Midkine is a dual regulator of wound epidermis development and inflammation during the initiation of limb regeneration
Figures
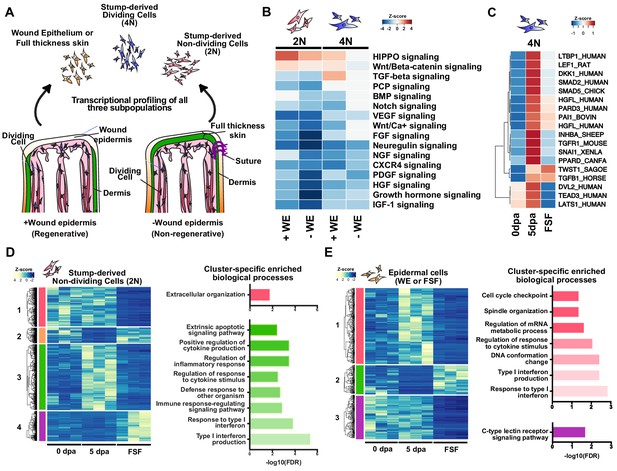
The early wound epidermis modulates inflammation, ECM remodeling, and tissue histolysis.
(A) Design of transcriptional profiling experiment. (B) Heatmap of Ingenuity Pathway Analysis (IPA) Z-score predictions for changes in signaling pathways in injured non-epithelial stump tissues (2N and 4N) in regenerative and full skin flap conditions. (C) Heatmap of normalized transcript levels of components/regulators of the canonical Wnt, TGF-beta, and HIPPO signaling pathways in dividing cells (4N). (D–E) Heatmaps and cluster-specific enriched biological processes of differentially expressed annotated transcripts in stump-derived non-dividing cells (2N) (D) and epithelial cells of the full skin flap (E) in regenerating vs. full skin flap conditions reveals global dysregulation of inflammation, ECM regulation, and tissue histolysis. Differentially expressed transcripts can be found in Supplementary files 1–3. Representative histological images of full skin flap sutured vs. normal regenerating limbs, PCA analysis of the samples, as well as enriched biological processes and related differentially expressed transcripts are shown in Figure 1—figure supplement 1. Z-score predictions shown in B from the IPA analysis are available in Figure 1—source data 1.
-
Figure 1—source data 1
Z-score predictions from the IPA analysis (for panel Figure 1B).
- https://cdn.elifesciences.org/articles/50765/elife-50765-fig1-data1-v1.csv

Sequencing of full skin flap sutured limbs reveals global dysregulation of ECM regulation, inflammation, and tissue histolysis.
(A–B’) Representative picro-mallory stained sections of normal regenerating and full skin flap sutured limbs. Full skin flap sutured limbs contain the extra dermal layer of skin tissue. Arrowheads demarcate the amputation plane. (C) Principal component analysis (PCA) of sequenced samples (5 dpa 2N, 5 dpa 4N, 5 dpa WE, FSF 2N, FSF 4N, FSF WE) based on the top 500 most highly expressed genes reveals separation along PC1 by tissue type and PC2 by condition. (D) Plot of enriched GO biological processes from all differentially expressed transcripts in non-dividing cells in full skin flap sutured limbs. (E) Heatmaps of normalized transcripts per million (TPM) values of annotated differentially expressed transcripts involved in ECM regulation, inflammation, or transcriptional regulation in non-dividing cells within regenerating stump tissues. (F) Heatmap of normalized TPM levels of annotated differentially expressed transcripts that are ECM regulators, immunomodulators, and transcriptional regulators in epithelial cells of full skin flap sutured limbs. dpa, days post-amputation, WE, wound epidermis, FSF, full skin flap, FDR, false discovery rate; ECM, extracellular matrix.
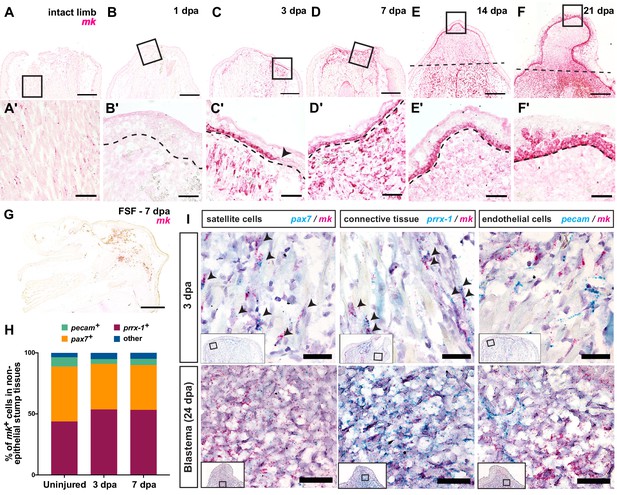
Midkine (mk) is highly expressed in the basal layers of the wound epidermis/AEC and blastemal progenitors.
(A–F’) Timecourse RNAscope in situ hybridization of midkine at 0 (intact), 1, 3, 7, 14, and 21 dpa. Insets in A-F are shown in A’-F’ at higher magnification. Arrowhead in C’ denotes the beginning of the wound epidermis. Dotted line marks amputation plane in E and F or wound epidermis/AEC boundary in B’-F’. (G) In situ hybridization of mk in full skin flap sutured limbs at 7 dpa. Axolotl MK protein expression can be found in Figure 2—figure supplement 1. (H) Breakdown of the percentages of mk+ cells that are pax7+, prrx-1+, and pecam+ in regenerating stump tissues during early stages of regeneration. At 3 dpa, a total N of 1579, 444, and 1180 cells were counted for pax7, prrx-1, and pecam double in situs, respectively. At 7 dpa, a total N of 456, 1043, and 274 cells were counted for pax7, prrx-1, and pecam double in situs, respectively (Figure 2—source data 1). (I) Representative images of RNAscope double in situ hybridization of mk with pax7 (left), prrx-1 (middle), or pecam (right) at early (3 dpa) and later blastema (24 dpa) stages. Insets depict where higher magnification images were taken. Black arrowheads mark double positive cells. More detailed analysis of the onset of mk expression in early stages of regeneration, representative images of double in situ hybridization of mk with cell type-specific markers in uninjured tissue, as well as the analysis of mk co-expression with the monocyte marker csf1r can be found in Figure 2—figure supplement 2. Scale bars, A-G: 500 µm, A’-F’: 100 µm, I: 50 µm. dpa, days post-amputation, FSF, full skin flap.
-
Figure 2—source data 1
Raw counts for mk double in situ hybridizations with cell-type specific markers.
- https://cdn.elifesciences.org/articles/50765/elife-50765-fig2-data1-v1.xlsx

MK protein is found throughout the wound epidermis/AEC.
(A) Normalized TPM levels of midkine (mk) in regenerating and full skin flap conditions. (B) Validation of custom polyclonal rabbit anti-MK antibody. Western blotting was performed with increasing volumetric ratios of MK antibody: blocking peptide ratios on 10 dpa protein extracts and revealed lower levels of staining with increased levels of peptide. (C–F’) Immunostaining of axolotl MK protein at 0 (C–C’), 5 (D–D’) and 14 dpa (E–E’). White arrowheads denote the amputation plane. Higher 20x magnification images of the wound epidermis or AEC (14 dpa) are shown in C’-F’. (F–F’) Immunostaining of axolotl MK protein in full skin flap sutured limbs at 7 dpa reveals lower levels of expression. Dotted lines demarcate full skin flap in F and the wound epidermis/stump boundary in C’-F’. Graphs are mean ± SD. **p<0.005, *p<0.05. Scale bars, B, E-H: 500 µm, E’-H’: 100 µm. FSF, full skin flap; dpa, days post-amputation.

Mk is expressed in satellite cells, connective tissue, and endothelial cells in intact tissues.
(A) RNAscope in situ hybridization of mk at 24, 56, and 72 hpa. Dotted line denotes wound epidermis boundary. Black arrowheads point to examples of mk+ cells. (B) Double RNAscope in situ hybridization of mk in red and pax7 (left) (N = 1073 cells counted), prrx-1 (middle) (N = 1723 cells counted), and pecam (right) (N = 1693 cells counted) in blue in non-regenerating intact limbs. Black arrowheads mark double positive cells. (B) Quantification of mk+ only, csf1r+ only, and co-positive cells. (C) Double RNAscope in situ hybridization of mk and csf1r at 5 dpa reveals discrete expression patterns (N = 582 cells counted). Black arrowheads denote mk+ only cells and black arrows denote csf1r+ only cells. Scale bars are 50 µm. hpa, hours post-amputation, dpa, days post-amputation.
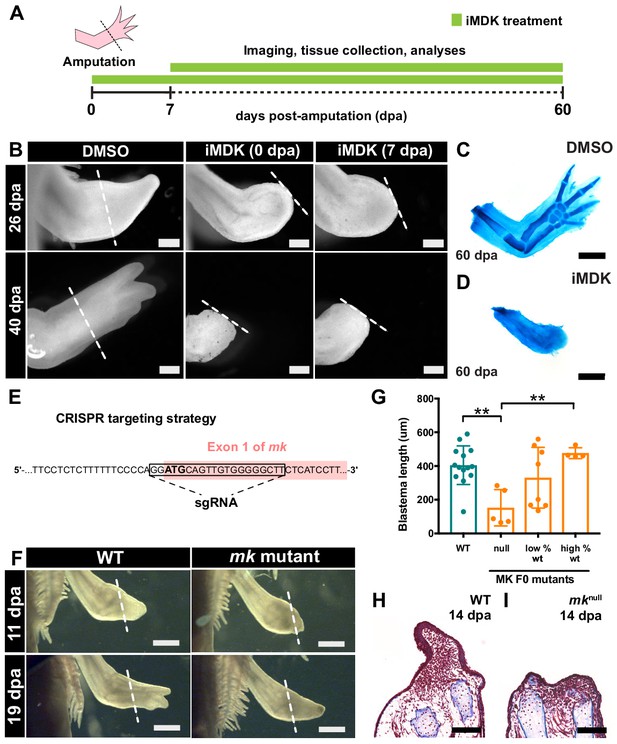
Chemical and genetic perturbations of mk impair limb regeneration.
(A) Experimental design. (B) Brightfield images of DMSO- or iMDK-treated limbs (N = 4/4 iMDK-treated in each condition did not regenerate). (C–D) Alcian blue staining of DMSO- or iMDK-treated limbs at 60 dpa. (E) CRISPR strategy to target the start codon of mk to generate mutants. Control animals were generated using a non-targeted tracrRNA/cas9 complex and were unmodified at the mk locus. (F) Brightfield images of regenerating limbs from control animals or mk F0 mutants. (G) Quantification of blastema length at 14 dpa in control or mk mutant limbs. The severity of the delay in regeneration segregated based on genotype of the animal as either control (mkWT), mosaic null (mknull), or partially modified (low or high % WT alleles) (N = 14 control mkWT, 5 mknull, 8 low % WT, 4 high % WT). Graph is mean ± SD. (H–I) Representative images of picro-mallory stained sections of regenerating limbs in control animals or mk null mutants. Example genotyping analyses and mk null mutant immunofluorescence validation can be found in Figure 3—figure supplement 1. **p<0.005, two-tailed unpaired Student’s t-test was employed. Each N represents one limb from a different animal. White dotted lines mark amputation plane. Scale bars, B-D, H-I: 500 µm, F: 1 mm. WT, wildtype, dpa, days post-amputation.
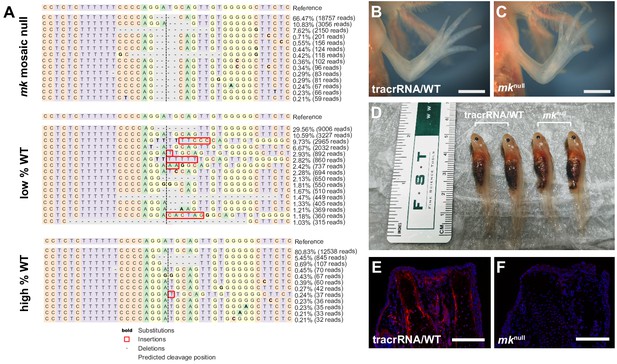
CRISPR generation of mk mosaic null mutants.
(A) Example genotyping results of fully modified (mosaic null, abbreviated mknull) or partially modified (low % or high % WT) animals. (B–C) Representative brightfield images of tracrRNA control and mknull forelimb of four-month old sibling axolotls. (D) Side-by-side brightfield image of tracrRNA control and mknull axolotls demonstrating no gross observational difference in body size. (E–F) Representative MK immunostaining of a tracrRNA control or mk mosaic null mutant at 7 dpa reveals a loss of staining. Scale bars, B-C: 2 mm, E-F: 500 µm. dpa, days post-amputation.
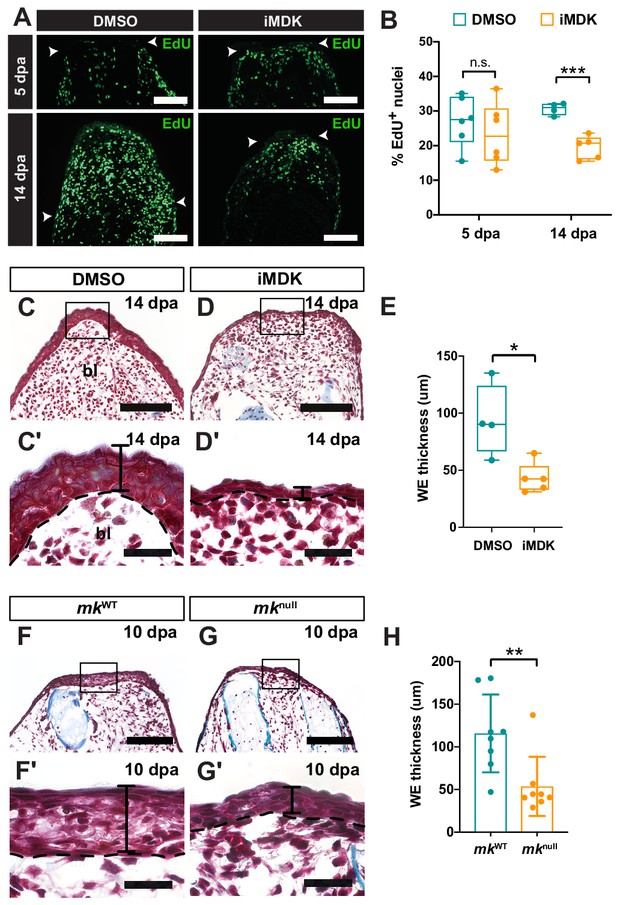
Mk perturbations result in defective AEC development.
(A–B) EdU staining (A) and quantification (B) of the total percentage of EdU+ nuclei of DMSO/iMDK-treated limbs at 5 dpa (N = 6 DMSO, 6 iMDK) and 14 dpa (N = 4 DMSO, 5 iMDK). White arrowheads denote the amputation plane in A. (C–D’) Picro-mallory stained sections from regenerating limbs from DMSO/iMDK-treated animals. Higher magnification images of the AEC are shown in C’-D’. (E) Quantification of wound epidermis (WE) thickness in DMSO/iMDK-treated regenerating limbs at 14 dpa (N = 4 DMSO, 5 iMDK). (F–G’) Picro-mallory stained sections from regenerating limbs from regenerating limbs of control WT (mkWT) or mknull animals. Higher magnification images of the developing AEC at 10 dpa are shown in F’-G’. (H) Quantification of WE thickness in regenerating limbs of mkWT or mknull animals at 14 dpa (N = 8 mkWT, 8 mknull). Picro-mallory staining of a 40 dpa iMDK-treated limb is shown in Figure 4—figure supplement 1. Each N represents a limb from a different animal. Two-tailed unpaired student’s t-test was used for statistical analysis. Graphs are mean ± SD. *p<0.05, **p<0.005, ***p<0.001. Scale bars, A, C-D, F-G: 200 µm, C’-D’, F’-G’: 100 µm. bl, blastema, dpa, days post-amputation.

iMDK-treated limbs do not regenerate.
Representative picro-mallory staining of a section from an iMDK-treated limb at 40 dpa. Re-formation of the blue collagen-thick dermal layer is commencing at the amputation plane (denoted with the black dotted line). Accumulation of fibroblastic blastemal cells can be seen at the amputation plane, although a blastema never forms. Scale bar, 200 µm.
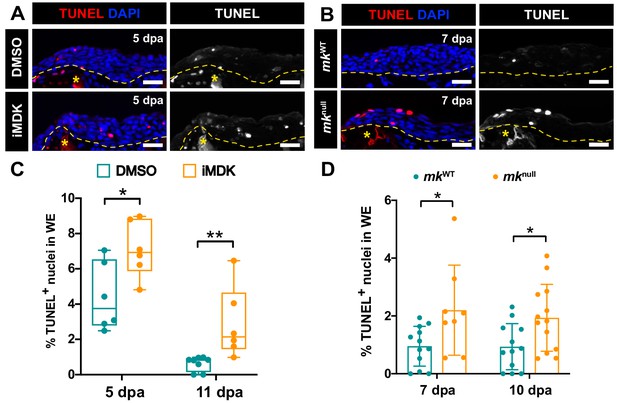
Mk acts as a critical survival factor during AEC development.
(A) Representative TUNEL-stained sections from limbs of DMSO and iMDK-treated limbs. (B) Representative TUNEL-stained sections from limbs of regenerating mkWT control or mknull mutants. (C) Quantification of the % TUNEL+ nuclei in the wound epidermis of DMSO or iMDK-treated limbs limbs at 5 dpa (N = 6 DMSO, 6 iMDK) or 11 dpa (N = 8 DMSO, 6 iMDK). (D) Quantification of the percentage of TUNEL+ nuclei in mkWT control and mknull mutants at 7 dpa (N = 12 mkWT, 8 mknull) and 10 dpa (N = 12 mkWT, 13 mknull). Asterisks mark auto-fluorescent bone. Each N represents a limb from a different animal. Quantification of levels of blastemal cell death in DMSO/iMDK-treated and mkWT/mknull regenerating limbs can be found in Figure 5—figure supplement 1. Two-tailed unpaired student’s t-tests were used for statistical analysis. Graphs are mean ± SD. *p<0.05, **p<0.005. Scale bars: 100 µm. dpa, days post-amputation.

Mk is not required for blastemal cell survival.
(A–B) Quantification of the percentage of TUNEL+ nuclei in blastemal cells of regenerating stump tissues in DMSO/iMDK-treated (5 dpa: N = 6 DMSO, 6 iMDK; 11 dpa: 8 DMSO, 6 iMDK) (A) or wildtype/mknull mutant (7 dpa: N = 12 mkWT, 8 mknull; 10 dpa: N = 12 mkWT, 13 mknull) (B) regenerating limbs reveals iMDK-treated limbs exhibit higher consistent levels of cell death, whereas mk mutant limbs are able to resolve cell death. Two-tailed unpaired t-tests were performed at each time point between the two conditions for statistical analysis. Graphs are mean ± SD. *p<0.05. n.s., not significant; dpa, days post-amputation.
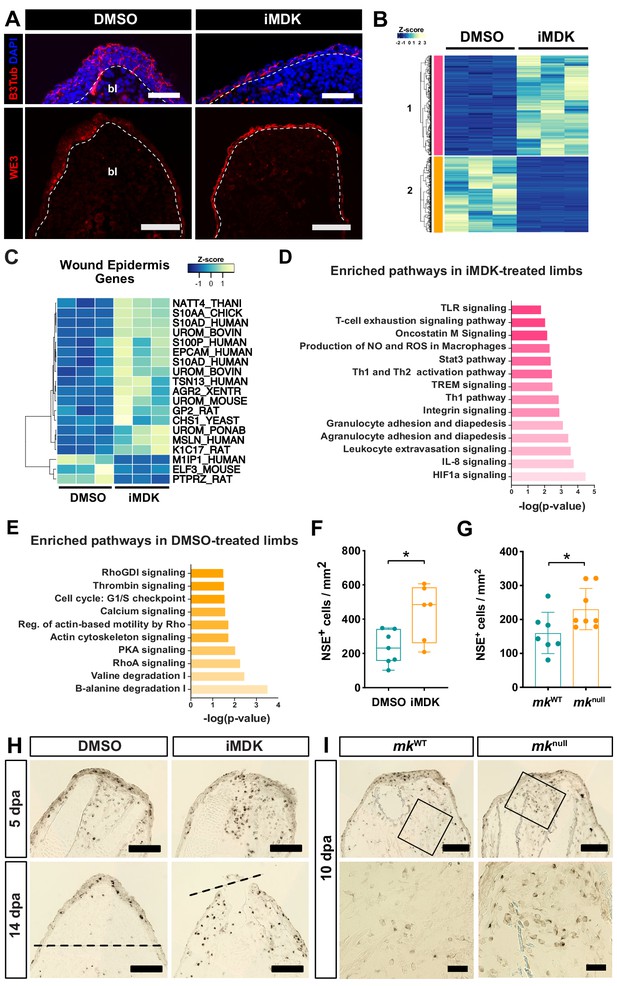
iMDK-treated and mk mutant regenerating limbs display dysregulated wound epidermis gene expression and persistent inflammation.
(A) Beta-III tubulin staining or WE3 staining of DMSO- or iMDK-treated limbs. White dotted line marks boundary of wound epidermis/AEC-blastema. (B) Heatmap of annotated differentially expressed transcripts in DMSO- and iMDK-treated limbs (N = 3 each) reveals two main clusters (colored pink and orange) of transcripts either enriched in DMSO or iMDK treatments. Transcript expression was normalized per row and plotted as a Z-score. Differentially expressed transcripts can be found in Supplementary file 4. (C) Heatmap of normalized TPM expression levels of wound epidermis genes in DMSO- or iMDK-treated regenerating limbs at 11 dpa. (D) Plot of enriched pathways in iMDK-treated limbs. (E) Plot of enriched pathways in DMSO-treated limbs. (F) Quantification of NSE+ monocytes at 5 dpa in DMSO/iMDK-treated limbs at 5 dpa (N = 7 DMSO, 6 iMDK). (G) Quantification of the density of NSE+ monocytes in mkWT control and mknull regenerating limbs at 10 dpa (N = 7 mkWT, 8 mknull). (H) NSE staining of DMSO- and iMDK-treated limbs at 5 dpa and 14 dpa. Dotted lines demarcate the amputation plane. (I) Representative NSE stained sections from regenerating limbs of mkWT control and mknull mutants at 10 dpa. Higher magnification insets are shown in bottom two panels. Each N represents a limb from a different animal. Data demonstrating rescue of mknull phenotypes via overexpression of mk during regeneration in mutant limbs as well as electroporation efficiency metrics can be found in Figure 6—figure supplements 1, 2 and 3. A two-tailed unpaired student’s t-test was used for statistical analysis. Graphs are mean ± SD. *p<0.05. Scale bars, A (top): 100 µm, A (bottom), H-I (top): 200 µm, I (bottom): 50 µm. bl, blastema, dpa, days post-amputation.

Mk overexpression in mknull regenerating limbs rescues delayed regeneration.
(A) Experimental schematic of rescue experiment. Either a pCAG-MK and/or pCAG-tdTomato control overexpression construct was injected and electroporated into the regenerating limbs of wildtype or mknull animals at 3 dpa. (B) Representative image of tdTomato fluorescence in a pCAG-MK/pCAG-tdTomato co-electroporated regenerating limb at 14 dpa (outlined in white dotted line). (C) Quantification of blastema length at 13 dpa. One-way ANOVA analyses were employed on each set of data for statistical analysis (N = 6 mkWT + tdT, 10 mknull+tdT, 7 mknull + MK). (D) Representative time course images of regenerating mkWT and mknull limbs electroporated with pCAG-tdTomato and/or pCAG-MK constructs. Two different rescue animals are shown here. Graphs are mean ± SD. **p<0.005, *p<0.05. Scale bars, B, D: 1 mm. n.s., not significant; dpa, days post-amputation.
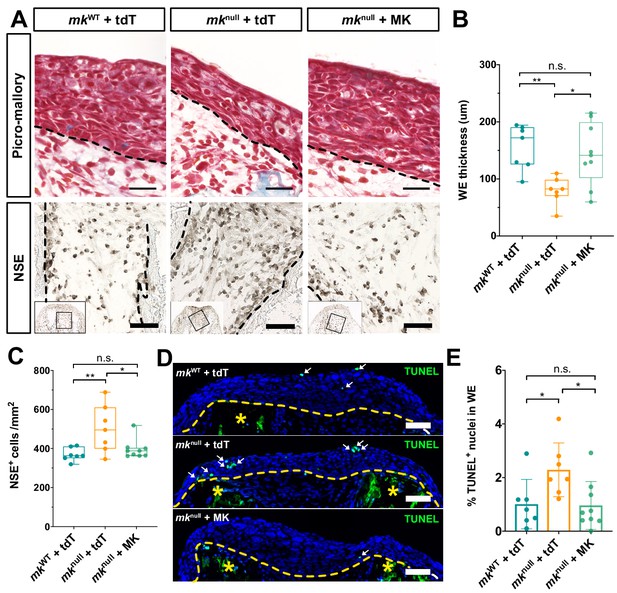
Mk overexpression in mknull regenerating limbs rescues mutant wound epidermis and monocyte density phenotypes.
(A) Representative images of picro-mallory-stained (top row) and NSE-stained (bottom row) sections from tdTomato and/or mk-overexpressing wildtype and mutant regenerating limbs. Black dotted lines mark the wound epidermis or bone boundary in the top and bottom rows, respectively. (B) Quantification of wound epidermis thickness at 10 dpa. (C) Quantification of NSE+ monocyte density at 10 dpa. (D) Representative images of TUNEL-stained sections, focusing on the wound epidermis. Yellow dotted line demarcates the boundary between the wound epidermis and regenerating stump tissues. Yellow asterisk marks the auto-fluorescent bone. White arrows point to green TUNEL+ nuclei. (E) Quantification of TUNEL+ nuclei in the wound epidermis at 10 dpa. One-way ANOVA analyses were employed on each set of data for statistical analysis (N = 7 mkWT + tdT, 7 mknull+tdT, 9 mknull + MK). Graphs are mean ± SD. **p<0.005, *p<0.05. Scale bars, B (top row), 50 µm, B (bottom row) and D, 100 µm. n.s., not significant; dpa, days post-amputation.
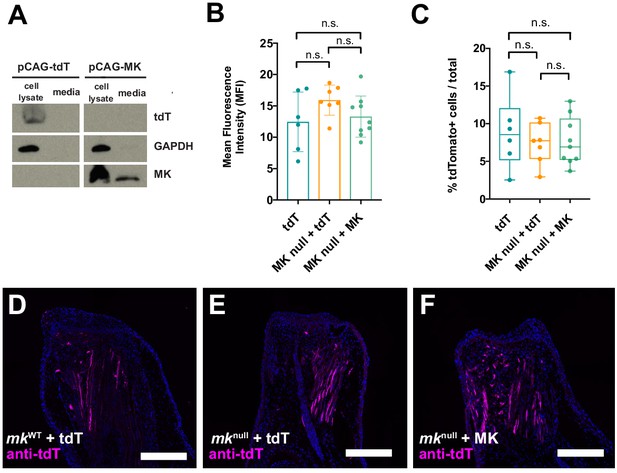
Electroporation efficiencies are similar between pCAG-tdTomato and pCAG-MK injected mknull mutant and mkWT regenerating limbs.
(A) Validation of pCAG-MK overexpression construct with MK antibody. Western blots of either pCAG-tdTomato or pCAG-MK transfected 293 T cells with the custom axolotl-MK antibody reveals the overexpression and secretion of axolotl MK in pCAG-MK, but not pCAG-tdTomato (tdT), transfected cell lysates and media. (B–C) Quantification of mean fluorescence intensity and the percentage of tdTomato+ expressing cells out of total DAPI+ cells in the distal-most 500 µm of regenerating tissue at 10 dpa. (D–F) Representative images of anti-tdTomato-stained sections from each condition. One-way ANOVA analyses were employed on each set of data for statistical analysis (N = 7 mkWT + tdT, 7 mknull+tdT, 9 mknull + MK). Scale bars, 500 µm. dpa, days post-amputation, n.s. not significant.
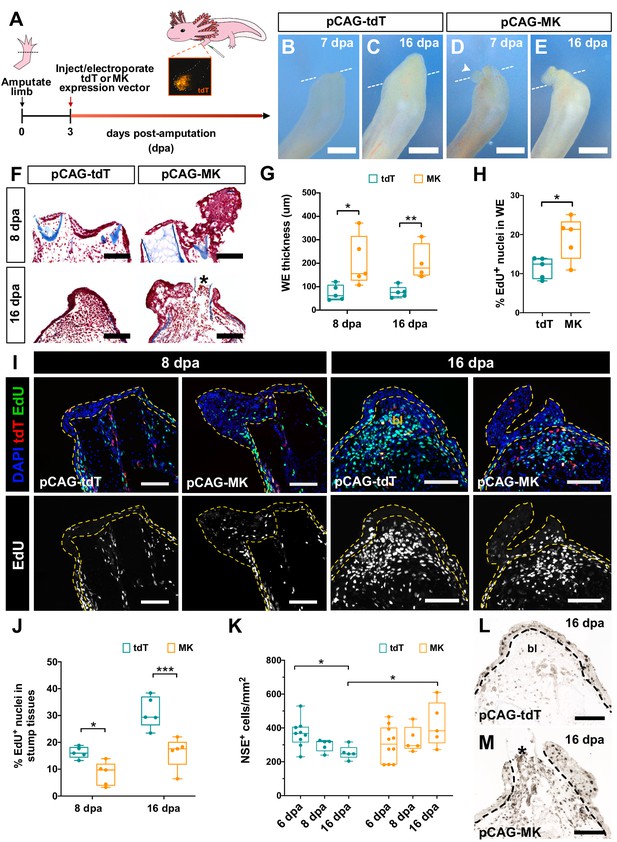
Overexpression of mk in regenerating limbs leads to wound epidermis expansion, decreased blastemal cell proliferation, and improper resolution of inflammation.
(A) Schematic of mk overexpression experiment. (B–E) Representative brightfield images of tdTomato (tdT)- (B–C) or mk-overexpressing (D–E) regenerating limbs at 7 and 16 dpa. White dotted line denotes amputation planes and white arrowhead in D denotes small aberrant skin growth. (F) Representative picro-mallory stained sections of tdT- or mk-overexpressing regenerating limbs. Asterisk marks protruding bone. (G) Quantification of wound epidermis thickness in tdTomato- or mk-overexpressing limbs at 8 dpa (N = 5 each) and 16 dpa (N = 5 tdT, N = 4 MK). (H) Quantification of the percentage of EdU+ nuclei in the wound epidermis of tdTomato- or mk-overexpressing limbs at 8 dpa (N = 5 each). (I) Representative images of EdU-stained sections of tdTomato- or mk-overexpressing limbs. Wound epidermis/AEC are outlined with yellow dotted lines. (J) Quantification of the percentage of EdU+ nuclei in regenerating stump tissues of tdT- or mk-overexpressing limbs. (K) Quantification of the density of NSE+ monocytes in tdT- or mk-overexpressing limbs. (L–M) Representative images of NSE-stained sections in tdT- or mk-overexpressing limbs. Black dotted line denotes wound epidermis boundary. Asterisk denotes protruding bone. Quantification of EdU+ cells in the wound epidermis and blastemal cells of DMSO/iMDK-treated and WT/mknull regenerating limbs is shown in Figure 7—figure supplement 1. Electroporation efficiency data can be found in Figure 7—figure supplement 2. Data showing that mk-overexpressing regenerating limbs display delayed regeneration is shown in Figure 7—figure supplement 3. Data demonstrating that overexpression of mk in non-regenerating intact limbs does not affect cellular proliferation or monocyte density can be found in Figure 7—figure supplement 4. Graphs are mean ± SD. Two-tailed unpaired student’s t-tests were used for statistical analysis. *p<0.05, **p<0.01, ***p<0.005. Scale bars, B-E: 500 µm, F, I, L-M: 200 µm. bl, blastema, dpa, days post-amputation.
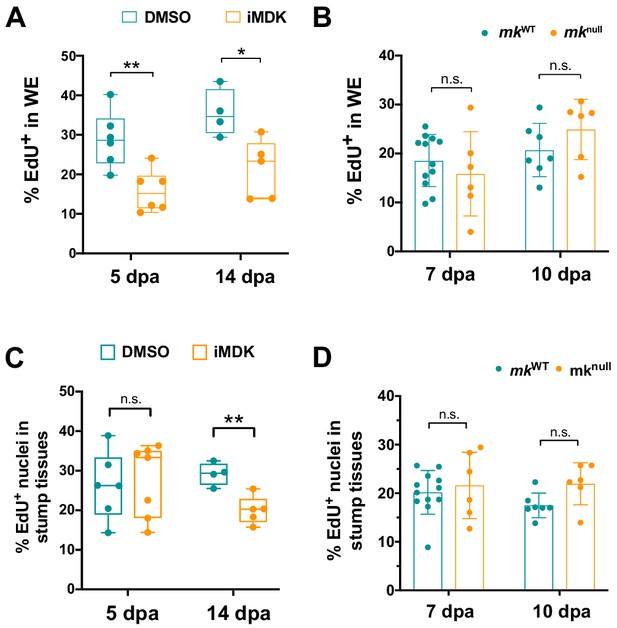
Mk is not required for proliferation in the wound epidermis or blastemal cells.
(A–B) Quantification of the percentage of EdU+ cells in the wound epidermis (WE) of DMSO/iMDK-treated limbs (5dpa: N = 6 DMSO, 6 iMDK; 14 dpa: N = 4 DMSO, 5 iMDK) (A) or mkWT/mknull limbs (7 dpa: N = 12 mkWT, 8 mknull; 10 dpa: N = 12 mkWT, 13 mknull) (B). (C–D) Quantification of the percentage of EdU+ blastemal cells in DMSO/iMDK-treated limbs (C) or mkWT/mknull limbs (D). Two-tailed unpaired t-tests were performed at each time point between the two conditions for statistical analysis. Graphs are mean ± SD. **p<0.005, *p<0.05. n.s., not significant; dpa, days post-amputation.

Electroporation efficiencies of pCAG-tdT and pCAG-MK injected regenerating limbs are similar.
(A–B) Quantification of mean fluorescence intensity (MFI) (A) and the percentage of tdTomato+ cells out of the total DAPI+ cells (B) in the distal most 500 µm of regenerating pCAG-tdT (N = 8) and/or pCAG-MK (N = 9) electroporated limbs at 6 dpa. (C–D) Representative immunostaining against tdTomato of tdTomato- or MK-electroporated limbs. Two-tailed unpaired t-tests were performed for statistical analysis. Scale bar, 250 µm. dpa, days post-amputation, n.s., not significant.
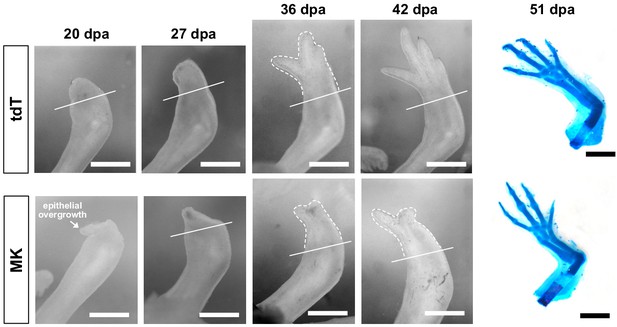
Mk-overexpressing limbs resolve epithelial overgrowth and undergo delayed regeneration.
Representative time course images of control tdTomato (top) or mk-overexpressing (bottom) regenerating limbs at 20, 27, 36, and 42 dpa (N = 9/12 animals exhibited a substantial delay in regeneration). Alcian-stained regenerated limbs at 51 dpa are shown on the far right. Scale bars, 1 mm. dpa, days post-amputation.
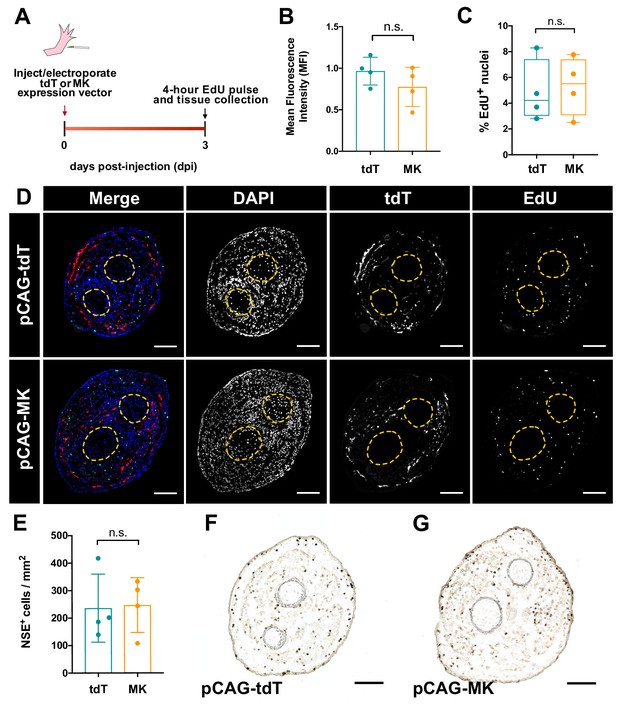
Overexpression of mk in non-regenerating intact limbs does not affect cellular proliferation or monocyte density.
(A) Experimental schematic of overexpression experiment in non-regenerating limbs. (B) Mean fluorescence intensity (MFI) quantification of electroporated limbs. (C) Quantification of the percentage of EdU+ cells in tdT- or mk-overexpressing intact limbs. (D) Representative images of tdT and EdU-stained cross-sections from electroporated limbs. (E) Quantification of monocyte density in overexpression limbs. (F–G) Representative images from NSE+ stained sections from non-regenerating overexpression limbs. Two-tailed unpaired t-tests were performed for statistical analysis. Graphs are mean ± SD. Scale bars, D, F-G: 200 um. n.s., not significant.
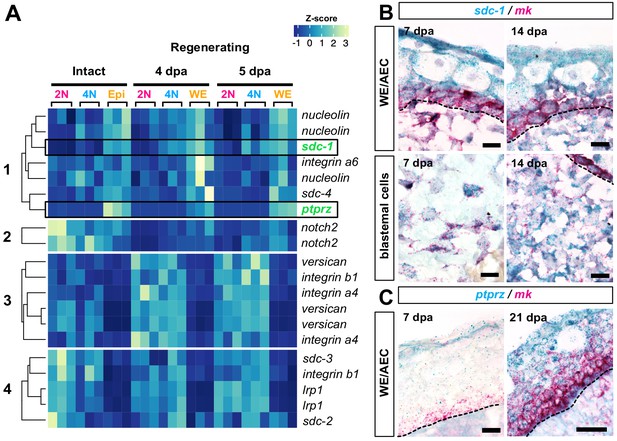
Mk receptors are expressed throughout regenerating tissues.
(A) Heatmap of normalized transcript levels for expressed mk receptors in intact and regenerating subpopulations from our RNA-seq dataset revealed four general patterns of expression (Figure 8—source data 1). (B–C) Double RNAscope in situ hybridization of mk (bright red puncta) with its cognate receptors sdc-1 (B) or ptprz (C) (dark blue puncta). Dotted black line denotes wound epidermis/AEC boundary. Since ptprz expression was low during early stages of regeneration, ptprz/mk in situs were performed without a hematoxylin counterstain to ease visualization of dark blue puncta in the wound epidermis at 7 dpa. Scale bars, B and C (left panel): 25 µm for 63x magnification, C (right panel): 50 µm for 40x magnification. WE, wound epidermis, AEC, apical epithelial cap, bl, blastema, dpa, days post-amputation.
-
Figure 8—source data 1
TPM values and transcript IDs for midkine receptors in Tsai et al. (2019).
- https://cdn.elifesciences.org/articles/50765/elife-50765-fig8-data1-v1.csv

Mk receptors are not highly expressed in myeloid cells during wound healing.
Publicly available single cell expression data of mk receptors from Leigh et al. (2018). Top left panel shows original clustering of single cell expression profiles at the wound healing stage of blastemal progenitors, wound epidermis, and myeloid cell populations circled. Top right panel shows t-SNE plot of apoeb expression, which was identified as highly enriched in myeloid cells, to give an example of a highly expressed gene in the myeloid cell populations. All panels below are t-SNE plots showing expression of mk receptors.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Ambystoma mexicanum) | Midkine (mk) | |||
| Strain, strain background (Ambystoma mexicanum) | Axolotl, White (d/d) | Ambystoma Genetic Stock Center (AGSC) (RRID:SCR_006372) | Cat. #AGSC_1015 | Subadults and juveniles, used for FSF transcriptional analysis and functional experiments |
| Strain, strain background (Ambystoma mexicanum) | Axolotl, Albino (a/a) | Ambystoma Genetic Stock Center (AGSC) (RRID:SCR_006372) | Cat. #AGSC_1025 | Subadults, used only for FSF transcriptional analysis |
| Genetic reagent (Ambystoma mexicanum) | Mknull mutants | This paper | Generated in d/d strain | |
| Cell line (Homo sapiens) | 293T HEK Cells | ATCC (RRID:SCR_001672) | Cat. #CRL-3216 (RRID:CVCL_0063) | For validation of MK overexpression construct |
| Antibody | Polyclonal goat anti-tdTomato | LS Bio (RRID:SCR_013414) | Cat. #LS-C340696 (RRID:AB_2819022) | WB: 1:200 |
| Antibody | Polyclonal chick anti-GAPDH | Millipore (RRID:SCR_008983) | Cat. #AB2302 (RRID:AB_10615768) | WB: 1:2000 |
| Antibody | Polyclonal rabbit anti-midkine (axolotl-specific) | This paper, NEPeptide | IF: 1:500, WB: 1:2000 | |
| Antibody | Monoclonal mouse Anti-WE3 | DSHB (RRID:SCR_013527) | Cat. #WE3 (RRID:AB_531902) | IF: 1:10 |
| Antibody | Monoclonal mouse Anti-Beta III Tubulin | Sigma Aldrich (RRID:SCR_008988) | Cat. #T8578 (RRID:AB_1841228) | IF: 1:200 |
| Recombinant DNA reagent | pCAG-tdTomato | Pathania et al., 2012 | Addgene: Cat. #83029 | Injected and electroporated at 500 ng/uL |
| Recombinant DNA reagent | pCAG-MK | This paper | Injected and electroporated at 500 ng/uL | |
| Sequenced-based reagent | Custom RNAscope axolotl prrx-1 probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl csf1r probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl pax7 probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl pecam probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl midkine probe (C2) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl ptprz probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Custom RNAscope axolotl sdc1 probe (C1) | This paper | In situ hybridization probe | |
| Sequenced-based reagent | Alt-R CRISPR-Cas9 crRNA (mk specific): 5’AAGCCCCCACAACTGCATCC −3’ | This paper | Midkine specific short guide RNA sequence | |
| Sequenced-based reagent | Alt-R CRISPR-Cas9 tracrRNA | Integrated DNA Technologies (IDT) | Cat. #1072534 | |
| Sequenced-based reagent | Mk forward genotyping primer: 5’-TTGCTTATTCCTTGTGATCATGC-3’ | This paper | Genotyping primer | |
| Sequenced-based reagent | Mk reverse genotyping primer: 5’- GGCACATTATTACACAGAAAGCTC-3’ | This paper | Genotyping primer | |
| Sequenced-based reagent | Mk nested PCR forward primer: 5’- tctttccctacacgacgctcttccgatctGAGGTTTGATTGGACCCTGA-3’ | This paper | Genotyping primer | |
| Sequenced-based reagent | Mk nested PCR reverse primer: 5’-tggagttcagacgtgtgctcttccgatctGGCACATTATTACACAGAAAGCTC-3’ | This paper | Genotyping primer | |
| Peptide, recombinant protein | Axolotl Midkine blocking peptide (amino acids 126 to 142) | This paper, NEPeptide | Used to validate custom polyclonal axolotl MK antibody | |
| Chemical compound, drug | iMDK (Midkine inhibitor) | Tocris Bio (RRID:SCR_003689) | Cat. #5126 | Used at 10 uM |
| Chemical compound, drug | EdU | Thermofisher Scientific (RRID:SCR_008452) | Cat. #A10044 | Used at 8 mg/mL concentration |
| Commercial assay or kit | In Situ Cell Death Detection Kit, TMR Red | Roche (RRID:SCR_001326) | Cat. #12156792910 | |
| Commercial assay or kit | In Situ Cell Death Detection Kit, Fluorescein | Roche (RRID:SCR_001326) | Cat. #11684795910 | |
| Commercial assay or kit | Click-iT EdU Cell Proliferation Kit for Imaging | Thermofisher Scientific (RRID:SCR_008452) | Cat. #C10337 | |
| Commercial assay or kit | α-Naphthyl Acetate Esterase (NSE) Kit | Sigma Aldrich (RRID:SCR_008988) | Cat. #91A | |
| Commercial assay or kit | RNAScope 2.5 HD Duplex Assay | ACD Bio | Cat. #322430 | |
| Commercial assay or kit | Miseq Reagent Nano Kit v2 (300-cycle) | Illumina (RRID:SCR_010233) | Cat. #MS-102–2002 | |
| Commercial assay or kit | Illumina Nextera XT DNA Library Prep Kit | Illumina (RRID:SCR_010233) | Cat. #FC-131–1024 | |
| Commercial assay or kit | Ovation RNA-seq System V2 | Integrated Sciences | Cat. #7102–32 | |
| Commercial assay or kit | PrepX ILM 32i DNA Library Prep Kit | Takara Bio | Cat. #400076 | |
| Software, algorithm | Trinity | Grabherr et al., 2011 | https://github.com/trinityrnaseq/trinityrnaseq/wiki | |
| Software, algorithm | Kallisto | Bray et al., 2016 | https://pachterlab.github.io/kallisto/ | |
| Software, algorithm | Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic | |
| Software, algorithm | DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | |
| Software, algorithm | WEB Gestalt | Wang et al., 2017 | http://www.webgestalt.org/ | |
| Software, algorithm | CRISPResso | Pinello et al., 2016 | http://crispresso.pinellolab.partners.org/ | |
| Software, algorithm | Complex Heatmaps | Gu et al., 2016 | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html | |
| Software, algorithm | ImageJ | Schindelin et al., 2012 | https://imagej.net/Fiji |
Additional files
-
Supplementary file 1
Annotated differentially expressed transcripts in dividing cells (4N) non-epithelial stump tissues in full skin flap sutured vs. normal regenerating limbs.
This excel table contains the list of differentially expressed transcripts in dividing cells (enriched for progenitors) in both conditions at 5 dpa. The respective fold change, blastx hit, and adjusted p-values are listed for each hit.
- https://cdn.elifesciences.org/articles/50765/elife-50765-supp1-v1.xlsx
-
Supplementary file 2
Annotated differentially expressed transcripts in non-dividing cells (2N) in non-epithelial stump tissues in full skin flap sutured vs. normal regenerating limbs.
This excel table contains the list of differentially expressed transcripts in non-dividing cells in stump tissues in both conditions at 5 dpa. The respective fold change, blastx hit, and adjusted p-values are listed for each hit.
- https://cdn.elifesciences.org/articles/50765/elife-50765-supp2-v1.xlsx
-
Supplementary file 3
Annotated differentially expressed transcripts in epithelial cells of full skin flap sutured vs. normal regenerating limbs.
This excel table contains the list of differentially expressed transcripts in epithelial cells of full thickness skin vs. wound epithelial cells at 5 dpa. The respective fold change, blastx hit, and adjusted p-values are listed for each hit.
- https://cdn.elifesciences.org/articles/50765/elife-50765-supp3-v1.xlsx
-
Supplementary file 4
Annotated differentially expressed transcripts in DMSO- vs. iMDK-treated regenerating limbs.
This excel table contains the list of differentially expressed transcripts in DMSO vs. iMDK-treated limbs at 11 dpa with the respective fold change, blastx hit, and adjusted p-values for each hit.
- https://cdn.elifesciences.org/articles/50765/elife-50765-supp4-v1.csv
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50765/elife-50765-transrepform-v1.pdf





