A genetic, genomic, and computational resource for exploring neural circuit function
Figures
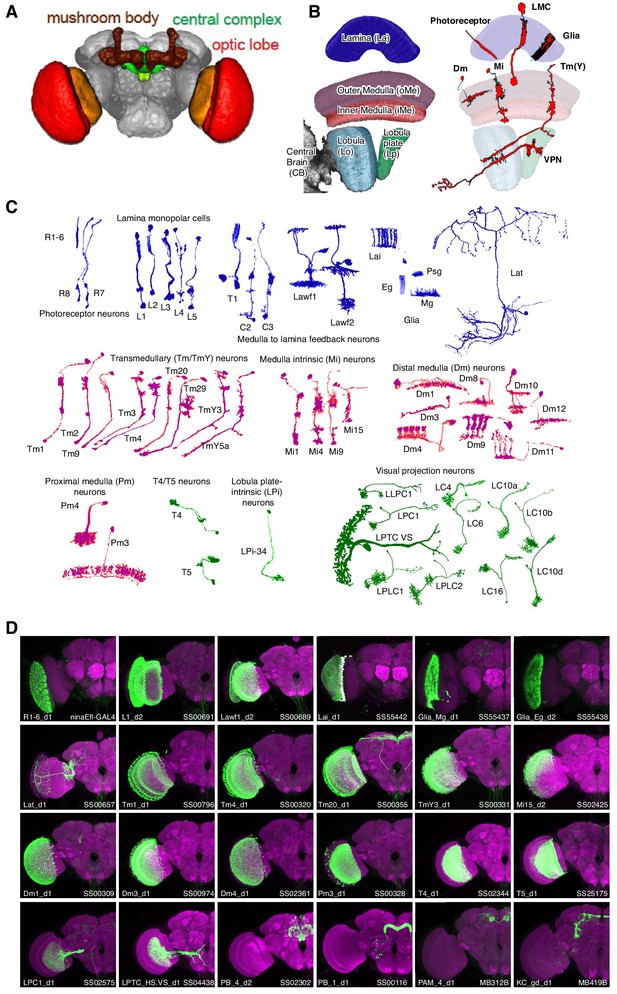
Genetic tools to access cell types in the Drosophila visual system.
(A) Major brain regions profiled in this study (brain image from Jenett et al., 2012). The optic lobes have a repetitive structure of ~750 retinotopically arranged visual columns of similar cellular composition. (B,C) Examples of single cells in the optic lobe. (B) Left, subregions of the fly visual system. Right, examples of layers and neuropil patterns of various classes of visual system neurons. (C) We profiled cell types arborizing in the lamina (blue), medulla (purple) and lobula complex (green) of the visual system. Many cells contribute to multiple neuropiles so other groupings are possible. Note, some cell types are present at one cell per column, while others are less numerous with cells that each contribute to several columns. For example, the main synaptic region of the first optic lobe layer, the lamina, contains processes of some 13,000 cells but these belong to only 17 main cell types: 14 neuronal and three glial (Figure 1C, top row). A small number of additional neurons (lamina tangential cells, Lat) project to a region just distal to the main lamina neuropile. (D) Representative expression patterns of driver lines that target specific cell types. Each image is a maximum intensity projection of a whole brain confocal stack (only one optic lobe is shown). In each image the brain is counter-stained (magenta) with a neuropil marker and both the targeted cell type and the driver are indicated in the lower left and right corner, respectively. Additional images (focusing on drivers first described in this study) are shown in Figure 1—figure supplements 1 and 2. Imaging parameters and brightness and contrast were adjusted individually for each image. For genotypes and image details see Supplementary file 1E.
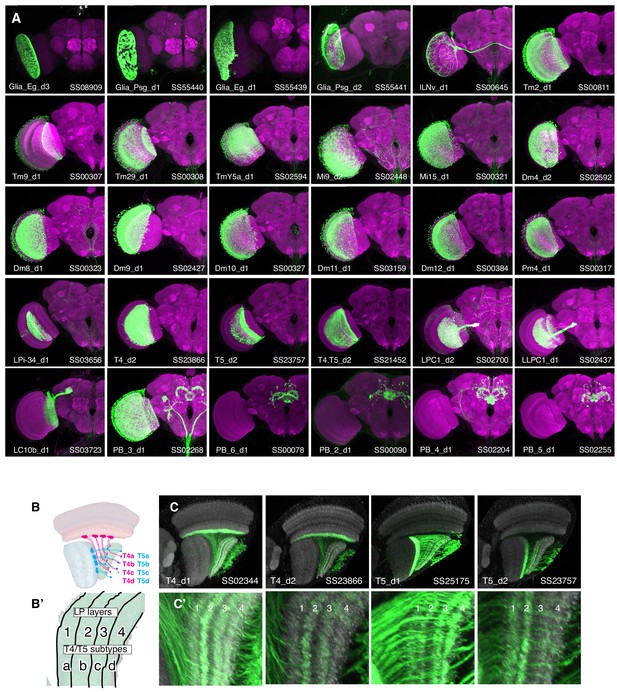
Whole brain expression patterns of new driver lines generated in this study.
(A) Maximum intensity projection of confocal stacks taken from whole fly brains (only one optic lobe is shown). Expression patterns of the driver lines (myristoylated-GFP) are in green and a neuropil marker is in magneta. Imaging parameters and brightness and contrast were individually adjusted for each sample. (B, B’) T4 and T5 cells comprise four subtypes (a,b,c,d) each of which project to specific layers of the lobula plate (B’). (C, C’). Individual T4 and T5 driver lines label combinations of subtypes but show preferential expression in some subtypes. Subtypes were identified by their projections to specific layers in the lobula plate (C,C’). For example, T5_d2 mainly labels lobula plate layers one and two, indicating expression in T5a and T5b. Each of the lower panels is a higher magnification view of the lobula plate region (C’). In (C) both the driver and the split identifier are indicated in the lower left and right corner respectively.
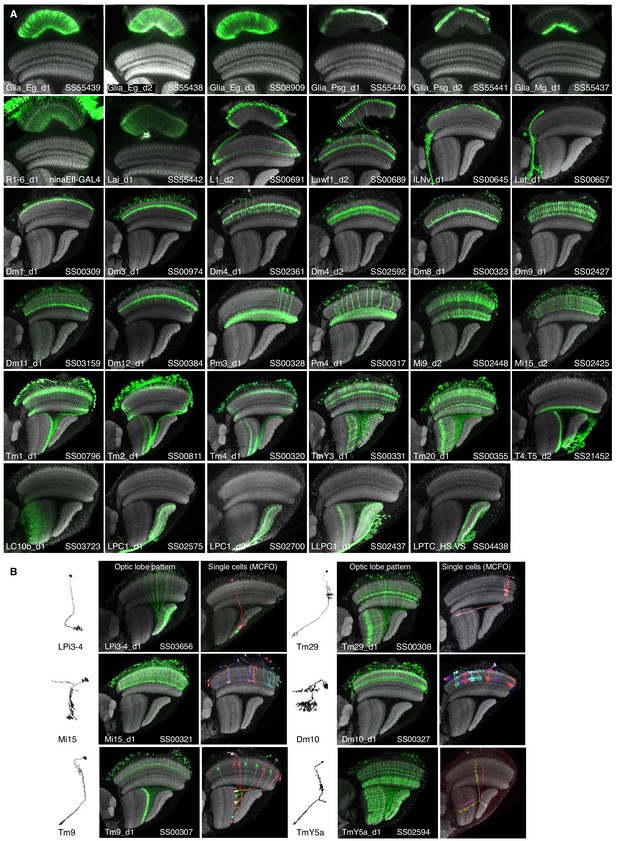
Optic lobe patterns of driver lines.
(A) Optic lobe expression patterns of new driver lines used in this study. All images orient the mediolateral axis of the brain along the lower left to upper right diagonal and are resampled substack projections generated from high resolution (63x) confocal stacks. (B) Examples of segmented single cells illustrating cell morphology (left), the complete optic lobe expression pattern (middle) or individual cells labeled by MultiColor FlpOut (MCFO, right). In all images the neuropil marker is in gray and both the targeted cell type and driver are indicated in the lower left and right corner respectively.
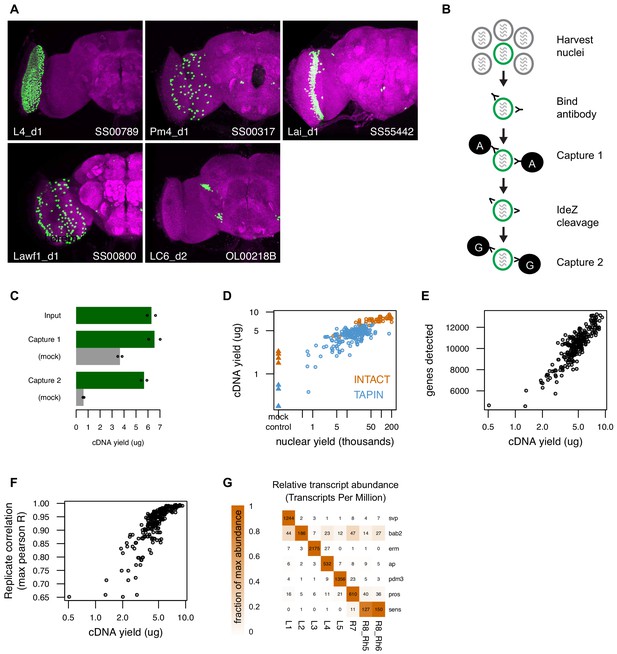
Tandem-affinity purification of INTACT nuclei (TAPIN) enables neuronal genomics.
(A) Cell type-specific drivers enable expression of the UNC84-2XGFP nuclear tag (green) in specific populations of cells. Both the targeted cell type and driver are indicated in the lower left and right corner, respectively. (B) Following nuclei harvest, two rounds of magnetic bead capture serially purify target nuclei. After the first round of protein A bead capture, bacterial protease IdeZ cleaves the anti-GFP antibody in the flexible hinge region, allowing a second round of bead capture with protein G, which recognizes the F(ab’)2 region. Protein G, unlike Protein A, can bind both the Fc and F(ab’)2 regions of an immunoglobulin. (C) Two capture rounds reduce the level of non-specific background (gray bars, mock IgG control) while maintaining the cDNA yield from the captured target nuclei (green bars). Bars represent the mean of two replicates (shown as points). (D) RNA-seq libraries created with more nuclei yield more cDNA (circles). TAPIN libraries had lower non-specific background than INTACT (blue vs orange triangles). (E) Libraries with more cDNA detect more genes. (F) Libraries with more cDNA have more reproducible transcript abundances. (G) Previously identified markers of lamina monopolar and inner photoreceptor neurons (Tan et al., 2015) are enriched in the expected cells.
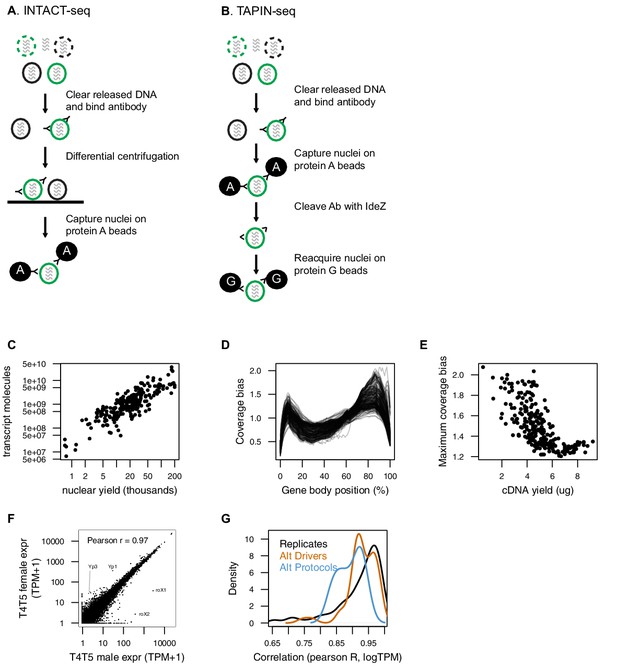
Two variants of nuclei capture, INTACT and TAPIN.
(A) We used an INTACT-seq variant, that we originally developed for mouse (Mo et al., 2015), that purifies nuclei by differential centrifugation. (B) TAPIN-seq replaces the space- and time-intensive centrifugation with a two-step capture enabled by antibody hinge cleavage with the bacterial protease IdeZ. Both protein A and protein G bind the Fc region, while only protein G is able to bind F(ab’)2. (C) Libraries built from more nuclei have more transcript molecules (estimated using synthetic spike-ins). (D) Nearly all libraries showed relatively unbiased positional coverage across gene bodies. (E) The maximum bias in positional coverage observed in each library was inversely correlated with cDNA yield, although with large variance in bias for lower yield libraries. (F) T4.T5 transcriptomes of female (y-axis) and male (x-axis) flies are well correlated, but also recover known sex-specific genes including RNA on X 1 (roX1) and roX2 (Amrein and Axel, 1997) and yolk protein 1 (Yp1) and Yp3 (Belote et al., 1985). (G) Estimated transcript abundances were reproducible as evaluated by Pearson correlation (of log-transformed transcript abundance) between biological replicates (black), alternative drivers for the same cell type (orange), or comparing TAPIN to INTACT profiles (blue).
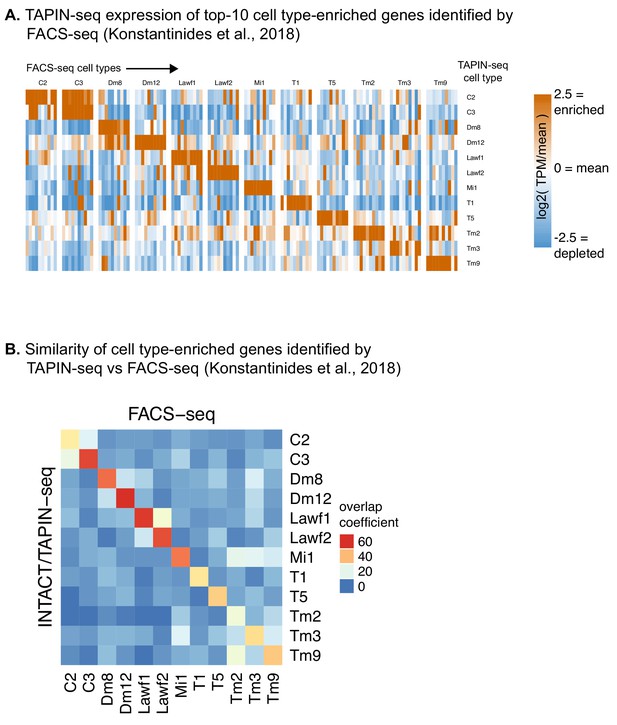
TAPIN-seq vs FACS-seq comparison.
(A) TAPIN-seq expression of marker genes identified from an independent FACS-seq dataset covering 12 cell types we also profiled (Konstantinides et al., 2018). We defined marker genes for each cell type as the top-10 most highly expressed genes relative to the mean of all cell types, requiring at least 4x higher abundance than the mean and a relative abundance of at least 50 TPM. (B) We more broadly compared the TAPIN-seq and FACS-seq datasets by first identifying cell type-enriched genes within each dataset (at least two-fold higher than mean expression; at least 50 TPM in one sample) and then quantifying the degree of overlap between datasets using the overlap coefficient: 100 * (# genes enriched in both TAPIN-seq AND FACS-seq)/minimum(# genes enriched in TAPIN-seq, # genes enriched in FACS-seq)).
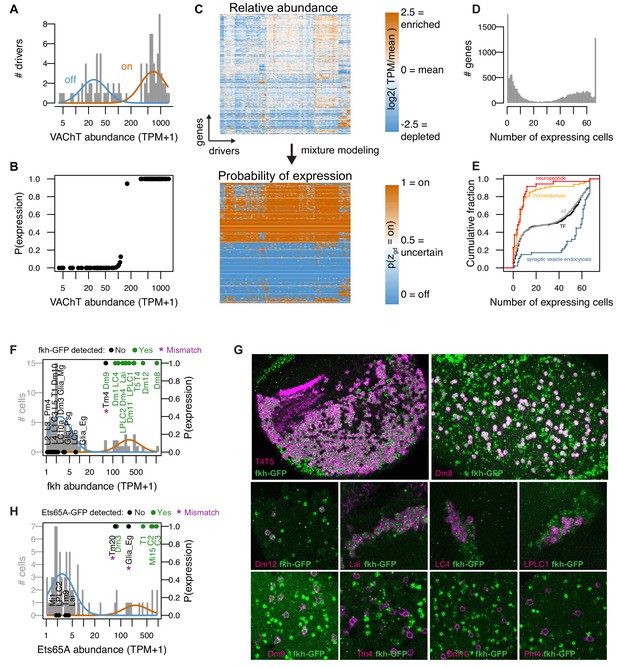
Mixture modeling accurately interprets TAPIN-seq measurements.
(A) The distribution of Vesicular acetylcholine transporter (VAChT) abundance fit with a mixture of two log-normal components. (B) Interpreting these components as ‘off’ and ‘on’ states unambiguously infers expression state in essentially all drivers. (C) Mixture modeling transforms our catalog of relative transcript abundances (top) to discretized expression states (bottom). (D) Histogram of expression breadth per gene. (E) Cumulative distributions of expression breadth for all genes (gray), transcription factors (black), homeobox TFs (orange; InterPro domain IPR001356), neuropeptides (red), and genes involved in synaptic vesicle endocytosis (blue). (F,G) The fkh modeling results were compared to its protein expression pattern as evaluated with a BAC transgenic (See Figure 3—figure supplement 2A). (F) Histogram bars represent raw abundance of all cells in our catalog. Blue and orange curves represent the inferred off and on components, respectively. Points represent the cells tested for transgene expression showing either detectable GFP (Green) or no signal (Black). The points’ vertical position reflect the estimated probability of gene expression. (G) Forkhead-GFP expression in selected cell types. Fkh-GFP (mainly nuclear, in green) and cell type-specific expression of a membrane marker (in magenta) are shown. Because of the wide range of fkh expression levels, imaging parameters and brightness and contrast adjustments are not identical for different panels. Cells with detectable nuclear GFP signal above the background in the same image were scored as expressing fkh. (H) As in J, to evaluate Ets65A modeling results (See Figure 3—figure supplement 2B).
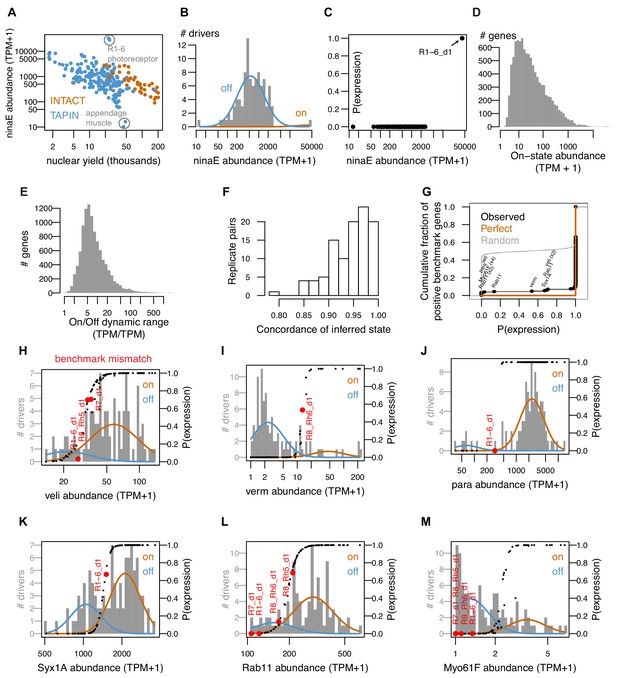
Overview of INTACT-seq and TAPIN-seq libraries.
(A) Libraries with fewer nuclei had greater carry-over of ninaE transcript, which encodes the abundant rhodopsin in the fly eye. The upper outliers are libraries made from R1-6 photoreceptors, the only cells that express ninaE. The lower outliers are appendage muscle libraries created after heads are removed from the fly bodies, effectively eliminating ninaE carry-over from photoreceptors. (B, C). Modeling the distribution of ninaE correctly distinguishes true expression by R1-6 from transcript carry-over in the remaining samples. (D) Distribution of mean on-state transcript abundance across all modeled genes. (E) Distribution of dynamic range across all modeled genes. (F) Concordance of inferred expression states between replicates. Concordance was computed as the number of genes predicted to express (p(on)>=0.8) or not (p(on)<=0.2) in both replicates divided by the number of genes predicted to express or not in either replicate. (G) Cumulative distribution of inferred expression probabilities for gene/cell pairs reported to express in FlyBase (n = 193 positive benchmark points). Our mixture model correctly inferred expression of 179 of the 193 gene/cell pairs. The 14 discordant pairs involved six genes (labeled in black). (H–M) Modeling results for the six genes with benchmark mismatches. The on and off components are represented as orange and blue curves, respectively. Black points represent the inferred probabilities of expression for all drivers. Red points highlight the drivers where the model results disagreed with the benchmark. The transcript abundance (x-axis) reflects the average of all ‘high quality’ replicates (minimum two per cell type).
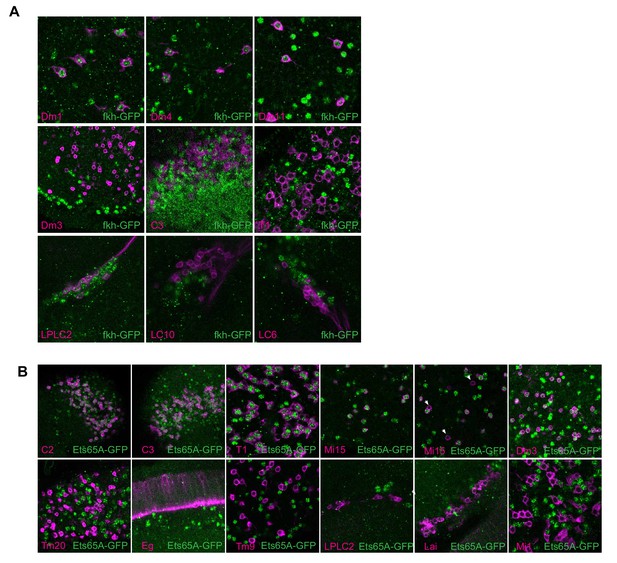
Validation of fkh and Ets65A model inferences.
(A, B) To evaluate our modeling results for fkh and Ets65A we evaluated protein expression in several cell types (related to Figure 3F–H) using GFP-fusion proteins (Materials and methods). The indicated cell types (lower left corner) were visualized with a membrane marker (magenta). The second Mi15 panel (in B) includes examples of occasional cells without detectable Ets65-GFP expression (arrows).
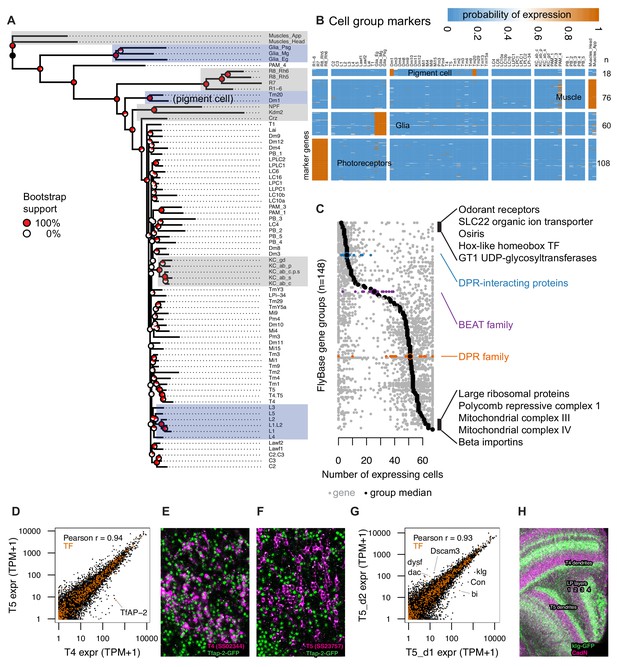
TAPIN-seq profiles identify genes enriched in cell types and groups.
(A) Cells grouped by a minimum evolution tree of their inferred expression states. (B) Heatmap of marker genes enriched in photoreceptors, glia, muscle, and pigment cells. (C) Distribution of expression breadth for genes in ‘terminal’ FlyBase gene groups with more than 10 members in our expression probability matrix. The least- and most- broadly expressed gene groups are labeled, along with the DPR-interacting, beat and DPR family of extracellular proteins. (D) TfAP-2 transcription factor distinguishes closely related cell types T4 and T5. (E,F) TfAP-2 protein is specifically expressed in T4 and not in T5, confirming this detection of differential expression levels. GFP-tagged TfAP-2 (mainly nuclear, in green; see Supplementary file 1E and Materials and methods) is shown together with a membrane marker (magenta) expressed in T4 (E) or T5 (F) cells. (G) Comparison of genes with differential expression in two driver lines for T5 neurons expressing in different subtypes, identify genes that differentially label layers of the lobula plate (corresponding to different subtypes of T5 cells). (H) Confirming the TAPIN-seq identification, klg protein (detected using a GFP tag (green); see Supplementary file 1E and Materials and methods) is expressed in T4/T5 cells with the expected layer specificity (layers 3 and 4) in the lobula plate (LP). A neuropil marker is shown in magenta.
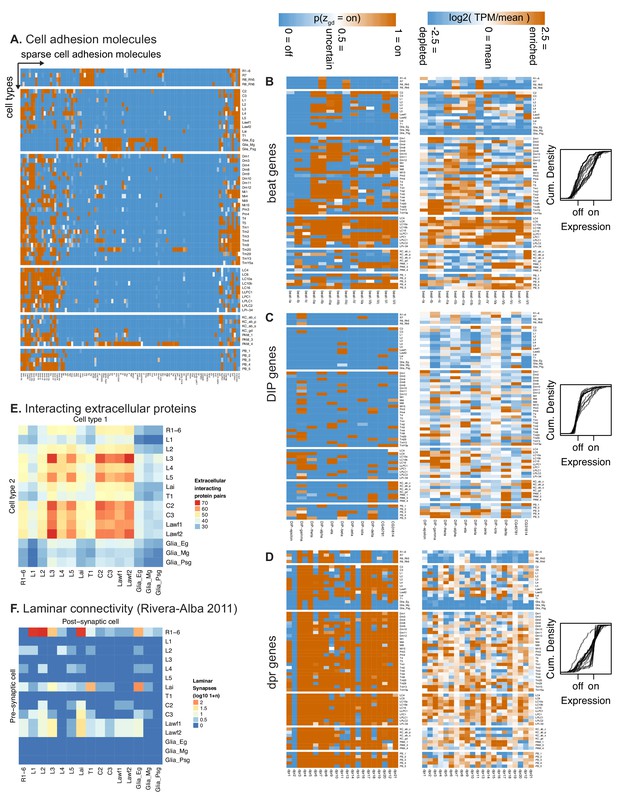
TAPIN-seq profiles identify genes enriched in cell types and groups.
(A) Cell adhesion molecules specifically expressed across our transcriptome catalog. (B–D) The expression pattern of all beat, DIP, and Dpr family members depicted as heatmaps of probabilities of expression (left), heatmaps of relative transcript abundance (middle), or cumulative density curves of normalized expression level (right). The density curves, each depicting a bimodally expressed gene in the gene family, show expression levels that were normalized using the mean expression levels of the modeled off an on states for each gene; normalized level = (logE - mu_off) / (mu_on - mu_off). The density curves illustrate that DIP genes are sparsely expressed, followed by beat and Dpr genes. Of the three families, the Dpr genes exhibit transcript abundance that appears more continuous between the estimated off and on states rather than discretely bimodal. (E,F) The number of interacting pairs of extracellular protein pairs (Özkan et al., 2013) expressed by pairs of cells in the lamina (E) is not sufficient to predict the synaptic connectivity of these cells F; data from Rivera-Alba et al. (2011). To match our expression data, we summed the synapse counts for the individual R1-R6 photoreceptors originally reported by Rivera-Alba et al. (2011). For the same reason, we also duplicated the subtype-unidentified Lawf synapse counts as separate Lawf1 and Lawf2 entries in the connectome matrix.
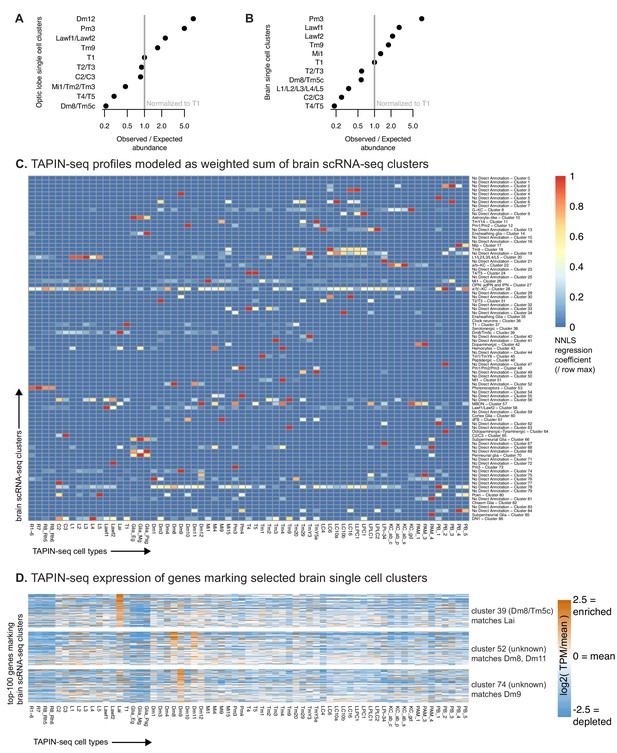
TAPIN-seq complements single cell RNA-seq profiling.
(A, B) We evaluated whether single cell RNA-seq of the optic lobe (A) (Konstantinides et al., 2018) and brain (Davie et al., 2018) proportionally represents cell types found in the optic lobe. By comparing the single cell cluster sizes to the true abundance of each cell type (estimated as described in the Materials and methods) we found that the scRNA-seq map can both under- and over-estimate the abundance of each cell type (assuming accurate cell type labels), or that the cell type is incorrectly assigned (i.e. contains different or additional cell types). To estimate the true cell count, we made use of known anatomy (for example, several cell types are known to be present exactly once in each of the ~2×750 medulla columns per brain) or relied on published counts. In addition, we performed some new counts. (See Materials and methods for details.) Observed/expected ratio = ((size of cluster labeled as cell type X/size of cluster labeled as T1) / (true abundance of cell type X/true abundance of T1)). (C) We used non-negative least squares regression to model each TAPIN-seq profile as a linear weighted sum of single cell clusters in the whole brain scRNA-seq map. The heatmap represents the regression coefficients of each single cell cluster (rows) contributing to the TAPIN-seq profile of each cell type, normalized within rows. (D) We evaluated expression of genes that mark selected single cell clusters (Davie et al., 2018) in our TAPIN-seq profiles of visual system neurons. (see Figure 5—figure supplement 2 for the complete heatmap).
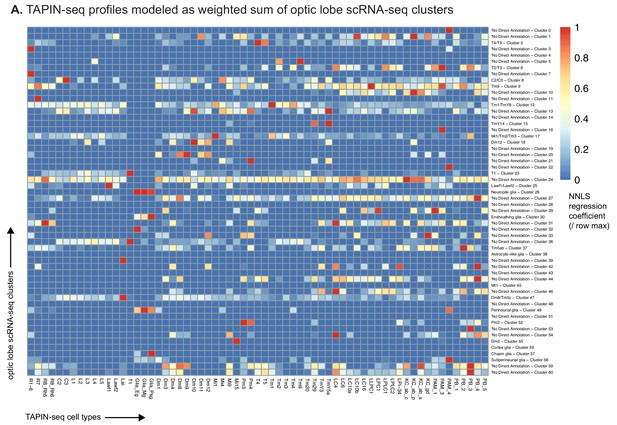
Regressing TAPIN-seq profiles against optic lobe single cell clusters.
(A) We used non-negative least squares regression to model each TAPIN-seq profile as a linear weighted sum of optic lobe single cell clusters (Konstantinides et al., 2018). The heatmap represents the regression coefficients of each single cell cluster (rows) contributing to the TAPIN-seq profile of each cell type, normalized within rows.
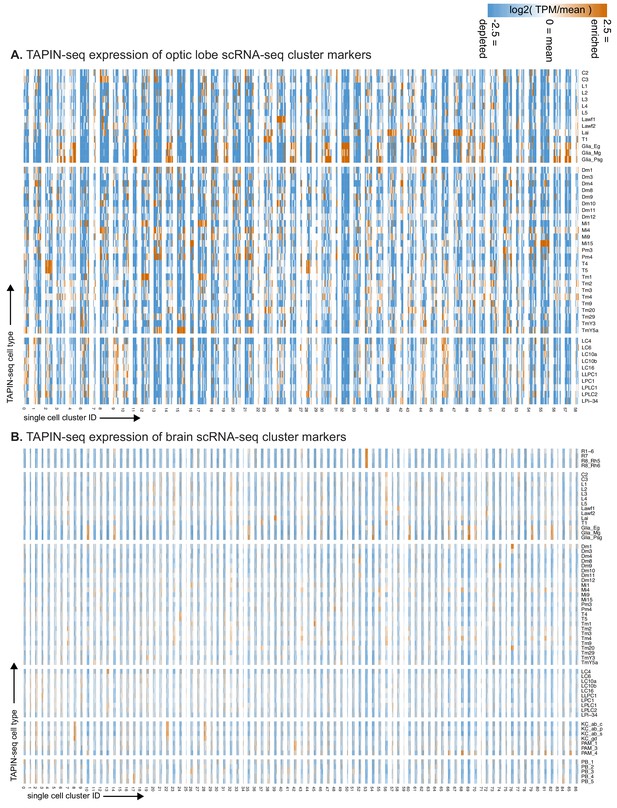
TAPIN-seq expression of genes marking single cell clusters.
(A) We evaluated expression of marker genes for each optic lobe single cell cluster (as reported in Konstantinides et al., 2018) in our TAPIN-seq profiles of visual system neurons. If a single cell cluster marker corresponds to one of our identified cell types, we expect to see its marker genes highly enriched in the corresponding cell type’s expression. Note that some of the single cell clusters with the best apparent cell type matches (e.g., cluster 15/TmY5a, cluster 55/Mi15) were originally reported with a different annotation. (B) Expression of marker genes for each brain single cell cluster (as reported in Davie et al., 2018), as in (A).
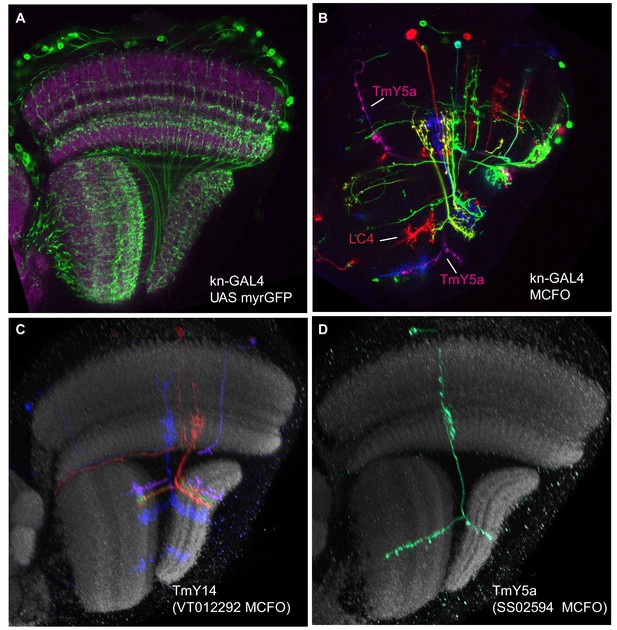
kn-GAL4 expression.
(A) kn-GAL4 driven expression of a membrane-targeted GFP (green) in the optic lobe. Single confocal section with a reference marker (anti-Brp) in magenta. TmY14 cell bodies are unusual in that they are found only in a subregion of the medulla cell body rind (see http://flweb.janelia.org/cgi-bin/view_flew_imagery.cgi?line=R10G02 for an image of a GAL4 line that in the optic lobe expresses mainly in TmY14). Both the cell body distribution and optic lobe layer pattern (compare cells in (C)) indicate that kn-GAL4 is expressed in optic lobe cell types other than TmY14. (B) Stochastic labeling of kn-GAL4 neurons using MCFO. A TmY5a cell and an LC4 cell are indicated. Other cell types, including several TmY14, are also labeled. Image shows a single confocal section without a reference marker. (C,D) Examples of TmY14 (C) and TmY5a (D) cells. Reconstructed views generated from confocal stacks of MCFO-labeled cells using the indicated driver lines.
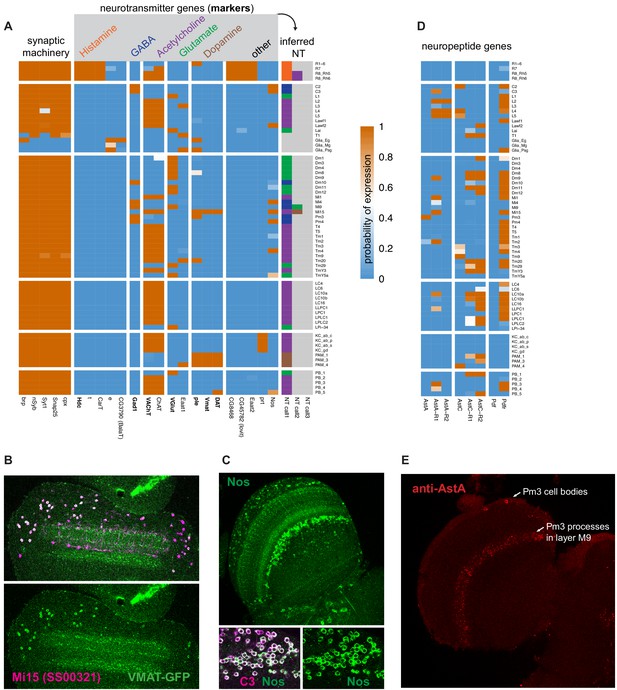
Expression of synthesis and transport genes indicate neurotransmitter phenotypes.
(A) Expression of neurotransmitter marker genes indicate the neurotransmitters produced in nearly all profiled cells. With few exceptions, nearly all cell types express only one fast neurotransmitter. (B, C) We confirm TAPIN-seq results at the protein level (green) for (B) Vesicular monoamine transporter (Vmat) expressed in Mi15 (magenta) and (C) Nitric oxide synthase (Nos) in C3 (magenta). Top panel in (C) shows a section through the optic lobe, lower panels C3 cell bodies. (D) Several neuropeptides and receptors also express specifically (examples). (E) Allatostatin A (AstA) protein expression in the medulla as an example of a neuropeptide with a very specific optic lobe expression pattern. The AstA distribution in the optic lobe matches the distribution and layer pattern of Pm3 cells, consistent with the TAPIN-Seq data.
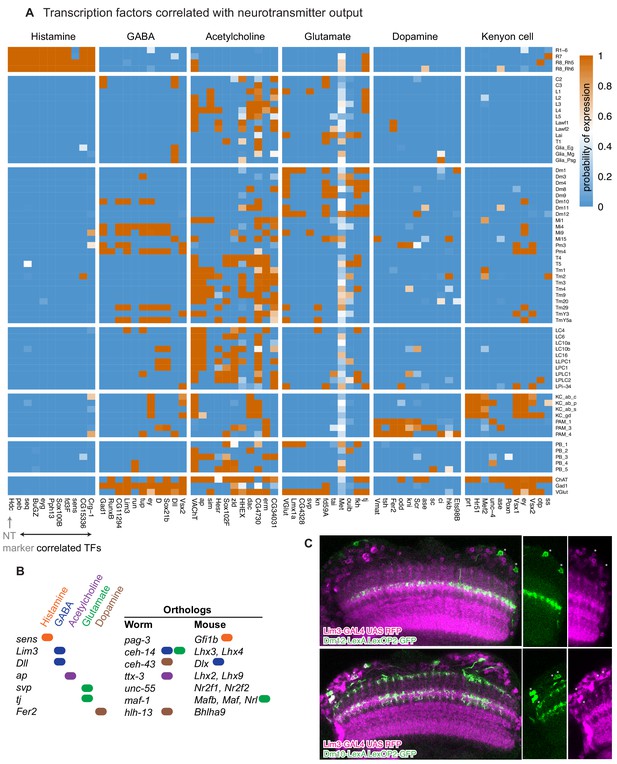
Transcriptional regulators of neurotransmitter identity.
(A) Transcription factors whose expression is predictive of neurotransmitter phenotype (i.e., P(neurotransmitter output | transcription factor expressed)). The ten most predictive transcription factors are shown for each neurotransmitter output marker. (B) Summary of orthologous transcription factors in worm and mouse and their association with specific neurotransmitter types. (C) The Gad1-associated gene Lim3 does not express in cholinergic Dm12 neurons, but does in the GABA-ergic Dm10 neurons. Double labeling using LexA-markers for Dm12 and Dm10 (green) with a Lim3 protein-trap-GAL4 driving RFP (magenta). This example highlights a case where Lim3 expression identifies a cell type in a group of similar cells (the Dm cells profiled in this study) that is GABAergic (all other Dms in this group are glutamatergic).
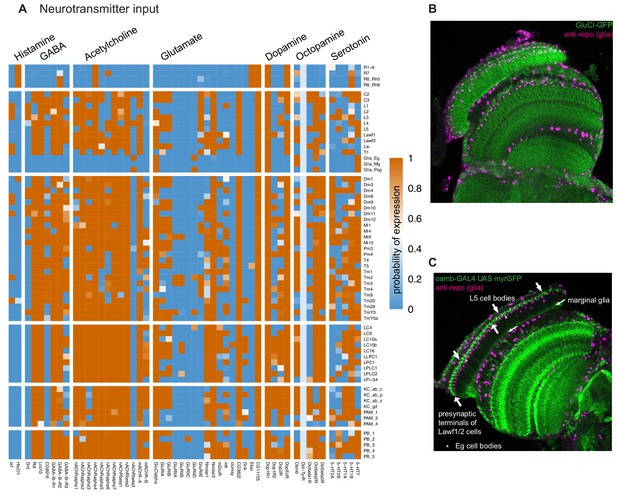
Patterns of neurotransmitter receptor expression.
(A) Neurotransmitter receptors are widely expressed in specific patterns. With the exception of histamine, most cells express receptors or receptor subunits for nearly all neurotransmitters. (B) Expression of the glutamate-gated chloride channel (GluClalpha), detected using a GFP-tag (green), in the optic lobe. The lamina pattern includes many neurons as well as proximal satellite, epithelial and marginal glia. A glia-specific nuclear marker (anti-repo) is shown in magenta. (C) Octopamine receptor (Oamb) expressing cells in the optic lobe detected with a protein-trap GAL4 driving expression of a membrane targeted GFP (green). Anti-repo (magenta). In the lamina (to the top and left of the image), Lawf1/2 and L5 neurons and marginal glia are recognizable.
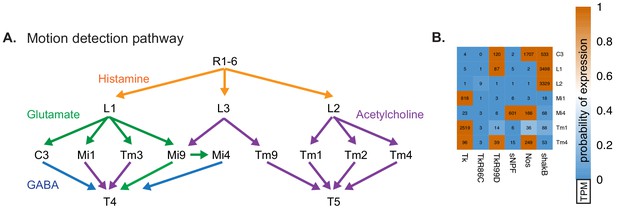
Patterns of neurotransmitter receptor expression complement connectomics.
(A) Transcriptomes reveal the neurotransmitters in core cell types of the ON and OFF components of the motion detection pathway. The ON and OFF motion detection pathways supply inputs to directionally sensitive T4 and T5 neurons, respectively (Takemura et al., 2017; Shinomiya et al., 2019). Our results show that all of the inputs to T5 (Tm1, Tm2, Tm4, and Tm9) are cholinergic, whereas the inputs to T4 are a mixture of GABAergic (C3, Mi4), cholinergic (Mi1, Tm3), and glutamatergic (Mi9), suggesting different input signs. Discovering the functional signs of inputs to the directionally selective neurons is an essential step in understanding the mechanism of this long-studied neuronal computation (Strother et al., 2017). In addition, our data reveals aspects of the motion pathway that have not yet been functionally examined, such as the identification of other signaling components (see B). (B) Examples of the expression of genes involved in neuropeptide, non-canonical small molecule (nitric oxide), or gap junction communication in the cell types in (A).
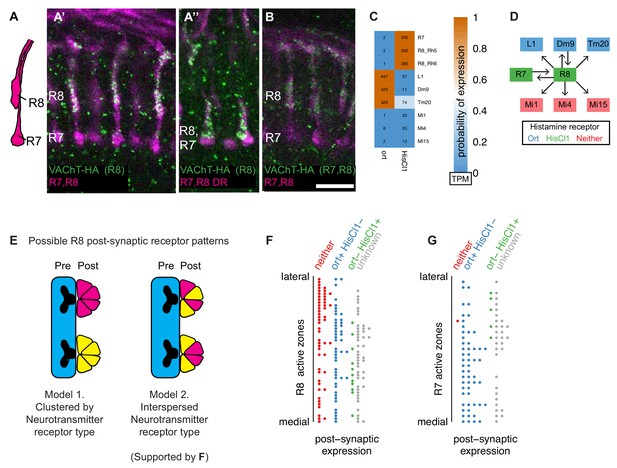
Molecular and connectomics analyses suggest R8 photoreceptors signal via both histaminergic and cholinergic neurotransmission.
(A, A’, A’’, B). Expression of VAChT in R8 cells. Expression of a HA-tagged VAChT was induced in R8 cells by recombinase-mediated excision of an interruption cassette from a modified genomic copy of the VAChT gene (Pankova and Borst, 2017). R7 and R8 cells project to different layers of the medulla (A, schematic). Single confocal sections show R7 and R8 cells in magenta and anti-HA immunolabeling in green. R7 and R8 cells (labeled with mAb 24B10) are shown in magenta. Stop-cassette excision in R8 photoreceptors (using sens-FLP) results in VAChT-HA labeling of R8 terminals in both the main medulla (A’) and the dorsal rim (where R7 and R8 cells project to very similar layer positions) (A’’). Stop-cassette excision in all photoreceptors (using ey3.5-FLP) also produces VAChT-HA labeling in R8 while expression in R7 was not detected (B). Scale bar, 10 µm. (C) Heatmap of receptor expression probabilities (color) and relative abundance (numbers; transcripts per million) in R8 targets identified by EM (at least five synapses in Takemura et al., 2013). (D) Connectivity network for R8 cells, overlaid with receptor expression. (E) Possible distributions of postsynaptic receptors at R8 synapses. Individual active zones can interact with multiple postsynaptic cells which could be grouped in distinct ways. (F) Classification of postsynaptic cells at individual R8 active zones (Takemura et al., 2013) based on histamine receptor expression. (G) Same analysis as in F but for an R7 cell.
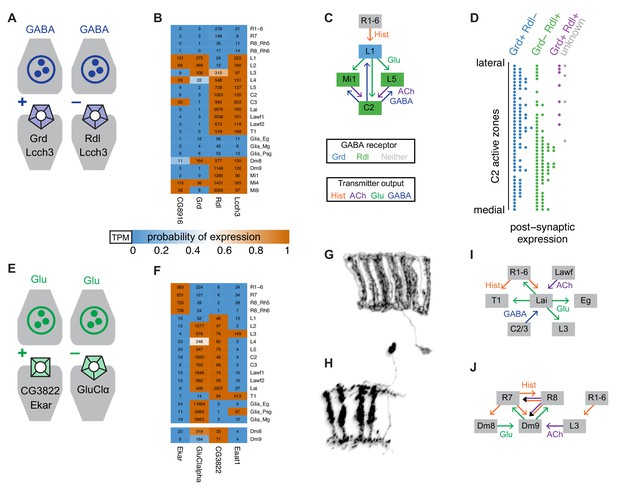
Using gene expression to functionally interpret circuit structure.
(A) Different properties of GABA-A receptors in Drosophila observed in in vitro studies. GABA-A receptor subunits can form either cation or anion channels depending on subunit composition. (B) Expression of GABA-A subunits in selected cell types. (C) L1 and two of its target cells form strong reciprocal connections with C2 neurons. (D) Distribution of Rdl and Grd expressing cells at individual C2 synapses. (E) Glutamate receptors can also be excitatory or inhibitory. (F) Examples of expression patterns for selected glutamate receptors and transporters. (G, H) Morphology of Lai (G) and Dm9 (H) cells. Illustrations based on MCFO images of single cells. (I, J) Analysis of the input and output pathways of Lai (I) and Dm9 (J) neurons suggests a potentially similar functional role for these cells. The predicted absence of GluCl-alpha in Dm9 suggests that glutamatergic input from Dm8 to Dm9 may be excitatory.
Additional files
-
Supplementary file 1
Supplemental tables.
(A) All drivers. (B) RNAseq samples. (C) Benchmark entries. (D) Marker genes. (E) Anatomy details. (F) TAPIN-seq buffers.
- https://cdn.elifesciences.org/articles/50901/elife-50901-supp1-v2.xlsx
-
Supplementary file 2
Key Resource Table.
List of key resources and reagents used in this study.
- https://cdn.elifesciences.org/articles/50901/elife-50901-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50901/elife-50901-transrepform-v2.pdf





