LRRK2 maintains mitochondrial homeostasis and regulates innate immune responses to Mycobacterium tuberculosis
Figures
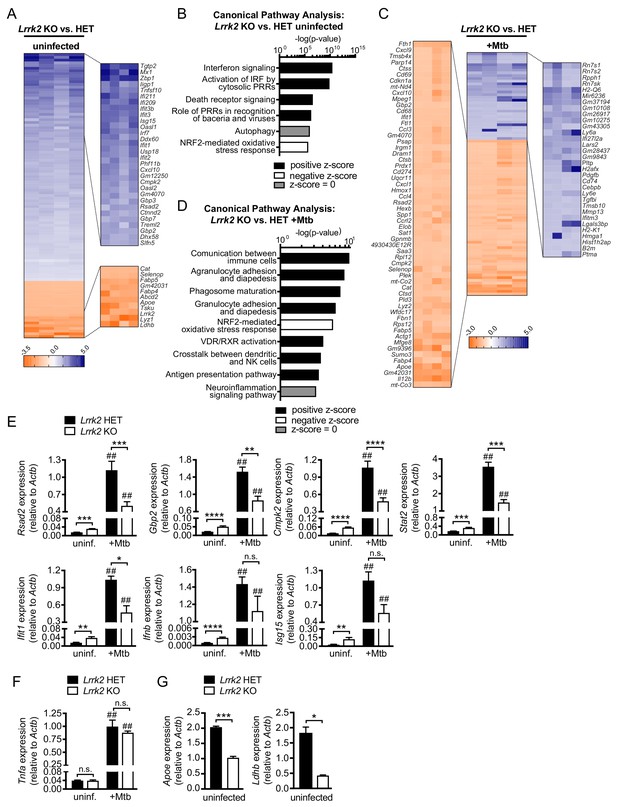
Global gene expression analysis reveals that Lrrk2 KO macrophages are deficient at inducing type I IFN expression and have higher basal levels of ISGs.
(A) Heatmap depicting significant gene expression differences (Log2 fold-change, p<0.05) between uninfected Lrrk2 KO and HET BMDMs. (B) IPA software analysis showing cellular pathways enriched for differentially expressed genes in uninfected Lrrk2 KO vs. HET BMDMs. (C) Heatmap depicting significant gene expression differences (Log2 fold-change) between Lrrk2 KO and HET BMDMs during infection with Mtb. (D) As in (B) but for pathways enriched for differentially expressed genes in Mtb-infected Lrrk2 KO and HET BMDMs, 4 hr post-infection. (E) RT-qPCR showing expression of Ifnb and IFN stimulated genes in uninfected and Mtb-infected Lrrk2 KO and HET macrophages. Data are shown as ISG/Actb. (F) RT-qPCR of Tnfa in Lrrk2 KO and HET BMDMs. (G) RT-qPCR of Apoe and Ldhb normalized to Actb in uninfected BMDMs. Throughout the manuscript, data are expressed as a mean of three or more biological replicates with error bars depicting SEM. Statistical tests used can be found at the end of the legend. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); ##p<0.001 (comparing stimulated to unstimulated of same genotype). In (E–F) a two-way ANOVA Tukey post-test was applied, and in (G) a two-tailed Student’s T test.
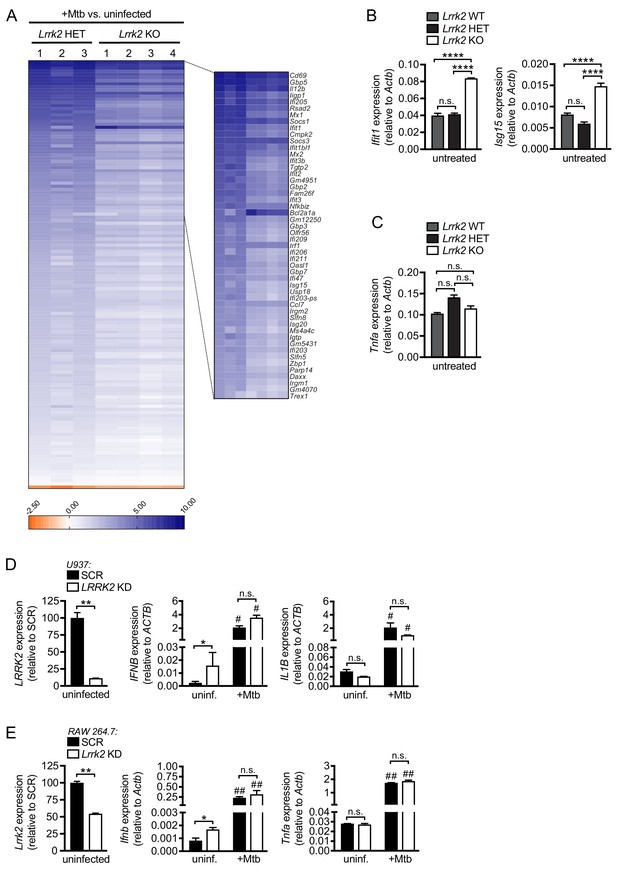
Lrrk2 KO macrophages fail to induce proper levels of ISG expression in response to Mtb infection.
(A) Heatmap depicting gene expression differences (Log2 fold-change) between Lrrk2 KO and HET BMDMs after infection with Mtb. (B) RT-qPCR of Ifit1 and Isg15 in Lrrk2 WT, HET, and KO BMDMs. (C) RT-qPCR of Tnfa expression in Lrrk2 WT, HET, and KO BMDMs. RT-qPCR showing expression of Ifnb and IFN stimulated genes in uninfected and Mtb-infected Lrrk2 KO and HET macrophages. Data are shown as ISG/Actb. (D) RT-qPCR of LRRK2 in uninfected SCR and LRRK2 KD U937 macrophage-like cells and gene expression of IFNB and IL1B in uninfected and Mtb-infected cells (MOI = 10, 4 hr). (E) RT-qPCR of Lrrk2 in uninfected SCR and Lrrk2 KD RAW 264.7 cells and gene expression of Ifnb and Tnfa in uninfected and Mtb-infected cells (MOI = 10, 4 hr). Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); ##p<0.001 (comparing stimulated to unstimulated of same genotype). (B–E) One-way and two-way ANOVA Tukey post-test.
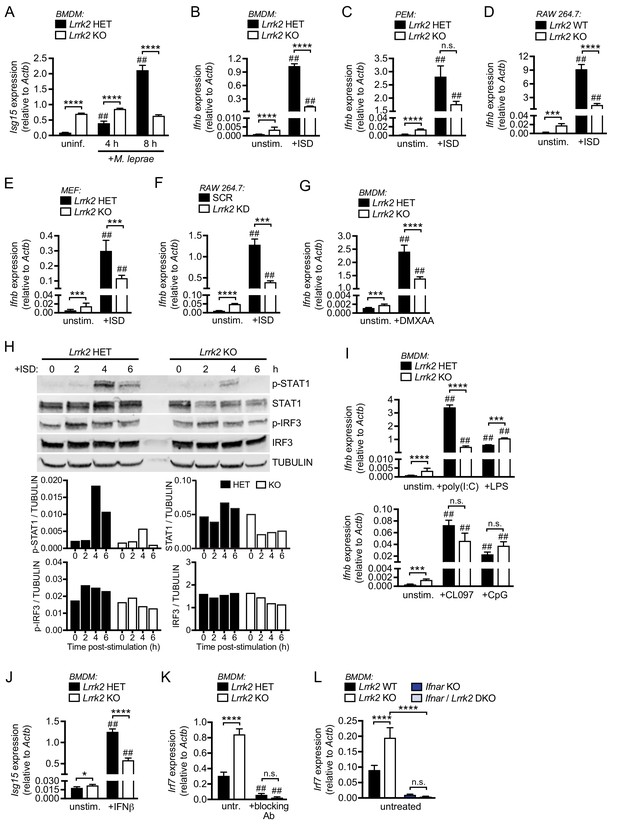
Lrrk2 KO macrophages exhibit blunted type I IFN expression in response to cytosolic nucleic acid agonists.
(A) RT-qPCR of Isg15 expression after 4 and 8 hr of infection with M. leprae (MOI = 50) in Lrrk2 KO BMDMs and HET controls. (B) RT-qPCR of Ifnb in unstimulated Lrrk2 KO and HET BMDMs alongside cells transfected with 1 μg/ml ISD (dsDNA) for 4 hr. (C) As in (B) but with peritoneal macrophages (PEMs) from Lrrk2 KO and HET mice elicited for 4 days with 1 ml 3% Brewer’s thioglycolate broth. (D) As in (B) but with RAW 264.7 Lrrk2 KO cells and WT controls. (E) As in (B) but with MEFs from day 14.5 Lrrk2 KO or HET embryos. (F) As in (B) but with RAW 264.7 Lrrk2 KD and scramble (SCR) controls cells. (G) RT-qPCR of Ifnb expression in uninfected Lrrk2 KO or HET BMDMs and in cells treated with 50 ng/ml DMXAA for 2 hr. (H) Western blot analysis and quantification of IRF3 phosphorylation (Ser396) and STAT1 phosphorylation (Tyr701) in BMDMs from HET and Lrrk2 KO mice compared to total IRF3 and STAT1 with tubulin as a loading control following transfection with 1 μg/ml ISD (dsDNA). (I) As in (G) but following transfection with 1 μg/ml poly(I:C), 100 ng/ml LPS, transfection with 10 μM CpG 2395, or stimulation with 1 μM CL097, all for 4 hr. (J) RT-qPCR of Isg15 expression after treatment with 200 IU IFN-β for 4 hr. (K) RT-qPCR of Irf7 gene expression in Lrrk2 HET and KO BMDMs with or without overnight treatment with IFN-β neutralizing antibody (blocking Ab, 1:250). (L) RT-qPCR of Irf7 gene expression in BMDMs from WT, Lrrk2 KO, Ifnar KO, and double knockout (Lrrk2/Ifnar DKO) mice. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); ##p<0.001 (comparing stimulated to unstimulated of same genotype). (A–L) two-way ANOVA Tukey post-test.
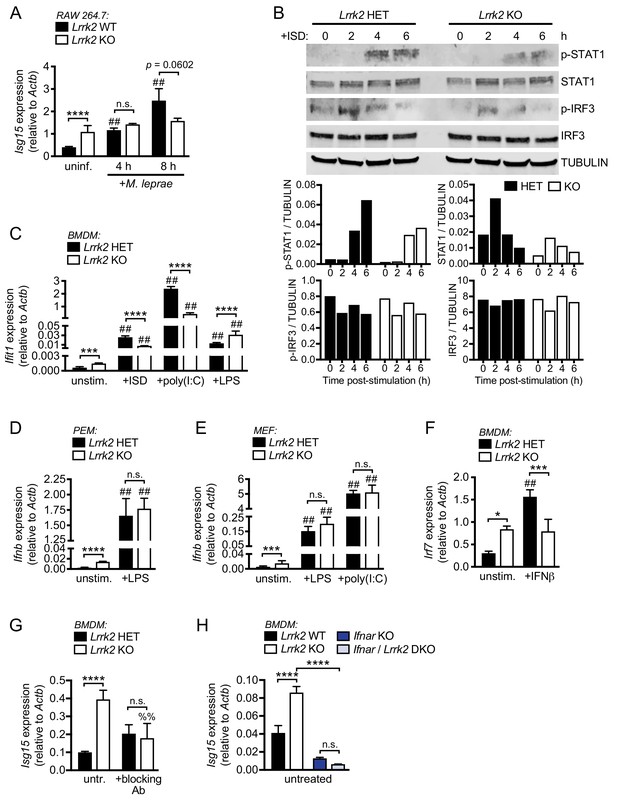
LRRK2-deficient macrophages are unable to properly induce type I IFN expression in response to nucleic acid agonists.
(A) RT-qPCR of Isg15 expression after 4 and 8 hr of infection with M. leprae (MOI = 50) in WT and Lrrk2 KO RAW 264.7. (B) Western blot analysis and quantification of IRF3 phosphorylation (Ser396) and STAT1 phosphorylation (Tyr701) in BMDMs from HET and Lrrk2 KO mice compared to total IRF3 and STAT1 with tubulin as a loading control following transfection with 1 μg/ml ISD (dsDNA). (C) RT-qPCR of Ifit1 in Lrrk2 HET and KO BMDMs transfected with 1 μg/ml ISD (dsDNA) or 1 μg/ml poly(I:C) or treated with 100 ng/ml LPS for 4 hr. (D) RT-qPCR of Ifnb expression in uninfected Lrrk2 KO or HET PEMs treated with 100 ng/ml LPS for 4 hr. (E) RT-qPCR of Ifnb expression in uninfected Lrrk2 KO or HET MEFs treated with 100 ng/ml LPS or transfected with 1 μg/ml poly(I:C) for 4 hr. (F) RT-qPCR of Irf7 expression in Lrrk2 HET and KO BMDMs after treatment with 200 IU IFN-β for 4 hr. (G) RT-qPCR of Isg15 expression in Lrrk2 HET and KO BMDMs with or without overnight blocking with IFN-β neutralizing antibody (blocking Ab, 1:250). (H) RT-qPCR of Irf7 gene expression in BMDMs from WT, Lrrk2 KO, Ifnar KO, and double knockout (Lrrk2/Ifnar DKO) mice. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); %%p<0.01, ##p<0.001 (comparing stimulated to unstimulated of same genotype). (A–H) two-way ANOVA Tukey post-test.
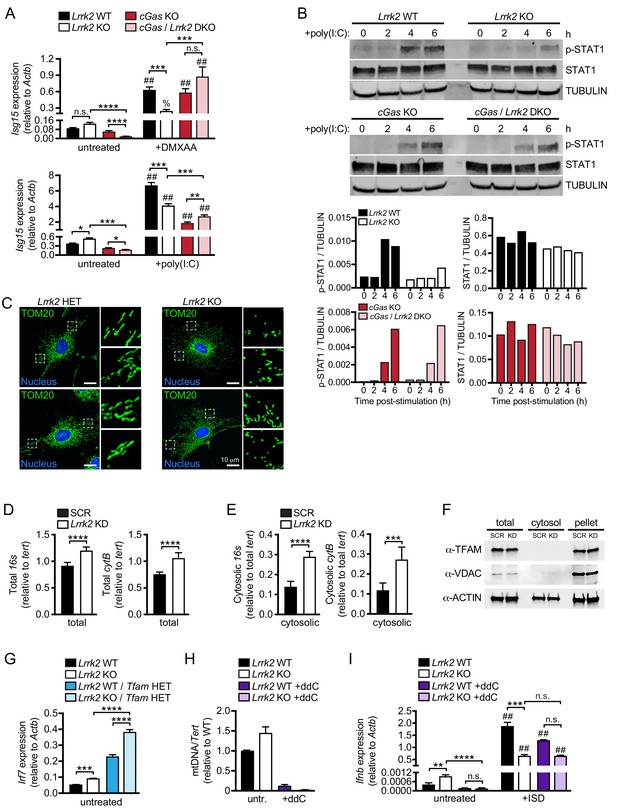
Cytosolic mtDNA drives basal type I IFN expression in Lrrk2 KO macrophages.
(A) Isg15 gene expression in Lrrk2 WT, Lrrk2 KO, cGas KO, and double KO (cGas/Lrrk2 DKO) BMDMs treated with 5 μg/ml DMXAA or transfected with 1 μg poly(I:C) for 4 hr. (B) Western blot analysis of STAT1 phosphorylation (Tyr701) in BMDMs from WT, Lrrk2 KO, cGas KO, and cGas/Lrrk2 double knockout (DKO) mice compared to total STAT1 with tubulin as a loading control. (C) Immunofluorescent images with anti-TOM20 antibody to visualize the mitochondrial network of Lrrk2 HET and KO MEFs. TOM20 (green), nucleus (DAPI, blue); Scale bar = 10 μm (D) qPCR of total 16s and cytB (mitochondrial DNA) relative to Tert (nuclear DNA). (E) As in (D) but cytosolic mitochondrial DNA. (F) Western blot of ACTIN, TFAM, and VDAC protein levels in total, cytosol, and pellet (organelle and membrane) fractions of Lrrk2 KD and SCR RAW 264.7 cells. (G) Irf7 gene expression normalized to Actb in untreated BMDMs from Lrrk2 WT, Lrrk2 KO, Tfam HET, and Lrrk2 KO/Tfam HET mice. (H) qPCR of dLoop (mitochondrial DNA) normalized to Tert (nuclear) to confirm mtDNA depletion in WT and Lrrk2 KO RAW 264.7 cells treated with 10 μM ddC for 4 days. (I) RT-qPCR of Ifnb gene expression in WT and Lrrk2 KO RAW 264.7 cells with or without ddC treatment, untreated and at 4 hr post-transfection with 1 μg/ml ISD. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); %p<0.05, ##p<0.001 (comparing stimulated to unstimulated of same genotype). (A and I) 3-way ANOVA, Tukey post-test; (D and E) two-tailed Student’s T test; (G and H) two-way ANOVA Tukey post-test.
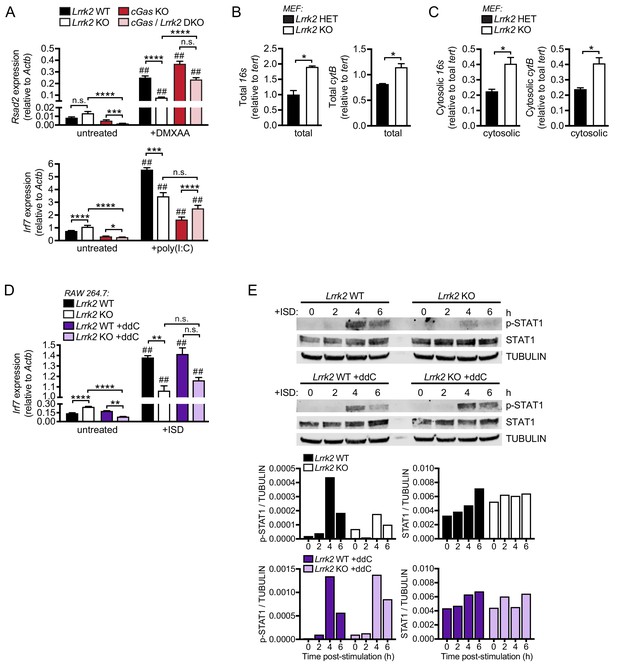
Higher levels of cytosolic mtDNA contribute to defective type I IFN responses in Lrrk2 KO macrophages.
(A) Rsad2 or Irf7 gene expression in Lrrk2 WT, Lrrk2 KO, cGas KO, and double KO (cGas/Lrrk2 DKO) BMDMs treated with 5 μg/ml DMXAA or transfected with 1 μg poly(I:C) for 4 hr. (B) qPCR of total 16s and cytB (mitochondrial DNA) relative to Tert (nuclear DNA) in Lrrk2 HET and KO MEFs. (C) As in (B) but measuring cytosolic mitochondrial DNA. (D) RT-qPCR of Irf7 gene expression in WT and Lrrk2 KO RAW 264.7 cells with or without ddC and transfected with 1 μg ISD for 4 hr. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); ##p<0.001 (comparing stimulated to unstimulated of same genotype). (A and D) 3-way ANOVA Tukey post-test; (B and C) two-tailed Student’s T test. *p<0.05, **p<0.01, ***p<0.005. (E) Western blot analysis of STAT1 phosphorylation (Tyr701) in BMDMs from wild-type and Lrrk2KO mice either untreated (top) or treated with ddC (bottom) at 0, 2, 4, and 6h post-ISD transfection. Total STAT1 and tubulin are shown as controls. Densitometry quantitation of shown under blots.
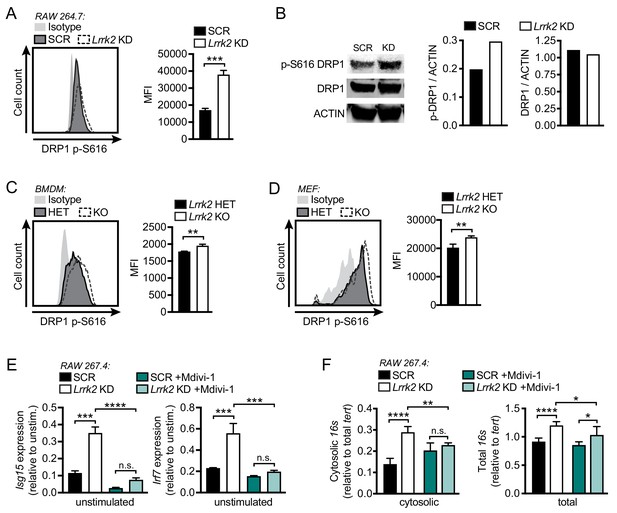
Mitochondrial fragmentation and increased DRP1 phosphorylation contribute to type I IFN dysregulation in Lrrk2 KO macrophages.
(A) Histograms showing counts of phospho-S616-DRP1 in SCR and Lrrk2 KD RAW 264.7 cells as measured by flow cytometry. (B) Western blot analysis and quantification of DRP1 phosphorylation (Ser616) in SCR and Lrrk2 KD RAW 264.7 cells compared to total DRP1 and actin as a loading control. (C) As in (A) but for BMDMs. (D) As in (A) but for MEFs. (E) Basal gene expression of Isg15 and Irf7 in SCR and Lrrk2 KD RAW 267.4 cells treated with Mdivi-1 50 μM for 12 hr. (F) qPCR of cytosolic and total 16s (mitochondrial DNA) relative to Tert (nuclear DNA) in SCR and Lrrk2 KD RAW 264.7 cells treated with 50 μM Mdivi-1 for 12 hr. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001. (A, C, and D) Two-tailed Student’s T test; (E and F) two-way ANOVA Tukey post-test.
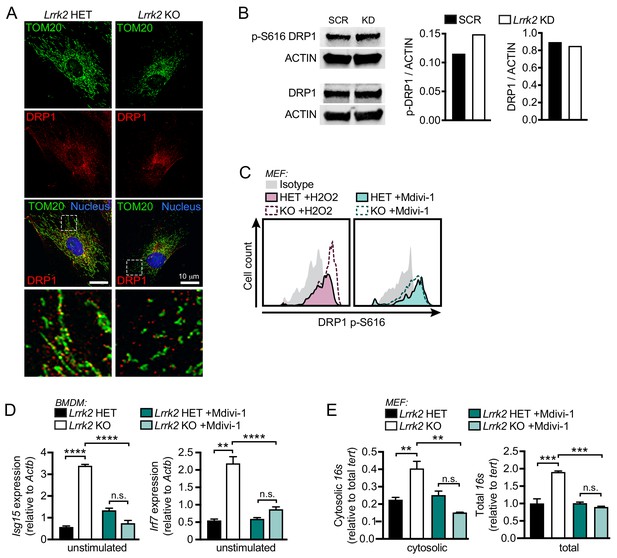
Hyperphosphorylation of DRP1 contributes to defects in type I IFN induction inLrrk2 KO macrophages.
(A) Representative immunofluorescence images of mitochondrial network TOM20 (green) and DRP1 (red) in MEFs from Lrrk2 HET and KO embryos. Nuclei are stained with DAPI (blue). Scale bar = 10 μm. (B) Western blot analysis and quantification of DRP1 phosphorylation (Ser616) in SCR and Lrrk2 KD RAW 264.7 cells compared to total DRP1 and actin as a loading control. (C) Histograms showing counts of phospho-S616-DRP1 in Lrrk2 HET and KO MEFs treated with 100 μM H2O2 or 50 μM Mdivi-1 for 4 hr. (D) RT-qPCR of Isg15 and Irf7 gene expression Lrrk2 KO and HET BMDMs treated with 50 μM Mdivi-1 for 12 hr. (E) qPCR of cytosolic and total 16s (mitochondrial DNA) relative to Tert (nuclear DNA) in Lrrk2 HET and KO MEFs treated with 50 μM Mdivi-1 for 12 hr. Statistical analysis: As in Figure 4; *p<0.05, **p<0.01, ***p<0.005.
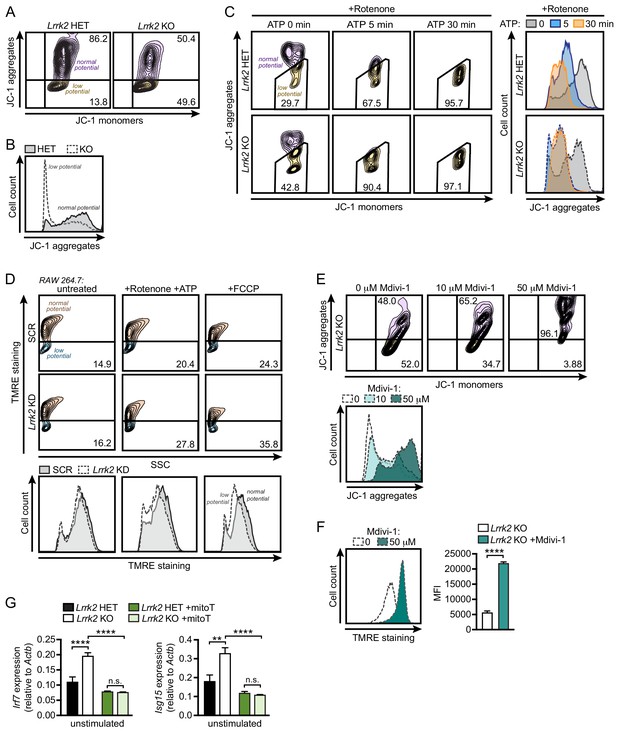
Lrrk2 KO macrophages are more susceptible to mitochondrial stress.
(A) Mitochondrial membrane potential in Lrrk2 HET and KO BMDMs as measured by JC-1 dye by flow cytometry. Aggregates (610/20) indicate normal mitochondrial membrane potential and monomers (520/50) indicate low membrane potential. (B) Histogram of (A) displaying cell counts of JC-1 aggregates for Lrrk2 HET and KO BMDMs. (C) JC-1 aggregates measured by flow cytometry in BMDMs treated for 3 hr with 2.5 μM rotenone followed by 5 μM ATP for 0, 5, or 30 min. Histogram of cell counts is on the right. (D) Flow cytometry of mitochondrial membrane potential measured by TMRE (585/15) in SCR and Lrrk2 KD RAW 264.7 cells treated for 3 hr with 2.5 μM rotenone followed by 5 μM ATP for 15 min or 50 μM FCCP for 15 min. (E) JC-1 aggregates measured by flow cytometry in Lrrk2 KO BMDMs treated with 10 or 50 μM Mdivi-1 for 4 hr. (F) The same as in (E) but with TMRE. (G) Basal gene expression of Irf7 and Isg15 in Lrrk2 HET and KO BMDMs treated overnight with 200 μM mitoTEMPO. JC-1 flow cytometry assays are representative of 3 independent experiments. Statistical analysis: **p<0.01, ***p<0.005, ****p<0.001. (F) Two-tailed Student’s T test; (G) two-way ANOVA Tukey post-test.
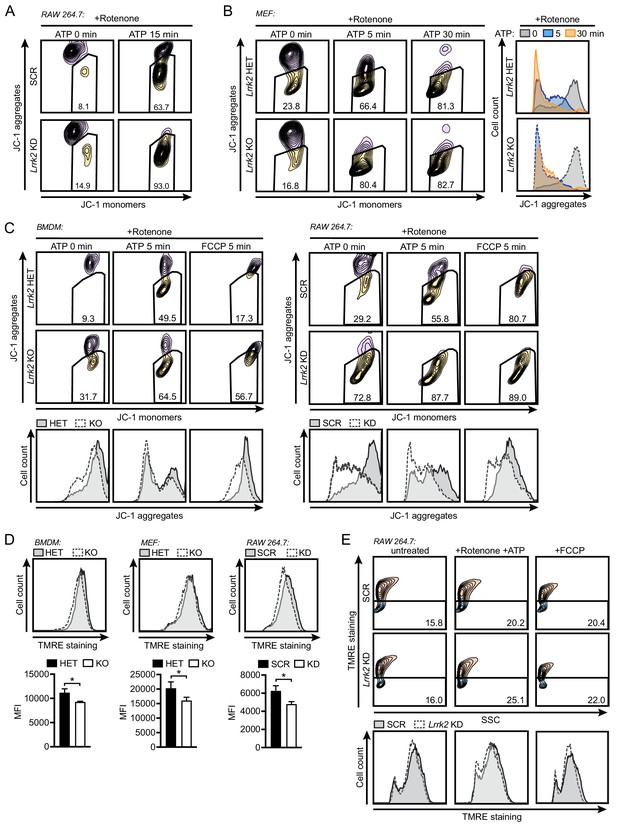
Lrrk2 KO macrophages are more prone in mitochondrial depolarization in response to mitochondrial stresses.
(A) Flow cytometry of SCR or Lrrk2 KD RAW 264.7 cells measuring mitochondrial membrane potential by JC-1 dye with 2.5 μM rotenone followed by 5 μM ATP for the indicated times. (B) As in (A) but with Lrrk2 HET and KO MEFs. Histogram of cell counts on the right. (C) As in (A) with 50 μM FCCP as a positive control in Lrrk2 HET and KO BMDMs (left) and SCR and Lrrk2 KD RAW 264.7 cells (right). Histogram of cell counts below. (D) Flow cytometry of mitochondrial membrane potential measured by TMRE (585/15) in BMDMs (left), MEFs (center), and RAW 264.7 cells (right). Histogram plots with quantifications below. (E) Flow cytometry of mitochondrial membrane potential measured by TMRE (585/15) in SCR or Lrrk2 KD RAW 264.7 cells treated with 2.5 μM rotenone followed by 5 μM ATP or 50 μM FCCP. JC-1 flow cytometry assays are representative of 3 independent experiments. Statistical analysis: As in Figure 5; *p<0.05.
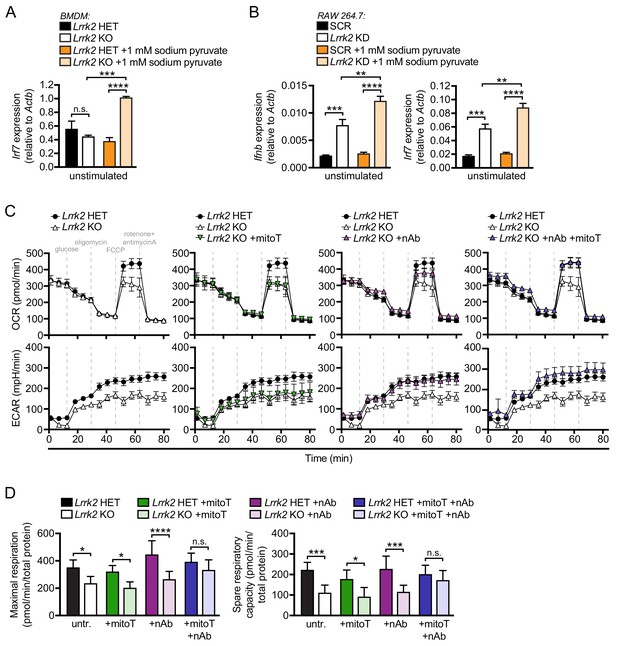
Lrrk2 KO macrophages are defective in oxidative phosphorylation and glycolysis.
(A) Irf7 gene expression in HET and Lrrk2 KO BMDMs cultured with or without 1 mM sodium pyruvate. (B) Ifnb and Irf7 gene expression in SCR and Lrrk2 KD RAW 264.7 cells cultured with or without 1 mM sodium pyruvate. (C) BMDMs from Lrrk2 HET and KO mice were treated with 200 μM mitoTEMPO, IFN-β blocking antibody, and the combination of both overnight followed by analysis of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measured using the Seahorse Metabolic Analyzer (Agilent). (D) Quantification of maximal respiration and spare respiratory capacity from (C). Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001. (A, B, and D) two-way ANOVA Tukey post-test.
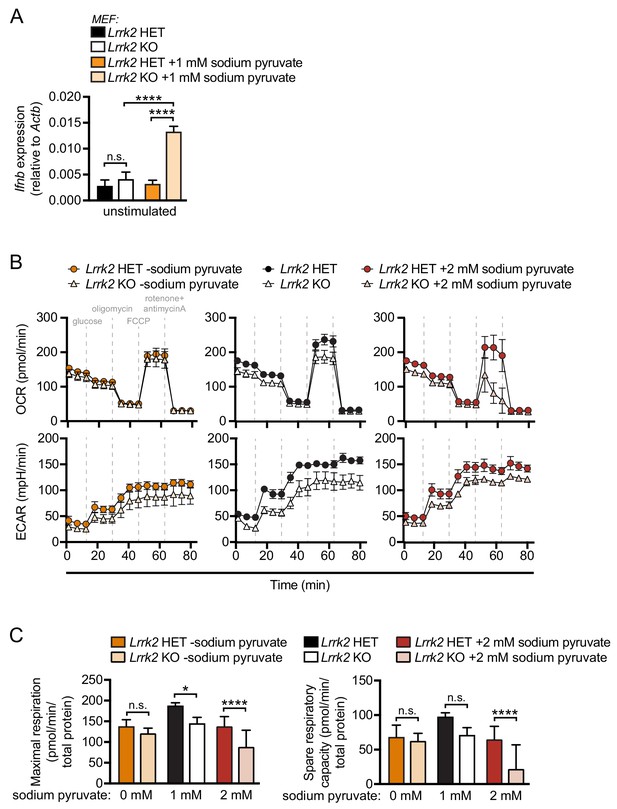
Increasing sodium pyruvate concentrations increases basal Ifnb expression and exacerbates metabolic defects in Lrrk2 KO macrophages.
(A) RT-qPCR of Ifnb expression in Lrrk2 HET and KO MEFs cultured with 1 mM sodium pyruvate for 24 hr. (B) Seahorse metabolic analysis of Lrrk2 HET and KO BMDMs treated with increasing concentrations of sodium pyruvate (0, 1, and 2 mM). (C) Quantitation of maximal respiration and spare respiratory capacity shown in (B). Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001. (A and C) two-way ANOVA Tukey post-test.
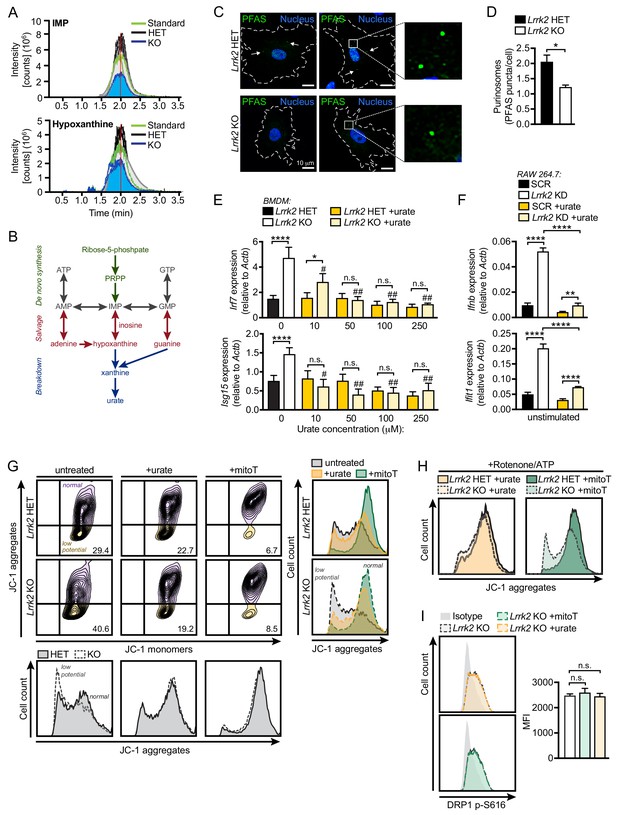
Reduced antioxidant pools in Lrrk2 KO macrophages result in mitochondrial stress.
(A) Chromatogram depicting targeted metabolomic analysis of Lrrk2 HET (n = 3) and KO BMDMs (n = 3) with pure molecular weight standard to IMP (top) and hypoxanthine (bottom). Replicate experiments are shown as individual lines (n = 2). Coefficient of variance (CV) for IMP = 8.8% (KO) and 21.7% (HET). CV for hypoxanthine = 9.1% (KO) and 14.0% (HET). (B) Diagram of key metabolites produced during purine metabolism oriented to the major steps of the pathway. De novo synthesis (green), salvage (red), breakdown (blue). (C) Representative immunofluorescence microscopy image of purinosome formation measured by PFAS puncta (green) in Lrrk2 HET and KO MEFs. Nuclei stained with DAPI (blue). (D) Quantification of number of PFAS puncta per cell. 100 cells were counted per coverslip from three coverslips. (E) RT-qPCR of Irf7 and Isg15 gene expression in Lrrk2 HET and KO BMDMs treated with increasing concentrations of urate (10, 50, 100, and 250 μM for 24 hr). (F) Basal gene expression of Ifnb and Ifit1 in SCR and Lrrk2 KD RAW 264.7 cells treated with 250 μM urate overnight. (G) JC-1 aggregate vs. monomer formation measured by flow cytometry in Lrrk2 HET and KO BMDMs treated with 100 μM urate or 200 μM mitoTEMPO overnight. Histograms shown below and merged histograms shown to the right. (H) Histograms of Lrrk2 HET and KO BMDMs treated with 100 μM urate or 200 μM mitoTEMPO overnight and then treated with 2.5 μM rotenone for 3 hr followed by 5 μM ATP for 15 min. (I) Histograms of DRP1 p-S616 flow cytometry analysis for Lrrk2 KO BMDMs following treatment with 100 μM urate or 200 μM mitoTEMPO. Quantification is shown on the right. JC-1 flow cytometry assays are representative of three independent experiments. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); #p<0.005, ##p<0.001 (comparing treated to untreated of same genotype). (D) Two-tailed Student’s T test; (E and F) two-way ANOVA Tukey post-test; (I) one-way ANOVA Tukey post-test.
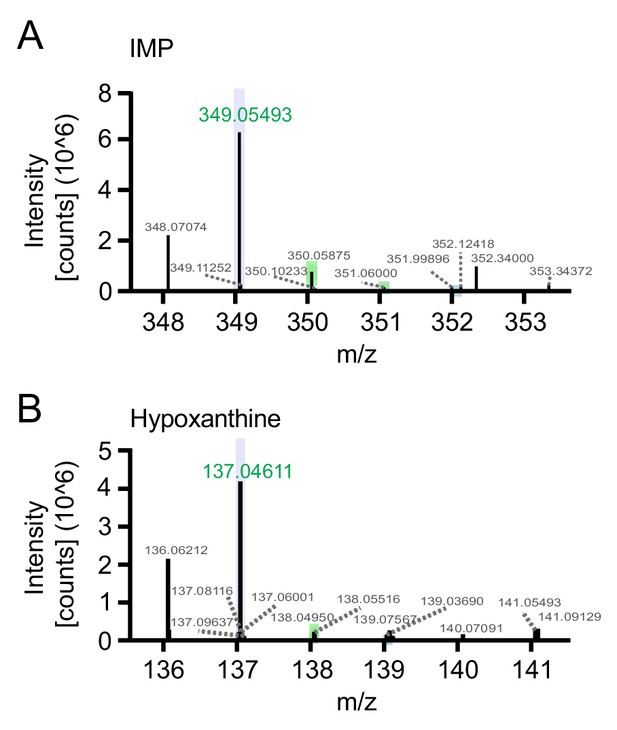
LC-MS/MS analysis identifies lower levels of IMP and hypoxanthine in Lrrk2 KO macrophages.
(A) LC-MS/MS analysis of BMDMS from Lrrk2 HET and KO mice showing IMP peak. (B) As in (A) but for hypoxanthine.
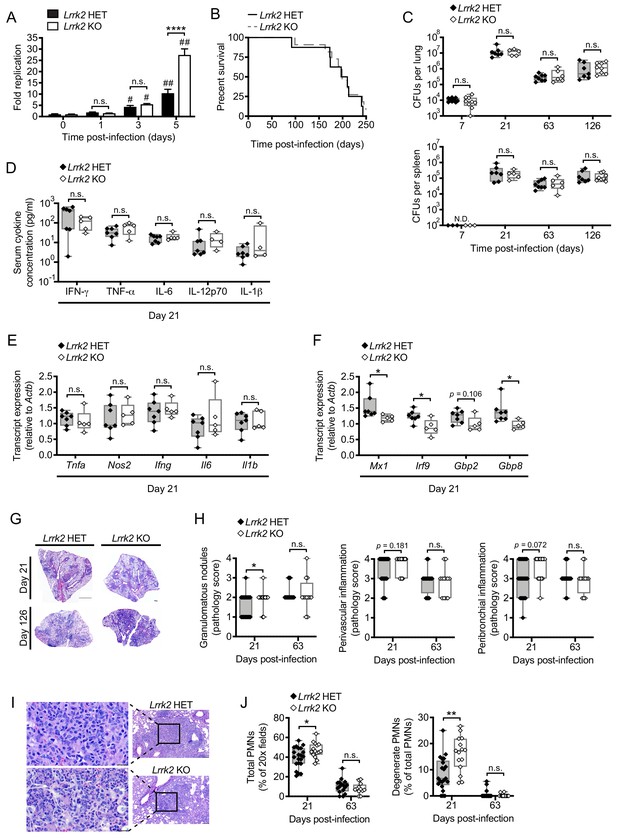
Lrrk2 KO mice exhibit increased lung inflammation during Mtb infection.
(A) Mtb colony forming units (CFUs) recovered from Lrrk2 HET and KO BMDMs over the course of 5 days (MOI = 1). (B) Survival curves for Lrrk2 HET (n = 8) and KO (n = 11) mice over a 250 day Mtb infection. Survival times not statistically different based on log-rank Mantel-Cox test. (C) CFUs recovered from lungs and spleens of Mtb-infected Lrrk2 HET and KO mice at Day 7, 21, 63, and 126 post-infection. (D) Circulating serum cytokines measured at Day 21 in Lrrk2 HET and KO mice. (E) RT-qPCR of inflammatory cytokines from total RNA recovered from lung homogenates from Day 21 Mtb-infected Lrrk2 HET and KO mice. (F) As in (E) but detecting ISGs. (G) Hematoxylin and eosin (H&E) stain of inflammatory nodules in the lungs of Lrrk2 KO and HET mice 21 days after infection with Mtb. Small scale bar, 500 μm; large scale bar 1 mm. (H) Semi-quantitative score of pulmonary inflammation with a score of 0, 1, 2, 3 or 4 assigned based on granulomatous nodules in none, up to 25%, 26–50%, 51–75% or 76–100% of fields, respectively. Perivascular and peribronchial inflammation was scored using an analogous scale based on percentage of medium-caliber vessels or bronchioles with adjacent inflammatory nodules. (I) H&E stain of neutrophils within an inflammatory nodule in the lung of Lrrk2 HET and KO mice 21 days after infection with Mtb. Left panel bar is 20 μm. Right panel bar is 200 μm. (J) Quantification of neutrophils in the lungs of Lrrk2 HET and KO mice infected with Mtb for 21 or 63 days. Total neutrophil scores were determined by the percentage of fields of view at 20X magnification containing neutrophils. Degenerate neutrophil scores were determined by the percentage of PMN positive fields containing degenerate neutrophils. Statistical analysis: *p<0.05, **p<0.01, ***p<0.005, ****p<0.001 (comparing indicated data points); #p<0.005, ##p<0.001 (comparing infected to uninfected of same genotype). (A) Two-way ANOVA Tukey post-test; (B) Mantel-Cox log-rank; (C–J) Mann-Whitney test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Lrrk2) | Lrrk2; LRRK2 | NA | MGI:1913975 | |
| Genetic reagent (Mycobacterium tuberculosis) | Mtb; Erdman | Watson et al., 2015; Watson et al., 2012 | ||
| Genetic reagent (Mycobacterium leprae) | Mlep | National Hansen's Disease Program | ||
| Genetic reagent (Mus musculus) | Lrrk2 KO; C57BL/6-Lrrk2tm1.1Mjff/J | Jackson Labs | 16121 Lrrk2 KO | |
| Genetic reagent (Mus musculus) | Ifnar KO; B6(Cg)-Ifnar1tm1.2Ees/J | Jackson Labs | 28288 Ifnar1 KO | |
| Genetic reagent (Mus musculus) | Tfam HET | obtained from A. P. West TAMHSC | Tfam HET | doi: 10.1016/j.ajpath.2011.10.003 |
| Genetic reagent (Mus musculus) | Cgas KO; B6(C)-Cgastm1d (EUCOMM)Hmgu/J | obtained from A. P. West TAMHSC | cGAS KO | |
| Cell line (Mus musculus) | RAW 264.7 Lrrk2 Parental; WT | ATCC | ATCC SC-6003 | |
| Cell line (Mus musculus) | RAW 264.7 Lrrk2 KO | ATCC | ATCC SC-6004 | |
| Cell line (Mus musculus) | RAW 264.7 | ATCC | ATCC TIB-71 | Cell line maintained in the Watson lab |
| Cell line (Homo sapiens) | U937 | ATCC | ATCC CRL-1593.2 | Cell line maintained in the Watson lab |
| Antibody | anti-pSTAT1 Rabbit monoclonal | Cell Signaling | (Tyr701) (58D6) #9167 | (1:1000 WB) |
| Antibody | anti-STAT1 Rabbit monoclonal | Cell Signaling | (D4Y6Z) #14995 | (1:1000 WB) |
| Antibody | anti-pIRF3 Rabbit monoclonal | Cell Signaling | (Ser396) (4D4G) #4947 | (1:1000 WB) |
| Antibody | anti-IRF3 Rabbit monoclonal | Cell Signaling | (D83B9) #4302 | (1:1000 WB) |
| Antibody | anti-Beta tubulin Rabbit polyclonal | Abcam | ab15568 | (1:5000 WB) |
| Antibody | anti-pDRP1 Rabbit polyclonal | Cell Signaling | (Ser616) #3455 | (1:75 FC), (1:1000 WB) |
| Antibody | anti-DRP1 Rabbit monoclonal | Abcam | ab184247 | (1:1000 WB) (1:200 IF) |
| Antibody | anti-IFNB Rabbit polyclonal | PBL Assay Science | 32400–1 | (1:250 neutralizing) |
| Antibody | anti-PFAS Rabbit polyclonal | Bethyl Laboratories | A304-219A | (1:200 IF) |
| Antibody | Anti-TOM20 Mouse monoclonal | Millipore, via A.P. West lab TAMHSC | MABT166 | (1:200 IF) |
| Antibody | anti-TFAM Rabbit polyclonal | Millipore, via A.P. West lab TAMHSC | ABE483 | (1:1000 WB) |
| Antibody | anti-VDAC Rabbit polyclonal | Protein Tech, via A.P. West lab TAMHSC | 55259–1-AP | (1:1000 WB) |
| Antibody | Goat anti Rabbit IgG | Licor | IRDye 800CW | (1:10000 WB) |
| Antibody | Goat anti Rabbit IgG | Licor | IRDye 680CW | (1:10000 WB) |
| Antibody | Goat anti Rabbit IgG AF 488 | Invitrogen | A32731 | (1:500 FC) |
| Sequence-based reagent | interferon stimulatory DNA; ISD | IDT | annealed in house | (1 ug/ml) |
| Sequence-based reagent | CpG 2395 | Invivogen | tlrl-2395 | (1 uM) |
| Peptide, recombinant protein | recombinant IFNB | PBL Assay Science | 12405–1 | (200 IU/mL) |
| Commercial assay or kit | Seahorse XF mito stress kit | Agilent | 103708–100 | |
| Commercial assay or kit | Direct-zol RNA mini prep | Zymo research | R2052 | |
| Commercial assay or kit | Mouse Cytokine Array | Eve Technologies | MD13 panel | |
| Chemical compound, drug | thioglycollate | Fisher Scientific | BD 211716 | |
| Chemical compound, drug | DMXAA | Invivogen | tlrl-dmx | (5 ug/ml) |
| Chemical compound, drug | urate | Sigma Aldrich | U2625 | |
| Chemical compound, drug | IMP | Sigma Aldrich | 57510 | |
| Chemical compound, drug | hypoxanthine | Sigma Aldrich | H9377 | |
| Chemical compound, drug | FCCP | Sigma Aldrich | C2920 | (50 uM) |
| Chemical compound, drug | CLO97 | Invivogen | tlrl-c97 | (100 ng/ml) |
| Chemical compound, drug | JC1 dye | Thermofisher | T3168 | (1 uM) |
| Chemical compound, drug | TMRE | Thermofisher | T669 | (25 ng/ml) |
| Chemical compound, drug | ATP | Invivogen | tlrl-atpl | (5 uM) |
| Chemical compound, drug | rotenone | Sigma Aldrich | R8875 | (2.5 uM) |
| Chemical compound, drug | mitoTEMPO | Santa Cruz Biotechnology | (CAS 1569257-94-8) | (200 uM) |
Additional files
-
Supplementary file 1
RNA-seq analysis of Lrrk2 KO and HET BMDMs.
Fold change and p-values separated into tabs. Tab 1: Lrrk2 HET vs. KO uninfected all genes fold change and p-value. Tab 2: Fold change values for genes shown in Figure 1A heatmap. All p-values<0.05. Tab 3: Lrrk2 HET vs. KO +Mth (4 hr time point) all genes fold change and p-value. Tab 4: Fold change values for genes shown in Figure 1C heatmap. All p-values<0.05. Tab 5: Fold change and p-values for all pairwise comparisons of RNA-seq data. Tab 6: Fold change and p-values for genes shown in Figure 1—figure supplement 1A heatmap.
- https://cdn.elifesciences.org/articles/51071/elife-51071-supp1-v2.xlsx
-
Supplementary file 2
LC/MS/MS metabolite identification.
Tab 1: All compounds identified and their relative amounts (group area) for each genotype (Lrrk2 KO and HET). Tab 2: Data for metabolites highlighted in the manuscript (IMP, hypoxanthine, oxidized glutathione).
- https://cdn.elifesciences.org/articles/51071/elife-51071-supp2-v2.xlsx
-
Supplementary file 3
Summary of statistical analyses.
Statistical test, n, and p-value for each data point in manuscript. Tabs labeled according to Figure.
- https://cdn.elifesciences.org/articles/51071/elife-51071-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51071/elife-51071-transrepform-v2.docx





