Sequential phosphorylation of NDEL1 by the DYRK2-GSK3β complex is critical for neuronal morphogenesis
Figures
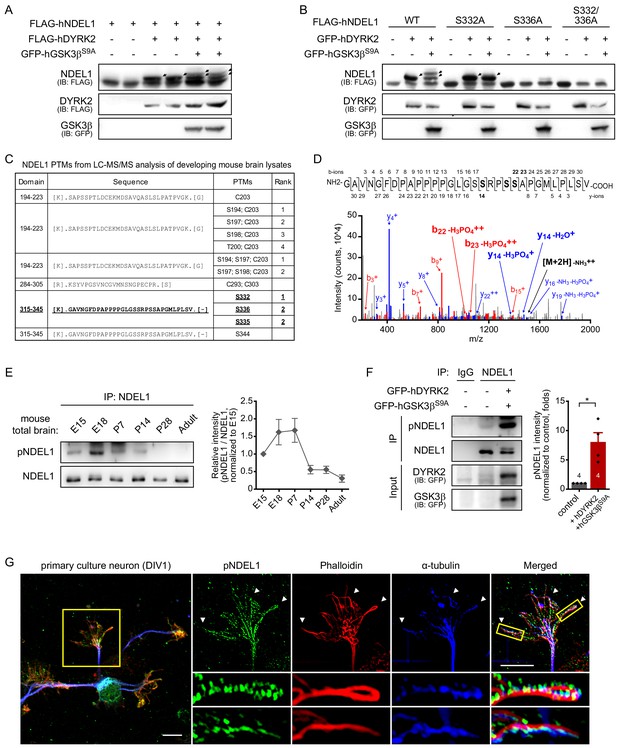
DYRK2 and GSK3β induce sequential phosphorylation of NDEL1 at S336 and S332.
(A) Identification of responsible kinases for NDEL1 phosphorylation. DYRK2, one of the positive candidates from human kinome screening for NDEL1 phosphorylation, and GSK3βS9A sequentially phosphorylated NDEL1. (B) Identification of NDEL1 S336 and S332 as target sites of DYRK2 and GSK3β. DYRK2 increased NDEL1 phosphorylation at S336 and GSK3βS9A additionally induced phosphorylation at S332. (C) List of NDEL1 PTMs identified from LC-MS/MS analysis of developing mouse brain lysates. Peptide containing S336 and S332 phosphorylation is indicated by bold and underlined letters. (D) MS/MS spectrum for the phosphorylated fragments of NDEL1 peptide including S332 and S336 residues. The sequence of the peptide (aa 315–345) and all detected fragment ions are shown above. The b- and y-ions annotated in the spectrum include the sizes of y14 and b22 ions indicative of phosphorylation at S332 or S336. (E) Phosphorylation levels of endogenous NDEL1 S336/S332 in the developing mouse brain. Amount of lysates subjected to IP was normalized by NDEL1 protein levels. N = 7 for E15, P7, P14, P28, and adult brain lysates and N = 6 for E18. All results are presented as means ± SEM. (F) Increased endogenous NDEL1 phosphorylation by DYRK2 and GSK3β. Transfected HEK293 cell lysates were IPed with pan-NDEL1 antibody followed by western blot with anti-pNDEL1 antibody. Over-expression of DYRK2 and GSK3βS9A increased the endogenous NDEL1 S336/S332 phosphorylation. The number of samples is shown at the bottom of the bar of the graph. All results are presented as means ± SEM. *p<0.05 from Student’s t-test. (G) Endogenous NDEL1 S336/S332 phosphorylation detected at the growth cone of the primary cultured mouse hippocampal neuron. Anti-pNDEL1 antibody signal was enriched at the growth cone and colocalized with both phalloidin and α-tubulin (indicated by arrowheads). Magnified confocal images show the strong overlap between pNDEL1, phalloidin, and α-tubulin. The scale bar represents 10 μm. See also Figure 1—figure supplements 1, 2 and 3 and Figure 1—source data 1 and 2.
-
Figure 1—source data 1
Source data for quantitation of endogenous pNDEL1 in developing mouse brain lysates.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Source data for quantitation of endogenous pNDEL1 with kinases over-expression.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig1-data2-v2.xlsx

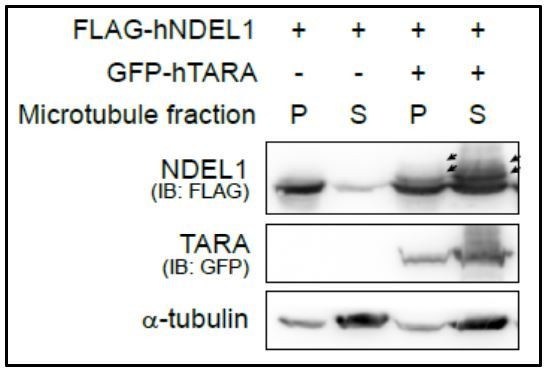
Figure of kinome library screening results for NDEL1 phosphorylation CCSB-Broad.
Human Kinase ORF collection screening to find responsible kinases for NDEL1 S336 phosphorylation. Among 301 serine/threonine kinases, 218 kinases were tested at least twice. TARA was used as a positive control to induce NDEL1 phosphorylation.
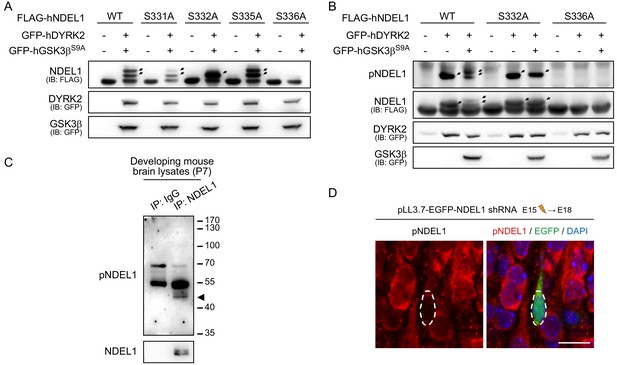
Figure related to Figure 1.
(A) Validation of target sites for NDEL1 phosphorylation by DYRK2-GSK3β. S332A and S336A mutations, in contrast to S331A and S335A mutations, changed the pattern of phosphorylation signals. (B) Characterization of anti-pNDEL1 antibody for immunoblotting. Anti-pNDEL1 antibody specifically detected NDEL1 phosphorylation at S332 and S336. (C) Detection of endogenous NDEL1 S336/S332 phosphorylation by anti-pNDEL1 antibody. Anti-pNDEL1 antibody detected the endogenous NDEL1 phosphorylation from P7 mouse brain lysates prepared. The arrowhead indicates NDEL1/pNDEL1 bands. (D) Validation of anti-pNDEL1 antibody for staining via immunohistochemistry. NDEL1 shRNA construct co-expressing EGFP marker was transfected via in utero electroporation to E15 ICR mouse and the brain was prepared at E18 and subjected to immunohistochemistry. NDEL1 knockdown significantly diminished the anti-pNDEL1 antibody signal. The scale bar represents 10 μm.

Table for the list of NDEL1 PTMs identified from LC-MS/MS analysis of developing mouse brain lysates.
PTMs also identified from NDEL1-TARA expressing HEK293 cell lysate are indicated by bold and underlined letters.
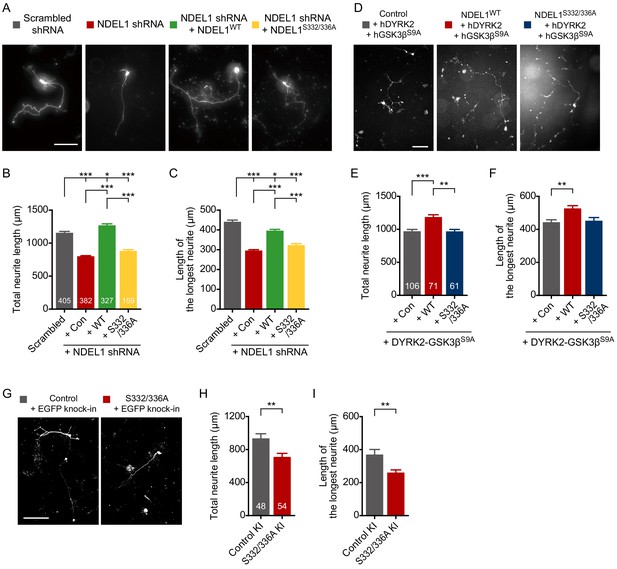
Phosphorylation of NDEL1 S336/S332 increases neurite outgrowth of primary cultured hippocampal neurons.
(A–C) Suppression of NDEL1 S336/S332 phosphorylation inhibited neurite outgrowth of hippocampal neurons. Each DNA construct was transfected at 9 hr after neuronal culture and neurites of transfected neurons were analyzed at DIV 3. All of NDEL1 over-expressing constructs here contain an shRNA-resistant mutation. (A) Representative images of transfected neurons. The total neurite length (B) and the longest neurite length (C) were measured by using ImageJ software. (D–F) NDEL1 S336/S332 phosphorylation induced by DYRK2-GSK3β co-expression is enough to increase neurite outgrowth of hippocampal neurons. Each DNA construct was transfected at DIV 2 and neurites of transfected neurons were analyzed at DIV 5. (D) Representative images of transfected neurons. The total neurite length (E) and the longest neurite length (F) were measured by using ImageJ software. (G–I) KI of phospho-deficient mutations resulted in neurite outgrowth defects. DNA constructs for KI were transfected at DIV 1 and neurites of transfected neurons were analyzed at DIV 4. (G) Representative images of transfected neurons. The total neurite length (H) and the longest neurite length (I) were measured by using ImageJ software. Each sample number is shown at the bottom of the bar of the graph. Scale bars represent 100 μm. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from one-way ANOVA for (B), (C), (E), and (F) and Student’s t-test for (H) and (I). All experiments were independently repeated at least three times. See also Figure 2—figure supplements 1 and 2 and Figure 2—source data 1, 2 and 3.
-
Figure 2—source data 1
Source data for axon/dendrite outgrowth of NDEL1 knockdown and rescue groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Source data for axon/dendrite outgrowth of NDEL1 and kinases over-expression groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Source data for axon/dendrite outgrowth assay of NDEL1 S332/336A KI.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig2-data3-v2.xlsx
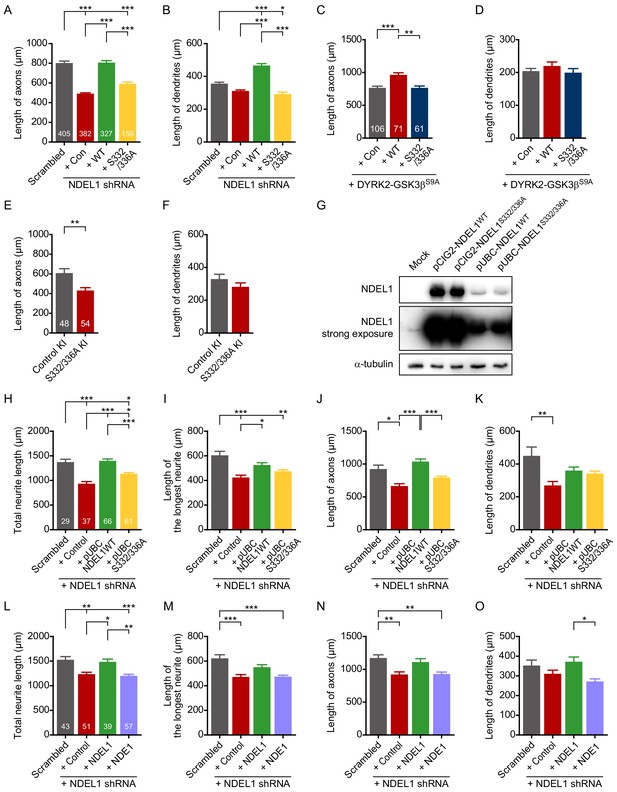
Figure for additional data on axon/dendrite outgrowth.
(A–B) Additional data related to Figure 2A–C. The axonal neurite length (A) and the dendritic neurite length (B) were measured by using ImageJ software. (C–D) Additional data related to Figure 2D–F. The axonal neurite length (C) and the dendritic neurite length (D) were measured by using ImageJ software. (E–F) Additional data related to Figure 2G–I. The axonal neurite length (E) and the dendritic neurite length (F) were measured by ImageJ software. (G) Comparison of NDEL1 expression under either CAG promoter or UBC promoter. Expression levels of NDEL1WT and NDEL1S332/336A were substantially lower under UBC promoter. (H–K) Moderate expression of NDEL1 rescue the decreased axon/dendrite outgrowth caused by NDEL1 knockdown. Each DNA construct was transfected at DIV 1 and neurites of transfected neurons were analyzed at DIV 4. The total neurite length (H), the longest neurite length (I), the axonal neurite length (J) and the dendritic neurite length (K) were measured by ImageJ software. (L–O) NDE1 failed to reverse the decreased axon/dendrite outgrowth caused by NDEL1 knockdown. Each DNA construct was transfected at DIV 1 and neurites of transfected neurons were analyzed at DIV 4. The total neurite length (L), the longest neurite length (M), the axonal neurite length (N) and the dendritic neurite length (O) were measured by ImageJ software. Each n number is shown at the bottom of the bar of the graph. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from one-way ANOVA for (A–D) and (H–O) and Student’s t-test for (E) and (F). All experiments were independently repeated for at least three times.
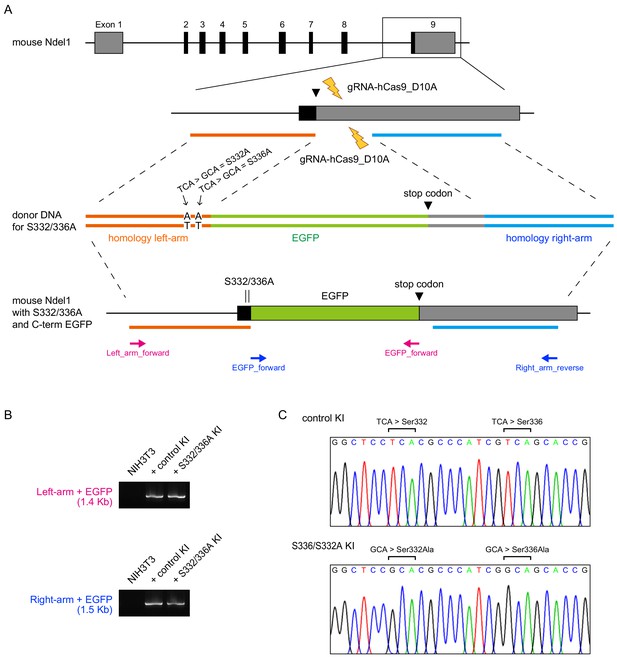
Schematic diagram of NDEL1 S332/336A KI strategy.
(A) A schematic diagram for the NDEL1 S332/336A KI targeting mouse Ndel1 locus. (B–C) Validation of S332/336A KI. NIH3T3 cells were transfected with hCas9_D10A, two guide RNAs, and donor plasmid DNA and EGFP-expressing cells. At 72 hr after transfection, genomic DNA was extracted and subjected to PCR with primer sets shown in (A). Acrylamide gel electrophoresis (B) and sequencing (C) analysis confirmed the successful KI of NDEL1 S332/336A or WT with EGFP.
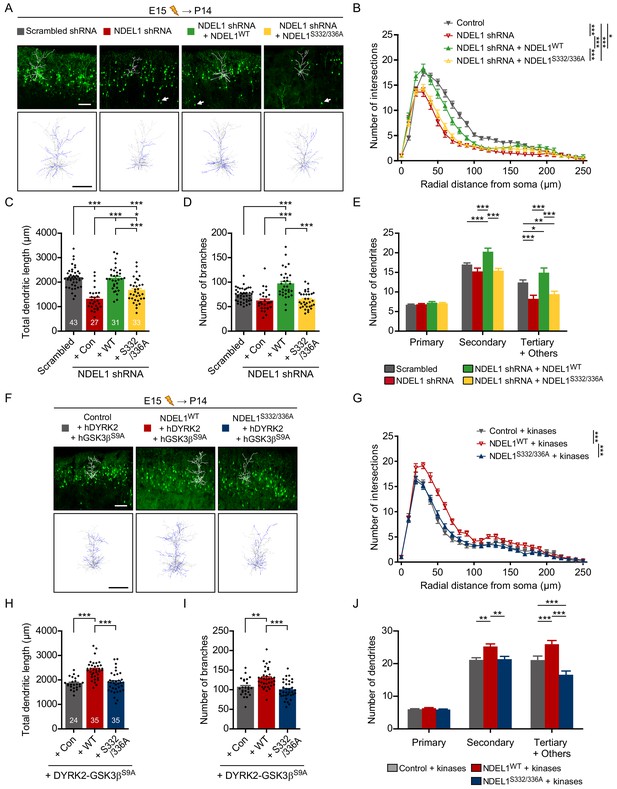
Phosphorylation of NDEL1 S336/S332 regulates dendritic arborization of cortical pyramidal neurons.
(A–E) Suppression of NDEL1 S336/S332 phosphorylation disrupted dendritic arborization of layer II/III pyramidal neurons. All constructs were electroporated in utero to E15 mouse brain and then P14 brain was subjected for analysis. All of NDEL1 over-expressing constructs here contain an shRNA-resistant mutation. (A) Representative images of the brain slices with the tracked neuron (above) and the overlapped dendritic structures of five independent neurons (bottom). Sholl analysis plots (B), the total length of dendrites (C), the total number of branches (D), and the number of primary/secondary/tertiary dendrites (E) were analyzed by using Simple neurite tracer plug-in of ImageJ software. White arrowheads in (A) indicate neurons with migration defect. (F–J) NDEL1 S336/S332 phosphorylation induced by DYRK2-GSK3β kinases increased dendritic arborization of layer II/III pyramidal neurons. All constructs were electroporated in utero to E15 mouse brain and P14 brain was subjected for analysis. (F) Representative images of the brain slices with the tracked neuron (above) and the overlapped dendritic structures of five independent neurons (bottom). Sholl analysis plots (G), the total length of dendrites (H), the total number of branches (I), and the number of primary/secondary/tertiary dendrites (J) were analyzed by using Simple neurite tracer plug-in of ImageJ software. Each n number is shown at the bottom of the bar of the graph. Scale bars represent 100 μm. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from one-way ANOVA for (C), (D), (H), and (I) and two-way ANOVA for (B), (E), (G), and (J). All brain samples for each group were collected from offspring of at least three independent in utero electroporation surgeries. See also Figure 3—figure supplement 1, Figure 3—videos 1 and 2, and Figure 3—source data 1 and 2.
-
Figure 3—source data 1
Source data for dendritic arborization of NDEL1 knockdown and rescue groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for dendritic arborization of NDEL1 and kinases over-expression groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig3-data2-v2.xlsx
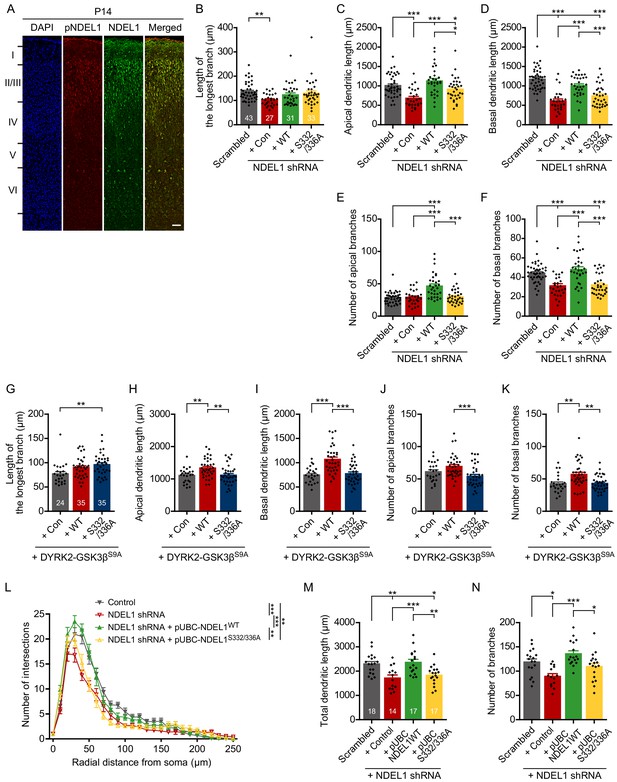
Figure for additional data on dendritic arborization.
(A) Immunohistochemistry for endogenous NDEL1 and its S336/S332 phosphorylation from the P14 mouse brain slice. Both NDEL1 and pNDEL1 were highly expressed from neurons in cortical layer II/III. (B–F) Additional data related to Figure 3A–E. The longest branch length (B), the apical dendrite length (C), the basal dendrite length (D), the number of apical branches (E), and the number of basal branches (F) were measured by using Simple neurite tracer plug-in of ImageJ software. (G–K) Additional data related to Figure 3F–J. The longest branch length (G), the length of apical dendrites (H), the length of basal dendrites (I), the number of apical branches (J), and the number of basal branches (K) were measured by using Simple neurite tracer plug-in of ImageJ software. (L–N) NDEL1 expression under UBC promoter was sufficient to rescue the decreased dendritic arborization of layer II/III pyramidal neurons caused by NDEL1 knockdown. All constructs were electroporated in utero into E15 mouse brain and P14 brains were subjected to analysis. Sholl analysis plots (L), the total length of dendrites (M), and the total number of branches (N) were analyzed by using Simple neurite tracer plug-in of ImageJ software. Each n number is shown at the bottom of the bar of the graph. The scale bar at (A) represents 50 μm All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from one-way ANOVA for (B–K), (M), and (N) and two-way ANOVA for (L). All brain samples for each group were collected from at least three independent in utero electroporation surgeries.
Video for 3D reconstructions of the dendritic structures of representative neurons, related to Figure 3A.
Video for 3D reconstructions of the dendritic structures of representative neurons, related to Figure 3F.

TARA recruits DYRK2 and GSK3β to induce sequential phosphorylation of NDEL1 S336/S332.
(A) In vitro phosphatase assay. Calf intestinal alkaline phosphatase (CIP) was treated in vitro after IP of FLAG-tagged NDEL1. Additional bands of NDEL1 disappeared upon CIP treatment indicating that these additional bands are caused by the multiple phosphorylation. (B) MS/MS spectrum for the phosphorylated fragments of NDEL1 peptide including S332 and S336 residues. NDEL1 proteins from HEK293 cells over-expressing NDEL1 and TARA were subjected to LC-MS/MS analysis. The sequence of the peptide (aa 306–345) and all detected fragment ions are shown above. The b- and y-ions annotated in the spectrum include the sizes of y12 and b33 ions indicative of a single phosphorylation at either S335 or S336. (C) TARA-induced NDEL1 phosphorylation S336 and S332. TARA increased sequential phosphorylation of NDEL1, first at S336 followed by S332. (D) The protein-protein interaction between DYRK2 and TARA. Endogenous DYRK2 was co-precipitated by IP of endogenous TARA from HEK293 cell lysates. Rabbit IgG was used as a negative control. At input lanes, 1% and 10% of lysates were loaded for anti-DYRK2 and anti-TARA blots, respectively. (E) The protein-protein interaction between GSK3β and TARA. Endogenous TARA was co-precipitated by IP of endogenous GSK3β proteins from HEK293 cell lysates. Mouse IgG was used as a negative control. (F) Co-IP among DYRK2, GSK3β, and TARA. Over-expression of TARA increased an amount of GSK3β proteins co-precipitated by IP of DYRK2, implying that TARA scaffolds these kinases to form a DYRK2-GSK3β-TARA tripartite complex. (G) Colocalization of DYRK2, GSK3β, and TARA in mouse hippocampal neurons. GFP-hDYRK2, FLAG-hGSK3β, and MYC-hTARA colocalized at the soma (above) and the growth cone (bottom) regions. See also Figure 4—figure supplements 1 and 2. The following figure supplement is available for Figure 4.
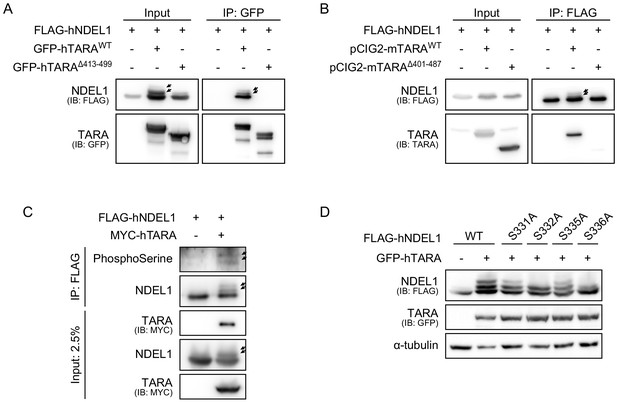
Figure related to Figure 4.
(A) Western blot band shift of NDEL1 depending on the interaction with hTARA. GFP-tagged hTARAWT or hTARAΔ413-499 was IPed with the anti-GFP antibody. Immunoblotting detected multiple bands of NDEL1 only from hTARAWT co-expressed lanes and all of these multiple bands were co-IPed with hTARAWT. (B) Western blot band shift of NDEL1 depending on the interaction with mTARA. mTARAΔ401-487 was absent for NDEL1 interaction and NDEL1 phosphorylation induction. (C) PhosphoSerine antibody blot for the increased NDEL1 phosphorylation by TARA over-expression. (D) Validation of target sites for TARA-dependent NDEL1 phosphorylation. S332A and S336A mutations, in contrast to S331A and S335A mutations, changed the pattern of phosphorylation signals.
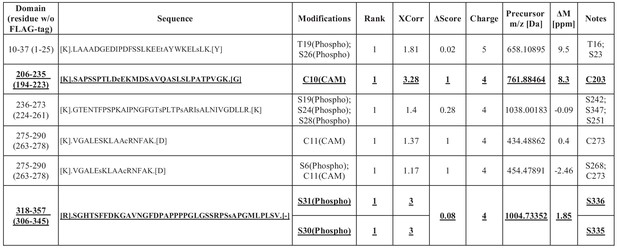
Table for the list of NDEL1 PTMs identified from LC-MS/MS analysis of lysates of HEK293 cells transfected with FLAG-hNDEL1 and GFP-hTARA.
PTMs also identified from developing mouse brain lysates are indicated by bold and underlined letters.
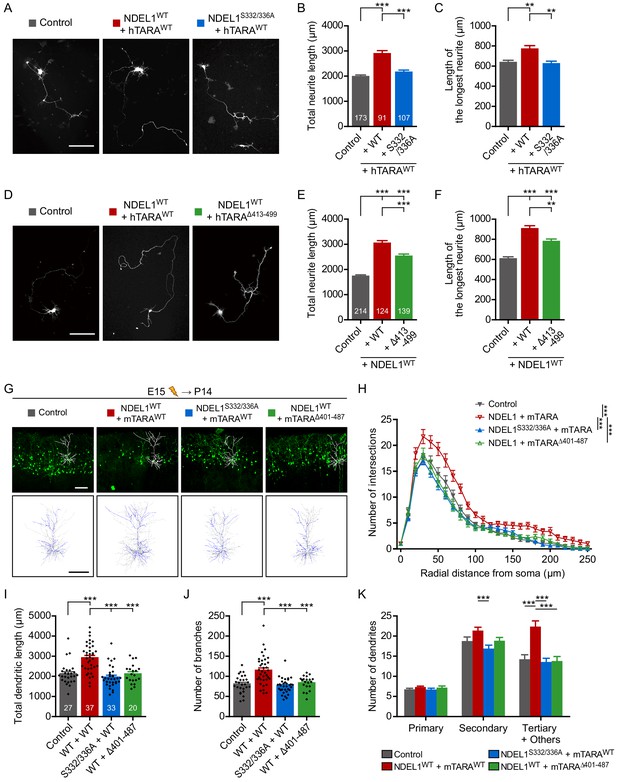
NDEL1 S336/S332 phosphorylation induced by TARA increases neurite outgrowth and dendritic arborization.
(A–C) Induction of NDEL1 S336/S332 phosphorylation by TARA increased neurite outgrowth. Each DNA construct was transfected at DIV 2 and neurites of transfected neurons were analyzed at DIV 5. (A) Representative images of transfected neurons. The total neurite length (B) and the longest neurite length (C) were measured by using ImageJ software. (D–F) TARAΔ413-499 could not increase neurite outgrowth. Each DNA construct was transfected at DIV 2 and neurites of transfected neurons were analyzed at DIV 5. (D) Representative images of transfected neurons. The total neurite length (E) and the longest neurite length (F) were measured by using ImageJ software. (G–K) Up-regulated NDEL1 S336/S332 phosphorylation increased dendritic arborization of layer II/III pyramidal neurons. All constructs were electroporated in utero to E15 mouse brain and P14 brain was subjected for analysis. (G) Representative images of the brain slices with the tracked neuron (above) and the overlapped dendritic structures of five independent neurons (bottom). Sholl analysis plots (H), the total length of dendrites (I), the total number of branches (J), and the number of primary/secondary/tertiary dendrites (K) were analyzed by using Simple neurite tracer plug-in of ImageJ software. Each n number is shown at the bottom of the bar of the graph. Scale bars represent 100 μm. All results are presented as means ± SEM. **p<0.01 and ***p<0.001 from one-way ANOVA for (B), (C), (E), (F), (I), and (J) and two-way ANOVA for (H) and (K). All neurite outgrowth experiments for (A–C) and (D–F) were independently repeated for at least three times. All brain samples for each group of (G–K) were collected from offspring of at least three independent in utero electroporation surgeries. See also Figure 5—figure supplement 1, Figure 5—video 1, and Figure 5—source data 1 and 2.
-
Figure 5—source data 1
Source data for axon/dendrite outgrowth of NDEL1 and TARA over-expression groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Source data for dendritic arborization of NDEL1 and TARA over-expression groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig5-data2-v2.xlsx
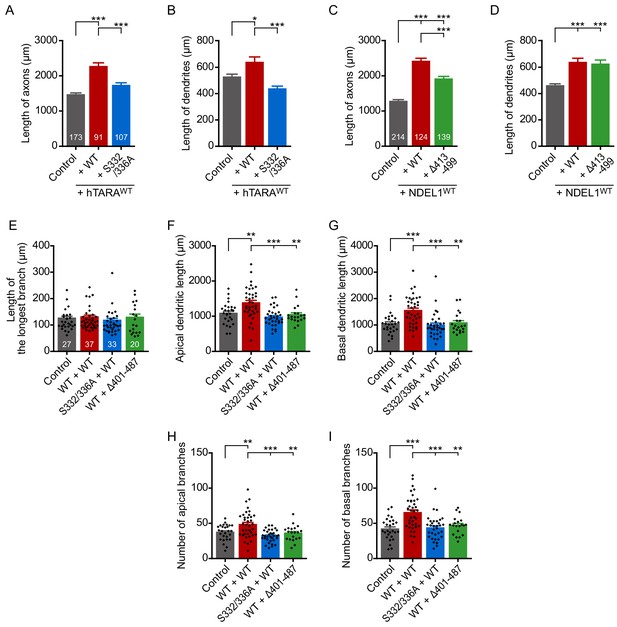
Figure for additional data on axon/dendrite outgrowth and dendritic arborization by TARA co-expression.
(A–B) Additional data related to Figure 5A–C. The axonal neurite length (A) and the dendritic neurite length (B) were measured by using ImageJ software. (C–D) Additional data related to Figure 5D–F. The axonal neurite length (C) and the dendritic neurite length (D) were measured by using ImageJ software. (E–I) Additional data related to Figure 5G–K. The longest branch length (E), the length of apical dendrites (F), the length of basal dendrites (G), the number of apical branches (H), and the number of basal branches (I) were measured by using Simple neurite tracer plug-in of ImageJ software. Each n number is shown at the bottom of the bar of the graph. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from one-way ANOVA. All neurite outgrowth experiments for (A–B) and (C–D) were independently repeated for at least three times. All brain samples for each group of (E–I) were collected from offspring of at least three independent in utero electroporation surgeries.
Video for 3D reconstructions of the dendritic structures of representative neurons, related to Figure 5G.

Phosphorylation of NDEL1 S336/S332 modulates F-actin dynamics.
(A) F-actin fractionation assay of NDEL1 and TARA. NDEL1 S336/S332 phosphorylation induced by TARA was observed at both G-actin in the supernatant fraction (S) and F-actin in pellet fraction (P). (B–F) Decreased F-actin dynamics by suppression of NDEL1 S336/S332 phosphorylation. (B) Representative time-lapse images of FRAP assay to measure F-actin dynamics at differentiating SH-SY5Y cells expressing RFP-LifeAct. All of NDEL1 over-expressing constructs here contain an shRNA-resistant mutation. A yellow-dashed circle indicates region-of-interest used for bleaching. Bleaching was given by stimulating with 10% 568 nm laser for 10 s. (C) Time-dependent fluorescence recovery graph. Comparisons of the area under FRAP curves (D), the percentage of mobile F-actin fraction calculated by the amount of eventual fluorescence recovery (E), and the average half-max (t1/2) of RFP-LifeAct fluorescence recovery (F). NDEL1 knockdown cells had decreased fluorescence recovery meaning more immobile fraction of F-actin and could not be rescued by NDEL1S332/336A implying its phosphorylation dependency. Each n number is shown at the bottom of the bar of the graph. The scale bar at (B) represents 10 μm. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from two-way ANOVA for (C) and one-way ANOVA for (D–F). See also Figure 6—figure supplements 1, 2 and 3, Figure 6—video 1, and Figure 6—source data 1.
-
Figure 6—source data 1
Source data for F-actin FRAP assay of NDEL1 knockdown and rescue groups.
- https://cdn.elifesciences.org/articles/50850/elife-50850-fig6-data1-v2.xlsx

Figure of raw data for F-actin FRAP assay plots and results for Microtubule FRAP assay.
(A–D) Raw data for FRAP assay plots (gray) with average (each color) of each group. RFP-LifeAct was co-expressed with scrambled shRNA (A), NDEL1 shRNA (B), NDEL1 shRNA with shRNA-resistant NDEL1WT (C), and NDEL1 shRNA with shRNA-resistant NDEL1S332/336A (D) in SH-SY5Y cells followed by differentiation and FRAP assay. (E–H) Microtubule dynamics analyzed from time-lapse imaging of FRAP assay with mCherry-α-tubulin. All NDEL1 constructs contain an shRNA-resistant mutation. (E) Time-dependent fluorescence recovery graph. Comparisons of the area under FRAP curves (F), the percentage of motile microtubule fraction calculated by the amount of eventual fluorescence recovery (G), and the half-max (t1/2) of mCherry-α-tubulin fluorescence recovery (H). (I–L) Raw data for FRAP assay plots (gray) with average (each color) of each group. mCherry-α-tubulin was co-expressed with scrambled shRNA (I), NDEL1 shRNA (J), NDEL1 shRNA with shRNA-resistant NDEL1WT (K), and NDEL1 shRNA with shRNA-resistant NDEL1S332/336A (L) in SH-SY5Y cells followed by differentiation and FRAP assay. Each n number is shown at the bottom of the bar of the graph. All results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from two-way ANOVA for (E) and one-way ANOVA for (F–H).
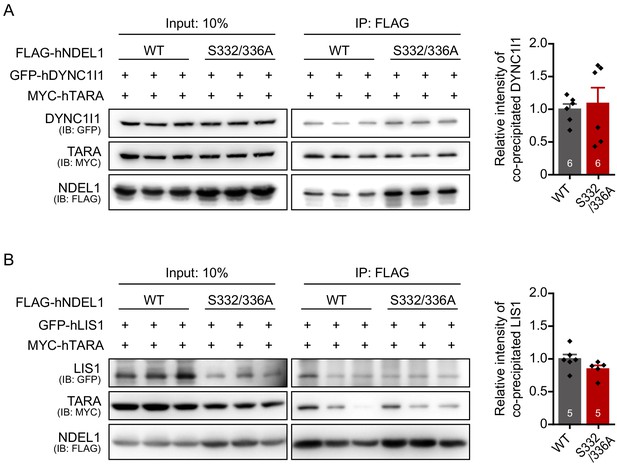
Figure for additional data on the effect of S336/S332 phosphorylation on the interaction of NDEL1 with DYNC1I1 and LIS1.
(A) The protein-protein interaction between NDEL1 and dynein intermediate chain. GFP-hDYNC1I1 was co-IPed with FLAG-NDEL1WT or FLAG-NDEL1S332/336A. No significant difference between wild-type and phospho-deficient mutant was detected. (B) The protein-protein interaction between NDEL1 and LIS1. GFP-hLIS1 was co-IPed with FLAG-NDEL1WT or FLAG-NDEL1S332/336A. No significant difference between wild-type and phospho-deficient mutant was detected. Each n number is shown at the bottom of the bar of the graph. All results are presented as means ± SEM.
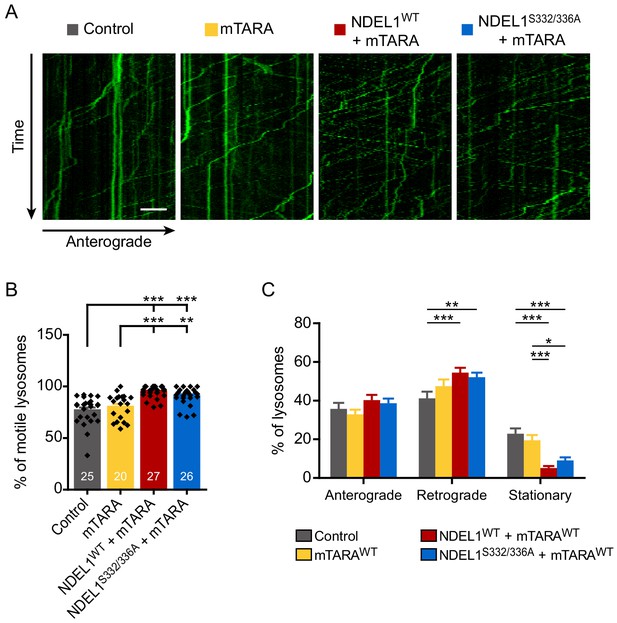
Figure for additional data on the effect of S336/S332 phosphorylation on the lysosomal trafficking.
(A–C) Lysosomal trafficking analyzed by time-lapse imaging of primary hippocampal neurons expressing GFP-LAMP1. (A) Representative kymographs. (B) Percentage of motile lysosomes. (C) Percentage of anterograde movement, retrograde movement, and stationary status of lysosomes. Each n number is shown at the bottom of the bar in the graph. The scale bar in (A) represents 10 μm. Results are presented as means ± SEM. *p<0.05, **p<0.01, and ***p<0.001 from one-way ANOVA for (B) and two-way ANOVA for (C).
Video for the time-lapse imaging series of the FRAP assay for representative cells, related to Figure 6B.
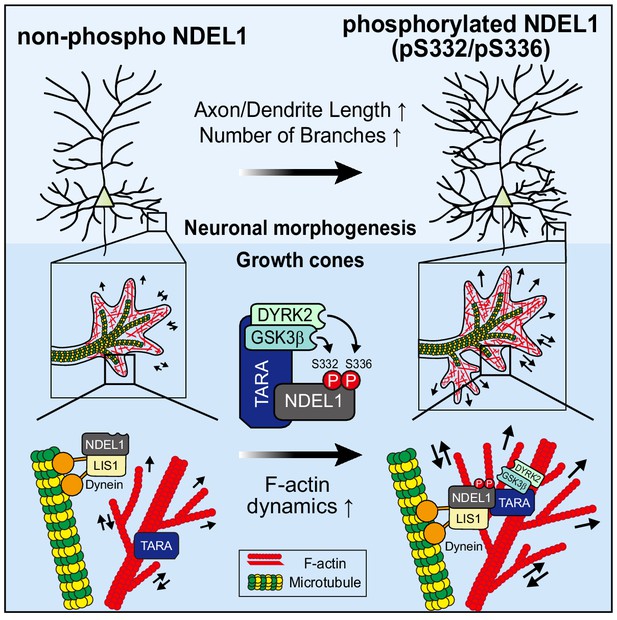
A model for the mechanism by which NDEL1 S336/S332 phosphorylation regulates neuronal morphogenesis.
The phosphorylation of NDEL1 at S336 by DYRK2 primes S332 phosphorylation by GSK3β. TARA mediates this process by recruiting DYRK2 and GSK3β to NDEL1 and forming a tripartite complex in association with F-actin. The phosphorylated NDEL1 enhances F-actin dynamics at the interface with microtubule cytoskeleton in growth cones, thereby facilitating axon/dendrite length and neuronal arborization.
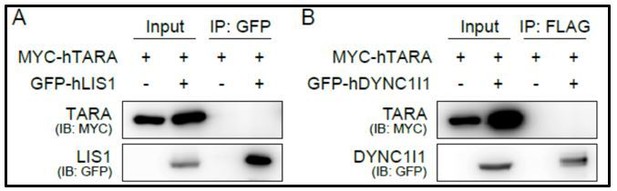
coIP results to examine interactions between TARA and LIS1 or TARA and dynein intermediate chain.
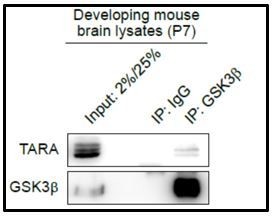
The protein-protein interaction between endogenous GSK3β and endogenous TARA in the developing mouse brain.
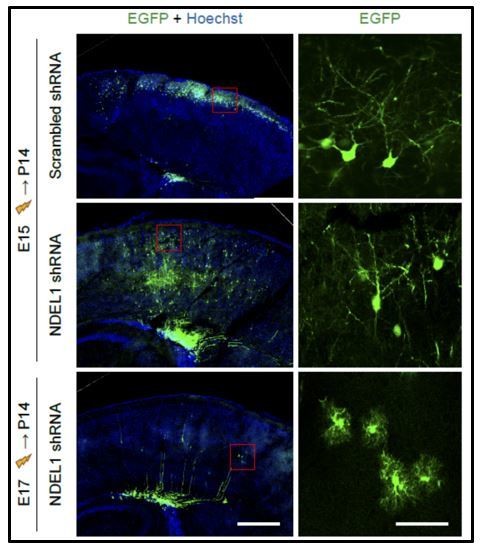
Comparison of cell-types affected by in utero electroporation at either E15 or E17.
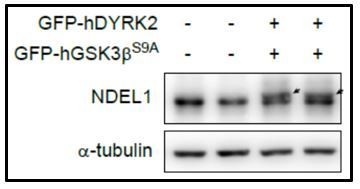
Existence of endogenous NDEL1 band shift induced by DYRK2-GSK3β over-expression.
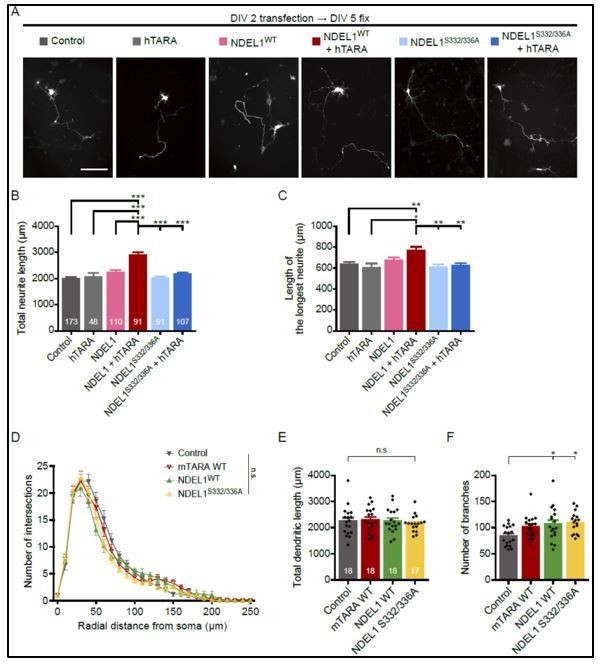
Neuronal morphogenesis with single over-expression of TARA, NDEL1WT, or NDEL1S332/336A.
Additional files
-
Supplementary file 1
Key resources table.
- https://cdn.elifesciences.org/articles/50850/elife-50850-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50850/elife-50850-transrepform-v2.docx