Endogenous siRNAs promote proteostasis and longevity in germline-less Caenorhabditis elegans
Figures
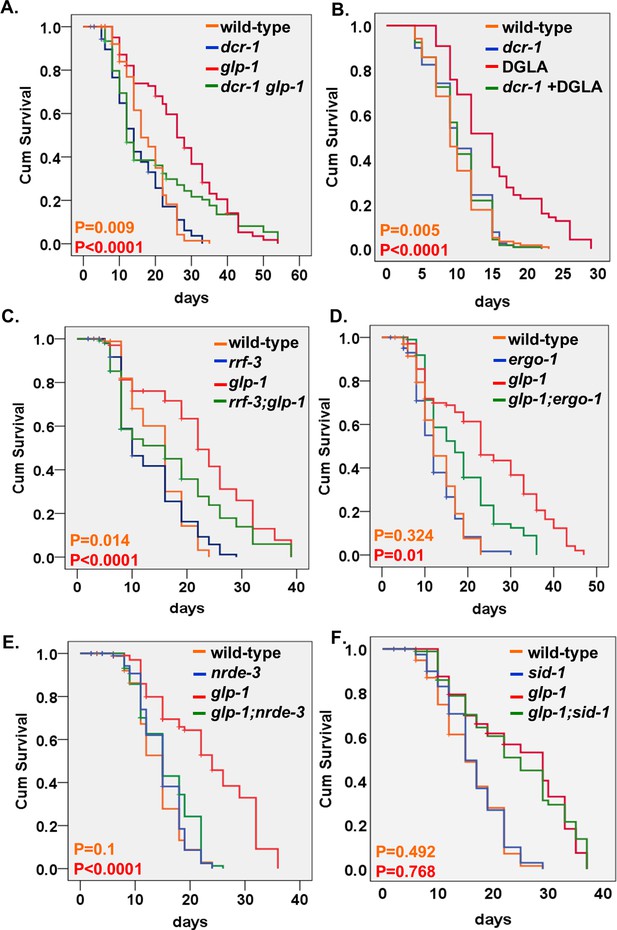
Endo-siRNAs are required for the longevity of GSC(-) animals.
(A,C–E) Impairment of the endo-siRNA pathway by dcr-1(mg375), rrf-3(pk1426), ergo-1(gg98), or nrde-3(gg66) compromises the longevity of glp-1 mutants. (B) The longevity conferred by germline depletion via DGLA supplementation is perturbed by the mg375 mutation in the dcr-1 helicase domain. (F) Impairment of the dsRNA channel sid-1(pk3321) does not compromise the longevity of glp-1 mutants. Breslow (Generalized Wilcoxon) P-values between endo-siRNA mutants and corresponding animals with intact endo-siRNA are indicated between GSC(+) animals (in orange) or between GSC(-) animals (in red). See Supplementary file 1. Note that the dcr-1 and nrde-3 mutants have not been outcrossed. This may affect their lifespan phenotypes.
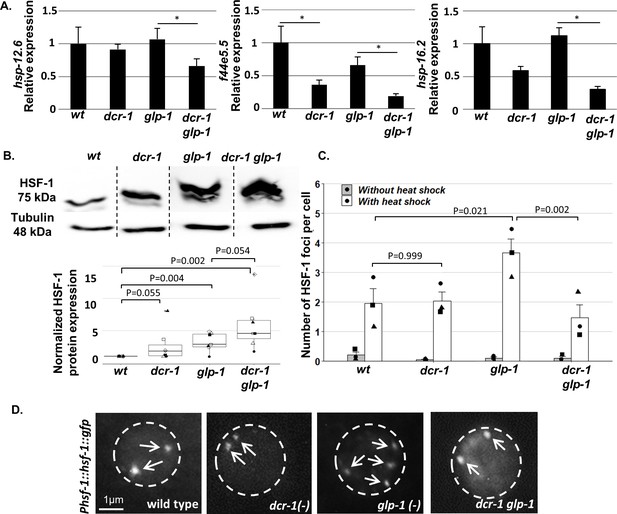
Endo-siRNAs are required for HSF-1 activation in GSC(-) animals.
(A) qRT-PCR of the indicated genes on day 1 of adulthood. Asterisks mark Student’s t-test values of p value<0.05 (N = 4). Note that the dcr-1 mutation reduced the transcript levels of all three chaperone genes in GSC(-) animals, consistent with the possibility that the activity of their upstream transcription factor HSF-1 has been compromised. Interestingly, the dcr-1 mutation also affected the levels of the f44e5.5 transcript in GSC(+) animals. Nevertheless, it did not significantly affect the transcript levels of the hsp-16.2 and hsp-12.6 chaperones in GSC(+) animals. Given that HSF-1 and some of its targets are expressed also in the germline (Ooi and Prahlad, 2017), to avoid biases due to the presence/absence of the germline tissue, comparisons should be made within GSC(-) animals or within GSC(+) animals (McCormick et al., 2012; Steinbaugh et al., 2015). See also Figure 2—figure supplement 1 and Supplementary file 2 and Supplementary file 3. (B) Representative western blot of endogenous HSF-1 in day one animals (upper panel) compared to loading control (lower panel). Boxplots represent the distribution of normalized HSF-1 levels per strain. Different shapes represent independent experiments (N = 7). P-values of One-Sample Test and One-Way ANOVA followed by Tukey's post hoc analysis across all seven experiments are indicated. See Supplementary file 8 for statistic details. (C) Bars represent mean of means of the number of HSF-1::GFP nuclear foci per hypodermal cell. Dots represent mean number of HSF-1 foci per cell with different shapes representing independent experiments. At least 140 cells per genotype were scored in a total of 3 independent experiments. P-values determined by One-Way ANOVA followed by Tukey's post hoc analysis are indicated. Data are presented as mean ± SEM. See Supplementary file 8 for statistic details. (D) Representative fluorescent micrographs of hypodermal cell nuclei in day three adults, harboring a single copy of the Phsf-1::hsf-1::gfp transgene upon heat shock stress. Exposures and contrast were adjusted for each picture independently to best emphasize foci amount. Nucleus boundaries are circled.
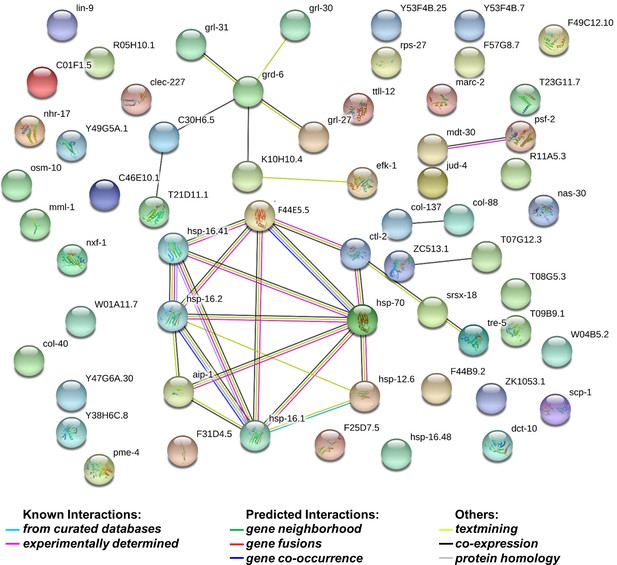
String analysis of 72 genes whose levels decreased by more than 1.5 fold in dcr-1 glp-1 double mutants compared to glp-1 mutant.

Endo-siRNAs are required for proteostasis maintenance in GSC(-) animals.
(A–B) Thermo-resistance was examined in age-synchronized animals subjected to heat shock (37°C, 9 hr) on day 2 of adulthood. Survival was assayed after 5 hr of recovery at 25°C (120 animals per treatment, N = 3). Asterisks mark Cochran-Mantel-Haenszel Test values of p<0.001. (C) Bars represent mean of the percentage of motile animals scored in age-synchronized day 5 Q35m or glp-1;Q35m animals (more than 45 animals per treatment, N = 3). Different shapes represent mean motility in independent experiments. Asterisks mark p-values<0.001 determined by One-Way ANOVA followed by Tukey's post hoc analysis. (D) Bars represent mean of means of the number of body bends per minute in age-synchronized day four unc-52(ts) animals. Animals were raised at 25°C till day 1 of adulthood, and shifted to the permissive temperature (15°C) thereafter. A total of 180 animals per strain were scored in three independent experiments. Different shapes represent mean motility in independent experiments. Asterisks mark Cochran-Mantel-Haenszel test values of p<0.001. Data are presented as mean ± SEM. See Supplementary file 8 for statistic details.

Inactivation of ptp-5.1 restores longevity in GSC(-) animals with perturbed endogenous siRNA.
(A) Scatter plot depicts comparisons of gene-by-gene siRNA counts from three paired glp-1 mutant and dcr-1 glp-1 double mutant samples. Gray- all genes. Orange- 132 genes whose levels increased by more than 1.5 folds in dcr-1 glp-1 double mutants compared to glp-1 single mutants at the transcriptome analysis. Purple- five overlapping genes between the transcriptome analysis and siRNA seq, which are candidate direct targets of endo-siRNA. Blue- ptp-5.1 (c24d10.1). See also Figure 4—figure supplement 1 and Supplementary files 4, 5, 6, and 7. (B) qRT-PCR of ptp-5.1 transcript on day 1 of adulthood. Asterisks mark Student’s t-test values of p value<0.05 of 4 independent experiments. Data are presented as mean ± SEM. The low levels of ptp-5.1 transcript in GSC(-) animals is consistent with the interpretation that in GSC(+) animals, most of the transcript is expressed in the germline. (C) Representative fluorescent images of dcr-1 glp-1 transgenic animals expressing an extrachromosomal array of a translational reporter fused to the ptp-5.1 gene, driven by ptp-5.1 upstream sequences. Note that the reporter is only detected in a fraction of the animals. Animals that expressed the reporter displayed a clear fluorescent signal specifically in two adjacent cells in the mid-intestine (see Figure 4—figure supplement 2B). A similar expression pattern of the transgene was observed in all genetic backgrounds (see Figure 4—figure supplement 2). Bars represent mean of the percentage of animals expressing the ptp-5.1::gfp transgene in the intestine. At least 250 animals per strain were scored in four independent experiments. Different colors represent independent experiments. Cochran-Mantel-Haenszel test P-values are indicated. Asterisks mark p<0.001. Somatic expression of the transgene was detected in all backgrounds in a fraction of the animals. Inactivation of the endo-siRNA pathway by the dcr-1 mutation increased the fraction of the animals expressing the reporter in their mid-intestine. (D) ptp-5.1(tm6122) extended the lifespan of dcr-1 glp-1 double mutants. Breslow (Generalized Wilcoxon) P-values for each mutant vs. the mutant; ptp-5.1(tm6122) double mutant are indicated. See Supplementary file 1 for additional lifespan data.
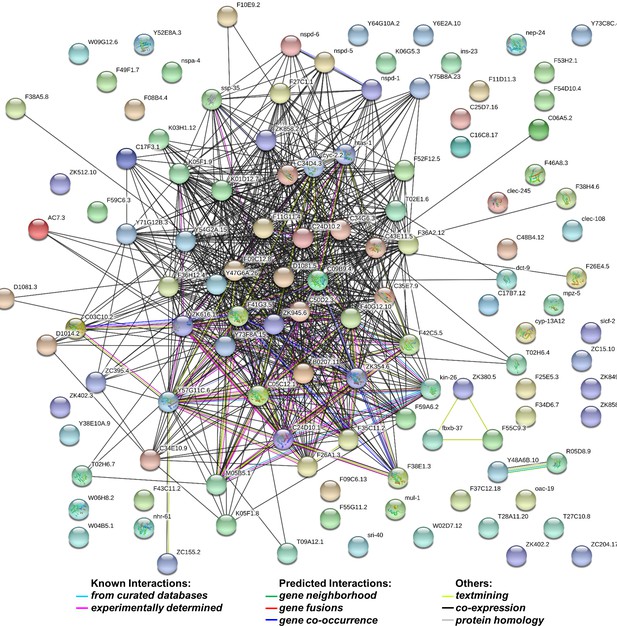
String analysis of 132 genes whose levels increased by more than 1.5 fold in dcr-1 glp-1 double mutants compared to glp-1 mutant.
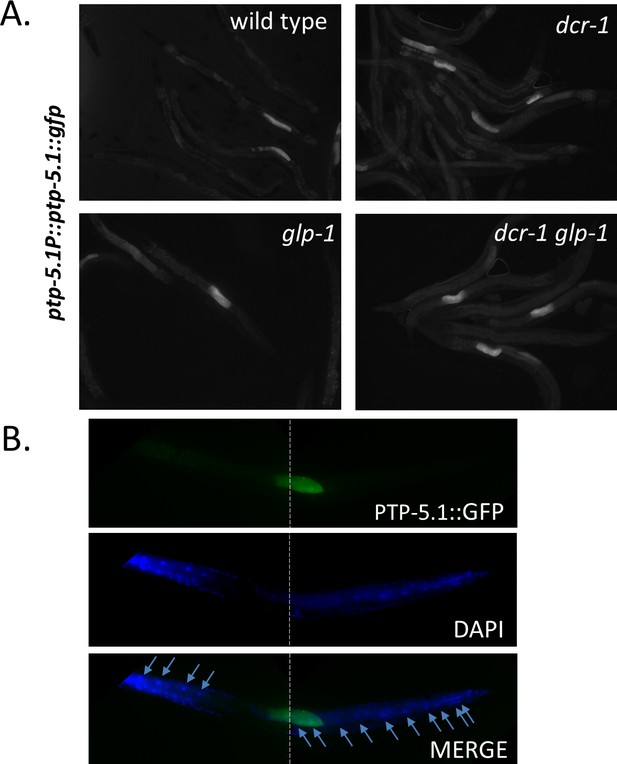
The ptp-5.1::gfp transgene is expressed in the intestine of a fraction of the animals.
(A) Only a fraction of the transgenic animals of the indicated genotypes express the transgene. However, all animals that express the transgene express it to a similar extent and almost always in two adjacent intestinal cells in the mid-body of the animal. The reporter is expressed both in the cytoplasm and in the nucleus. (B) Representative X200 images of Day one dcr-1 glp-1 transgenic animal stained with the nuclear dye DAPI while maintaining the fluorescence of the GFP. DAPI staining and GFP fluorescence were individually captured and overlaid in the merged image, to demonstrate the localization of the GFP-expressing cells relative to the intestinal nuclei. Anterior part of the animals is to te left. Arrows indicate DAPI-stained intestinal nuclei.
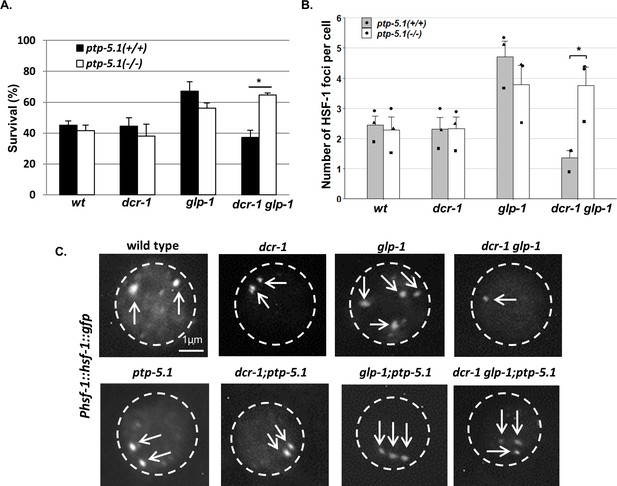
Inactivation of ptp-5.1 improves proteostasis in GSC(-) animals with perturbed endogenous siRNA.
(A) Thermo-resistance of age-synchronized animals subjected to heat shock (37°C, 9 hr) on day 2 of adulthood upon 5 hr of recovery at 25°C (120 animals per treatment, N = 3). Asterisks mark Cochran-Mantel-Haenszel test values of p<0.001. (B) Bars represent mean of mean number of HSF-1::GFP nuclear foci per hypodermal cell. At least 140 cells per genotype were scored in a total of 3 independent experiments. Dots represent mean number of HSF-1 foci per cell with different shapes representing independent experiments. Asterisk marks p-value<0.05 determined by One-Way ANOVA followed by Tukey's post hoc analysis. Data are presented as mean ± SEM. See Supplementary file 8 for statistic details. (C) Fluorescence micrographs of representative hypodermal cells in day three adults, harboring a single copy of the Phsf-1::hsf-1::gfp transgene upon exposure to heat shock. Nuclear boundaries are circled. Exposures and contrast were adjusted for each picture independently to best emphasize foci amount.
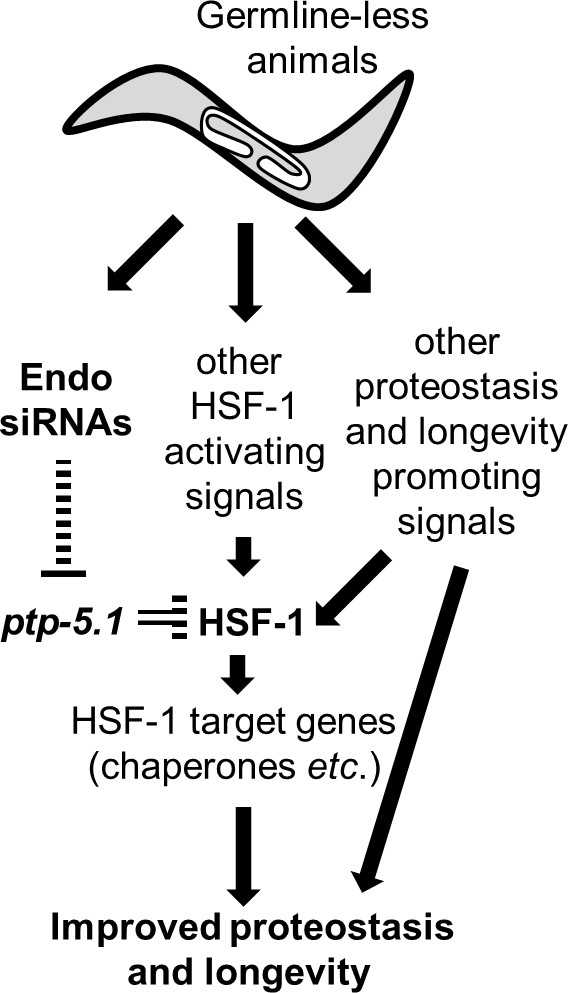
Endo-siRNAs improve proteostasis and promote longevity of GSC(-) animals by enabling HSF-1 activation.
Model: Germline-less animals extensively remodel their transcriptome to promote longevity and proteostasis. HSF-1 is one of the central transcription factors that transcribe proteostasis and longevity-promoting genes. We find that endo-siRNAs are critical for HSF-1 activity in GSC(-) animals, and consequently for their longevity and improved proteostasis. These endo-siRNAs are important because they indirectly limit the level of the tyrosine phosphatase ptp-5.1. The inhibition of this tyrosine phosphatase is critical for HSF-1 activation in proteostasis challenging settings such as heat-shock and aging. Whereas the release of HSF-1 from ptp-5.1 inhibition is required for the proteostasis and longevity benefits in GSC(-) animals, it is not sufficient. To achieve effective remodeling of the proteostasis and longevity promoting networks, germline removal must coordinate between the removal of ptp-5.1-dependent inhibition of HSF-1 and additional cellular events that promote HSF-1 activity such as reducing the repressive chromatin marks at HSF1-regulated stress-responsive genes.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (C. elegans) | N2 | Caenorhabditis Genetics Center | Wild Type | |
| Strain, strain background (C. elegans) | CF1903 | Caenorhabditis Genetics Center | glp-1(e2144) | outcrossed three times in C Kenyon's lab |
| Strain, strain background (C. elegans) | YY470 | Caenorhabditis Genetics Center | dcr-1(mg375) | an outcrossed version of YY11 dcr-1(m9375) |
| Strain, strain background (C. elegans) | SHK77 | This paper | dcr-1(mg375) glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | CF3152 | Cynthia Kenyon lab | rrf-3(pk1426) | outcrossed three times in C Kenyon's lab |
| Strain, strain background (C. elegans) | SHK55 | This paper | rrf-3(pk1426);glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK80 | This paper | ergo-1(gg98) | Strain outcrossed two times in S Henis-Korenblit lab. Total eight outcrosses |
| Strain, strain background (C. elegans) | SHK87 | This paper | glp-1(e2144);ergo-1(gg98) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | YY158 | Caenorhabditis Genetics Center | nrde-3(gg66) | |
| Strain, strain background (C. elegans) | SHK328 | This paper | glp-1(e2144); nrde-3(gg66) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK53 | This paper | sid-1(pk3321) | Strain outcrossed four times in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK56 | This paper | glp-1(e2144);sid-1(pk3321) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | OG497 | Caenorhabditis Genetics Center | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+] | |
| Strain, strain background (C. elegans) | SHK299 | This paper | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+];glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK300 | This paper | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+];dcr-1(mg375) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK301 | This paper | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+];dcr-1(mg375) glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | AM140 | Caenorhabditis Genetics Center | rmIs132 [unc-54p::Q35::YFP] | |
| Strain, strain background (C. elegans) | SHK409 | This paper | rmIs132 [unc-54p::Q35::YFP];glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK412 | This paper | rmIs132 [unc-54p::Q35::YFP];dcr-1 (mg375) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK410 | This paper | rmIs132 [unc-54p::Q35::YFP];dcr-1 (mg375) glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | HE250 | Caenorhabditis Genetics Center | unc-52(e669su250) | |
| Strain, strain background (C. elegans) | SHK574 | This paper | unc-52(e669su250);glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK575 | This paper | unc-52(e669su250);dcr-1(mg375) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK576 | This paper | unc-52(e669su250);dcr-1(mg375) glp-1(e2144) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK415 | This paper | ptp-5.1(tm6122) | Strain outcrossed three times in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK470 | This paper | glp-1(e2144);ptp-5.1(tm6122) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK469 | This paper | dcr-1(mg375);ptp-5.1(tm6122) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK471 | This paper | dcr-1(mg375) glp-1(e2144);ptp-5.1(tm6122) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK405 | This paper | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+];glp-1(e2144);ptp-5.1(tm6122) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK406 | This paper | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+];dcr-1(mg375);ptp-5.1(tm6122) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK407 | This paper | unc-119(ed3);drSi13[hsf-1p::hsf-1::GFP::unc-54utr;Cb-unc-119+];dcr-1(mg375) glp-1(e2144);ptp-5.1(tm6122) | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK619 | This paper | biuEx63[Pptp-5.1::genomic ptp-5.1::gfp+rol-6] | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK622 | This paper | dcr-1(mg375); biuEx63[Pptp-5.1::genomic ptp-5.1::gfp+rol-6] | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK623 | This paper | glp-1(e2144); biuEx63[Pptp-5.1::genomic ptp-5.1::gfp+rol-6] | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK624 | This paper | glp-1(e2144); biuEx63[Pptp-5.1::genomic ptp-5.1::gfp+rol-6] | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK620 | This paper | dcr-1(mg375) glp-1(e2144); biuEx63[Pptp-5.1::genomic ptp-5.1::gfp+rol-6] | Strain created in S Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK621 | This paper | dcr-1(mg375) glp-1(e2144); biuEx63[Pptp-5.1::genomic ptp-5.1::gfp+rol-6] | Strain created in S Henis-Korenblit lab |
| Sequence-based reagent | act-1 FW | This paper | qPCR primers | CCAATCCAAGAGAGGTATCCTTAC |
| Sequence-based reagent | act-1 BW | This paper | qPCR primers | CATTGTAGAAGGTGTGATGCCAG |
| Sequence-based reagent | F44E5.5 FW | This paper | qPCR primers | CAGAATGGAAAGGTTGAGATCCTCGCC |
| Sequence-based reagent | F44E5.5 BW | This paper | qPCR primers | ACTGTATTCTCTGGATTACGAGCTGCTTGA |
| Sequence-based reagent | hsp-16.2 BW | This paper | qPCR primers | CTCTCCATCTGAGTCTTCTGAGATTGTTAACA |
| Sequence-based reagent | hsp-16.2 FW | This paper | qPCR primers | CAATTCTTGTTCTCCTTGGATTGATAGCGT |
| Sequence-based reagent | hsp-12.6 BW | This paper | qPCR primers | GATGGAGTTGTCAATGTCCTCGACGAC |
| Sequence-based reagent | hsp-12.6 FW | This paper | qPCR primers | TTGTGCTCCATATGGATTTCAAGAAGTTCTCC |
| Sequence-based reagent | ptp-5.1 FW | This paper | qPCR primers | AAGGCTCCGTCTCCTGCACT |
| Sequence-based reagent | ptp-5.1 BW | This paper | qPCR primers | TCCAGAGACACTTGTTGCTATCGGAG |
| Sequence-based reagent | bw_kpni_ptp-5.1_cds | This paper | cloning primers | GACAATGGTACCTTTCCAGGTCCCATCATACT |
| Sequence-based reagent | fw_PstI_ptp-5.1_Prom | This paper | cloning primers | ATGCCTGCAGCACC TACATTACGCCTGCGC |
| Antibody | anti-HSF-1, rabbit polyclonal Antibody | Abcam | ABE1044 | WB(1:1,000) |
| Antibody | anti-Tubulin mouse monoclonal ascites fluid B-5-1-2 | SIGMA-ALDRICH | T5168 | WB(1:6000) |
| Antibody | anti-Tubulin, mouse monoclonal | DHSB | AA4.3 | WB(1:2,000), RRID:AB_579793 |
| Commercial kit | RNA spike-in kit | Agilent | 5188–5279 | |
| Commercial kit | miRVana miRNA isolation kit (w/phenol) | Ambion | AM1560 | |
| Commercial assay | C. elegans microarray 4 × 23,000 | Agilent | G2519F-020186 | |
| Chemical compound | TRIzol | Ambion | 15596026 | |
| Chemical compound | Linoleic acid sodium salt | Sigma | L8134 | |
| Chemical compound | Maxima SYBR GREEN | Thermo Scientific | K0221 | |
| Instrument | microarray scanner | Agilent | G2565BA | |
| Instrument | CFX-96 real time system | BioRad | ||
| Software, algorithm | Agilent Feature Extraction software | Agilent | version 9.5.1.1 | Agilent Technologies, RRID:SCR_014963 |
| Software, algorithm | Partek Genomics Suite software | Partek | version 6.6 | RRID:SCR_011860 |
| Software, algorithm | DAVID | RRID:SCR_001881 | ||
| Software, algorithm | STRING | RRID:SCR_005223 | ||
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 |
Additional files
-
Supplementary file 1
Lifespan analysis of mutants with defective processing of endo-siRNA and inactivation of ptp-5.1.
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp1-v2.xlsx
-
Supplementary file 2
72 genes whose levels decreased by more than 1.5 fold in dcr-1 glp-1 double mutants compared to glp-1 single mutants (p-value<0.05).
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp2-v2.xlsx
-
Supplementary file 3
GO analysis of 72 genes whose levels decreased by more than 1.5 fold in dcr-1 glp-1 double mutants compared to glp-1 mutants (p-value<0.05).
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp3-v2.xlsx
-
Supplementary file 4
132 genes whose levels increased by more than 1.5 fold in dcr-1 glp-1 double mutants compared to glp-1 single mutants (p-value<0.05).
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp4-v2.xlsx
-
Supplementary file 5
Expression of secondary siRNAs of dcr-1 glp-1 vs. glp-1 mutants (Padj <0.05).
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp5-v2.xlsx
-
Supplementary file 6
DAVID analysis of 132 genes whose levels increased by more than 1.5 fold in dcr-1 glp-1 double mutants compared to glp-1 mutants (p-value<0.05).
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp6-v2.xlsx
-
Supplementary file 7
RNAi lifespan screen of dcr-1 glp-1 double mutant.
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp7-v2.xlsx
-
Supplementary file 8
Statistical data.
- https://cdn.elifesciences.org/articles/50896/elife-50896-supp8-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50896/elife-50896-transrepform-v2.docx





