 zone and inactive Rho from the trialing edge of the Rho zone.
Additional files
-
Supplementary file 1
Apparent affinities and maximum velocities determined by hyperbolic fits to the RhoGDI extraction rates of membrane-bound RhoGTPases.
- https://cdn.elifesciences.org/articles/50471/elife-50471-supp1-v2.docx
-
Supplementary file 2
Primers.
- https://cdn.elifesciences.org/articles/50471/elife-50471-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50471/elife-50471-transrepform-v2.pdf