Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18
Figures
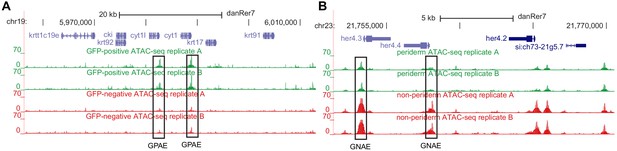
Identification of zebrafish GFP-positive active enhancers (zGPAEs) by integrating ATAC-seq and H3K27Ac ChIP-seq.
(A) Transverse section of an 11 hpf (4-somite stage) Tg(krt4:gfp) embryo, showing GFP is confined to the superficial layer of cells, and workflow of ATAC-seq in periderm and non-periderm cells. (B) Density plots of ATAC-seq results. Each line is centered on a nucleosome free region (NFR) with significantly more ATAC-seq reads in GFP-positive or GFP-negative cells; the majority of ATAC-seq peaks were not enriched in either cell type. Density plots also show H3K27Ac ChIP-seq signal in whole embryos at eight hpf and/or at 24 hpf data from Bogdanovic et al. (2012) at each of the GFP-positive NFRs; the latter are sorted in to those that overlap (or are flanked by within 100–1500 bp) peaks of H3K27Ac signal (cluster 1, 4301 elements) and or not (cluster 2, 7952 elements). (C) UCSC Genome browser tracks showing the ATAC-seq peaks in GFP-positive and GFP-negative cells, and H3K27Ac signal from whole embryos at eight hpf and at 24 hpf data from Bogdanovic et al. (2012) at the cldne locus. Boxes, examples of cluster one elements, also known as zebrafish GFP-positive active enhancers (zGPAEs). Elements are cldne+6 kb (zv9 : chr15:2625460–2625890), cldne +3 kb (chr15:2629012–2629544), cldne −8 kb (chr15:2639873–2640379), cldne −11 kb (chr15:2643578–2644160), and cldne TSS (chr15:2631981–2632513). (D) Plot of average density of H3K27Ac ChIP-seq signal (purple) and ATAC-seq signal (green). (E) GO enrichment for term ‘Gastrula:Bud 10–10.33 hr; periderm’ among NFRs enriched in GFP-positive cells with normalized fold change greater than 2 (ATAC(FC >2)) and 4 (ATAC(FC >4)), NFRs enriched in GFP-positive cells flanked or overlapped by 24hpf and 80% epiboly H3K27Ac ChIP-seq peaks (cluster 1) and or not (cluster 2), NFRs enriched in GFP-positve cells flanked or overlapped by 24hpf and 80% epiboly H3K4me1 ChIP-seq peaks (cluster 1) or not(cluster 2), NFRs enriched in GFP-positive cells flanked or overlapped by 24hpf H3K27Ac ChIP-seq peaks (cluster 1) or not (cluster 2), and NFRs enriched in GFP-positive cells flanked or overlapped by 80% epiboly H3K27Ac ChIP-seq peaks (cluster 1) or not (cluster 2). (F), (G) Lateral views of wild-type embryos at 11 hpf injected at the 1-cell stage with GFP reporter constructs built from (F) cldne +6 and (G) cldne transcription start site (TSS) elements. Left panels are stack views of the embryo, and right panels are surface plot for the embryos indicating most GFP signal is from the surface (periderm) of the embryos. Number in parentheses is the ratio of embryos with at least 10 GFP-positive periderm cells over injected embryos surviving at 11 hpf. (H) Volcano plot of RNA seq data, showing the expression of genes associated (by GREAT) with zGPAEs (green dots) or with zGNAEs (pink dots) in GFP-positive cells (beta-value >0) or in GFP-negative cells (beta-value <0). (I) Plot of accessibility scores of elements with differential accessibility (i.e., both zGPAEs and zGNAEs) associated with genes that are differentially expressed in GFP-positive and GFP-negative cells, showing that elements with increased accessibility in GFP-positive cells tend to be associated with genes whose expression is enriched in GFP-positive cells, and vice versa.
-
Figure 1—source data 1
Density plot for ATAC-seq and H3K27Ac ChIP-seq, as plotted in Figure 1D.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig1-data1-v2.csv
-
Figure 1—source data 2
Barchart for GO enrichment, as plotted in Figure 1E.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Scatter plot for the genes near GPAEs and GNAEs, as plotted in Figure 1H.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig1-data3-v2.csv
-
Figure 1—source data 4
Box plot for the normalized chromatin accessibility of periderm- and non-periderm enriched genes in GFP positive or negative cells, as plotted in Figure 1I.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig1-data4-v2.csv
Correlation of zebrafish periderm ATAC-seq two biological replicates.
(A) ATAC-seq summit centered heatmap of ATAC-seq signals from two biological replicates. (B) Scatter plots showing the ATAC-seq signal correlation between two biological replicates.

Annotation of ATAC-seq peaks relative to transcription start sites.
(A) Histogram of read density of ATAC-seq in 10 kb flanking transcription start sites (TSS). (B) Pie chart showing the genomic location of GFP-positive NFRs (from ATAC-seq biological replicate 1).

Average Vertebrate PhastCons Score (danRer7 genome) at different distances from the center of nucleosome free regions (NFRs) in GFP-positive and GFP-negative (flow through) cells sorted from Tg(krt4:gfp) embryos at 11 hpf.
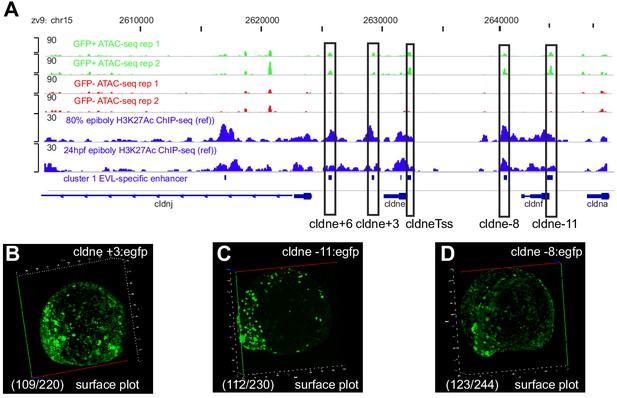
Transient reporter assay validation for cldne +3, cldne −11, and cldne −8 elements.
(A) Genome browser screenshot for cldne +3, cldne −11, and cldne −8 elements. (B–D) Surface plots for wild-type embryos at 11 hpf injected at the 1 cell stage with GFP reporter constructs for cldne +3, cldne −11, and cldne −8 elements. Number in parentheses is the ratio of embryos with at least 10 GFP-positive periderm cells over injected embryos surviving at 11 hpf.
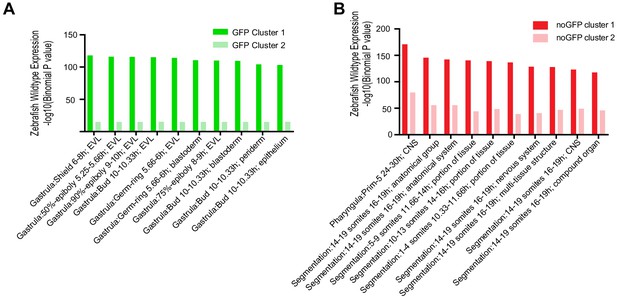
GO enrichment analysis for different clusters of GFP-positive or GFP-negative specific NFRs.
(A) Barchart showing GO enrichment analysis for two clusters of GFP-positive specific NFRs. (B) Barchart showing GO enrichment analysis for two clusters of GFP-negative specific NFRs.
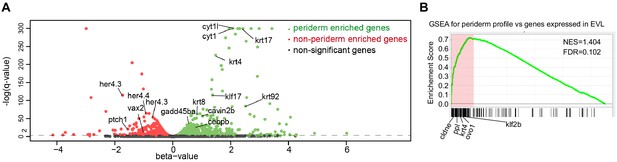
Summary for RNA-seq for krt4:GFP-positive and krt4:GFP-negative cells at 4-somite stage.
(A) Volcano plot for genes expressed in GFP-positive (in green) and –negative (in red) cells. (B) GSEA for genes expressed in GFP-positive cells using EVL gene set (www.zfin.org).
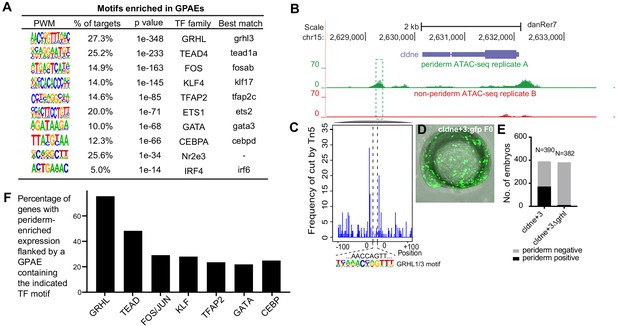
Features of zGPAEs.
(A) Enriched motifs in zGPAEs. PWM, position weighted matrix. TF, transcription factors. Best match, transcription factor in the indicated family with highest expression in GFP-positive cells, whether or not the expression is enriched in GFP-positive cells in comparison to GFP-negative cells. (B) Genome browser view showing a GFP-positive nucleosome free region (NFR) about 3 kb downstream of the transcription start site of cldne gene. (C) Schematic of frequency of Tn5 cleavage sites at within this NFR, indicating reduced frequency of cleavage at a motif matching the GRHL binding site relative to in flanking DNA. (D) Confocal image of a wild-type embryo at 10 hpf (2-somite stage) injected at the one-cell stage with a reporter construct containing this NFR. (E) Bar chart showing number of embryos positive for GFP signal in the periderm after being injected with the intact reporter or one in which the GRHL motif was deleted. (F) Bar chart showing the percentage of genes whose expression is higher in GFP-positive cells than in GFP-negative cells that are flanked by a zGPAE possessing the indicated binding site.

Different clusters of H3K27Ac ChIP-seq at different developmental stages in zGPAEs.
(A) ATAC-seq summit centered heatmap of H3K27Ac ChIP-seq at 4.5 hpf, 8hpf and 24hpf data from Bogdanovic et al. (2012), cluster performed by k-mean. (B) Motif enriched in zGPAEs with high H3K27Ac at 4.5hpf. (C) Motifs enriched in zGPAEs with high H3K27Ac at 24 hpf.
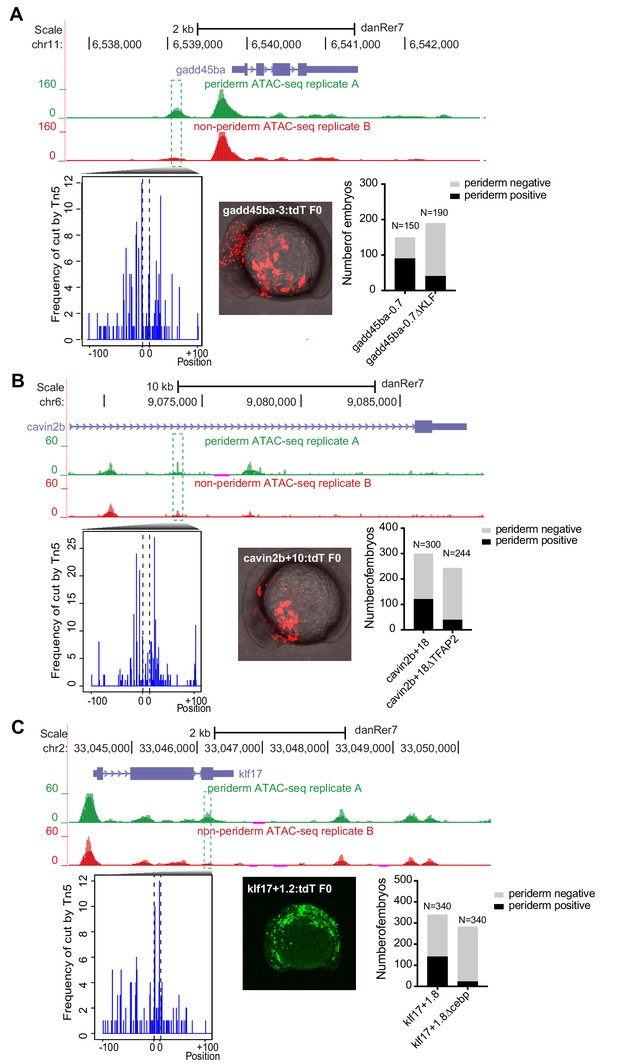
Transient reporter assay of (A) gadd45ba-3 with or without KLF motif, (B) cavin2b-+18 with or without TFAP2 motif and (C) klf17-+1.2 with or without C/EBP motif.

Putative regulatory interactions of major periderm-enriched transcription factors governing transcriptomic state in periderm cells at 4-somite stage.
Depending on the expression level in periderm cells (GFP-positive cells) most enriched transcription factors with the relevant motifs are in hexagon while other enriched transcription factors with the relevant motifs are in round. Each TF node is colored according to the normalized expression z-score (related to periderm genes). The thickness of each edge represents the number of motifs located in the all nearby enhancers to each TF (within 100kbp to the transcription start site).
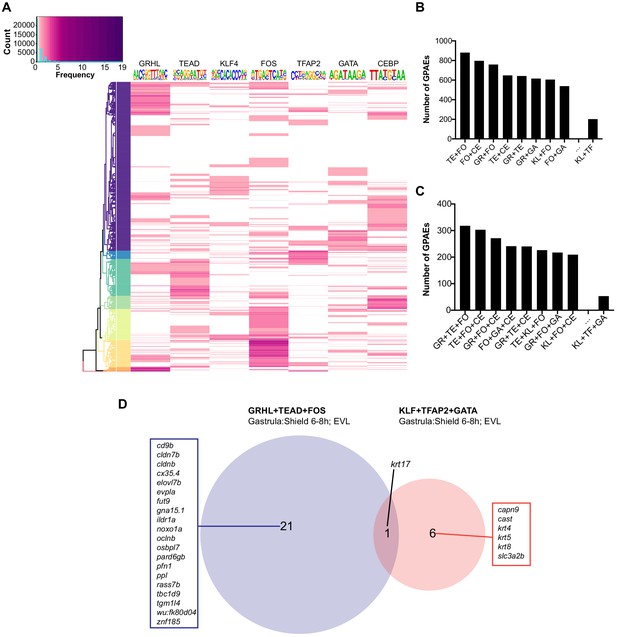
Motif combination in GPAEs.
(A) Hierarchy clustering for the number of enriched motifs in all GPAEs. ‘Count’ in the color key indicates the sum for different number of each motif ‘frequency’. (B) Bar chart for the number of GPAEs with different two-motif combination. (C) Bar chart for the number of GPAEs with different three-motif combination. (D) Nearest EVL genes (within 100.0 kbp) of the GPAEs with ‘GRHL+TEAD+FOS’ and ‘KLF+TFAP2+GATA’ combination. GR: GRHL, TE: TEAD, FO: FOS, TF: TFAP2, GA: GATA, CE: CEBP, KL: KLF.
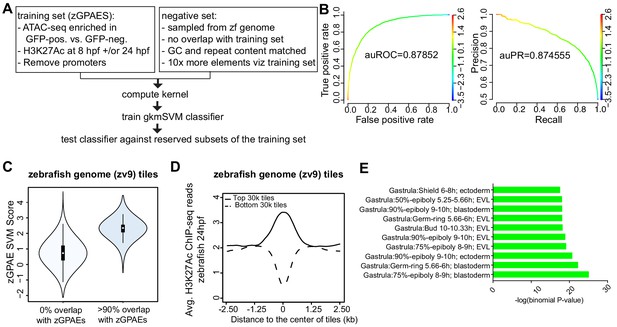
Training a gapped kmer support vector machine (gkmSVM) classifier trained on zGPAEs.
(A) Pipeline for training and cross-validation of gkmSVM classifier on zebrafish periderm enhancer candidates. (B) Receiver Operating Characteristic (ROC) and Precision-Recall (PR) curves using the gkmSVM trained on zGPAEs. au, area under. Color of curves corresponds to SVM scores. (C) Violin plots showing SVM scores of zebrafish genome tiles with 0% or at least 90% overlapped with the training set (GPAEs). (D) Average H3K27Ac ChIP-seq reads at the 30,000 elements with the highest or lowest scores from the gkmSVM trained on zGPAEs. (E) GO enrichment assay for genes associated with the top-scoring tiles 10,000 tilesincluding those that overlap the training set.
-
Figure 3—source data 1
Density plot for H3K27Ac ChIP-seq reads, as plotted in Figure 3D.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig3-data1-v2.csv
-
Figure 3—source data 2
Barchart for GO enrichment assay, as plotted in Figure 3E.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig3-data2-v2.xlsx

GO enrichment assay of gene expression for the top-scoring 10 K tiles that do not overlap zGPAEs.
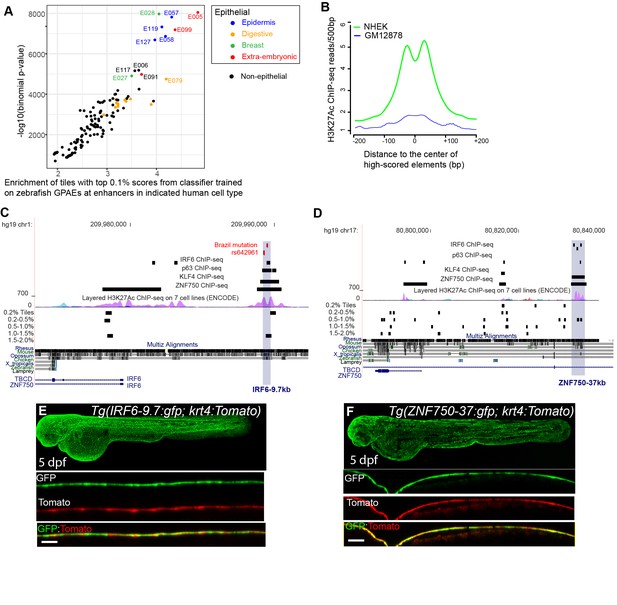
A classifier trained on zGPAEs applied to the human genome.
(A) Enrichment of human genome tiles that receive a top 0.1% bin score using a zGPAE-trained classifier at enhancers active in the indicated cell type, as revealed by ChIP-seq to chromatin marks in the Roadmap Epigenomics project (Visel et al., 2008). -Such tiles are significantly enriched within a variety of epithelial enhancers. [E05: H1 BMP4 Derived Trophoblast Cultured Cells; E027: Breast Myoepithelial Primary Cells; E028: Breast variant Human Mammary Epithelial Cells; E057, E058: Foreskin Keratinocyte Primary Cells; E079: Esophagus; E091: Placenta; E099: Placenta amnion; E119, Mammary Epithelial Primary Cells (HMEC); E127:NHEK-Epidermal Keratinocyte Primary Cells]. (B) Average density of H3K27Ac ChIP-seq signal in NHEK and GM12878 cells (Visel et al., 2008) at top 0.1% tiles using a zGPAE-trained classifier. (C) Genome browser view focused on IRF6-9.7, also known as IRF6 multispecies conserved sequence 9.7 (MCS9.7) (hg19 chr1:209989050–209989824). A SNP within it, rs642961 (chr1: 209989270), is associated with risk for non-syndromic orofacial cleft. Brazil mutation refers to a rare mutation reported in a patient with Van der Woude syndrome (Fakhouri et al., 2012). This element has peaks of H3K27Ac, IRF6, TP63, and KLF4 ChIP-seq in normal human keratinocytes. Multiz Alignments of 100 vertebrate species shows it is conserved among mammals but not in zebrafishNonetheless it possesses tiles in the top 1.0-1.5% and 0.2% bins using a zGPAE-trained classifier (D) Genome browser view focused on ZNF750-37 (hg19 chr17:80832267–80835105). This element has similar ChIP-seq signature as IRF6-9.7, and like it is not overtly conserved to fish but posseses high-scoring tiles using the zGPAE-trained classifier. (E) GFP expression pattern of Tg(IRF6-9.7:gfp; krt4:Tomato) at five dpf. (F) GFP expression pattern of Tg(ZNF750-37:gfp; krt4:Tomato) at five dpf. Both of these human non-coding elements have periderm enhancer activity in zebrafish embryos.
-
Figure 4—source data 1
Scatter plot for the enrichment of top scoring human genome tiles, as plotted in Figure 4A.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig4-data1-v2.csv
-
Figure 4—source data 2
Density plot for H3K27Ac ChIP-seq in NHEK and GM12878 cells within the top scoring human genome ties, as plotted in Figure 4B.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig4-data2-v2.csv
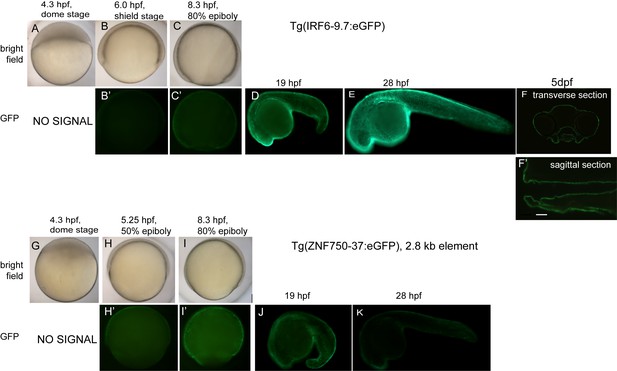
Detailed description of enhancer activity pattern of Tg(IRF6-9.7:gfp) and Tg(ZNF750-37:gfp).
(A–K) Lateral views of (A–C, G–I) bright field or (B’, C’, D, E, H’, I’, J, K) epifluorescence images of (A–F) Tg(IRF6-9.7:gfp) or (G–K) Tg(ZNF750-37:eGFP) transgenic zebrafish embryos at the indicated stage. Both strains exhibit GFP expression in the periderm. (F) Transverse and (F’) sagittal sections Tg(IRF6-9.7:gfp) larvae at 5 dpf showing GFP expression in oral epithelium.
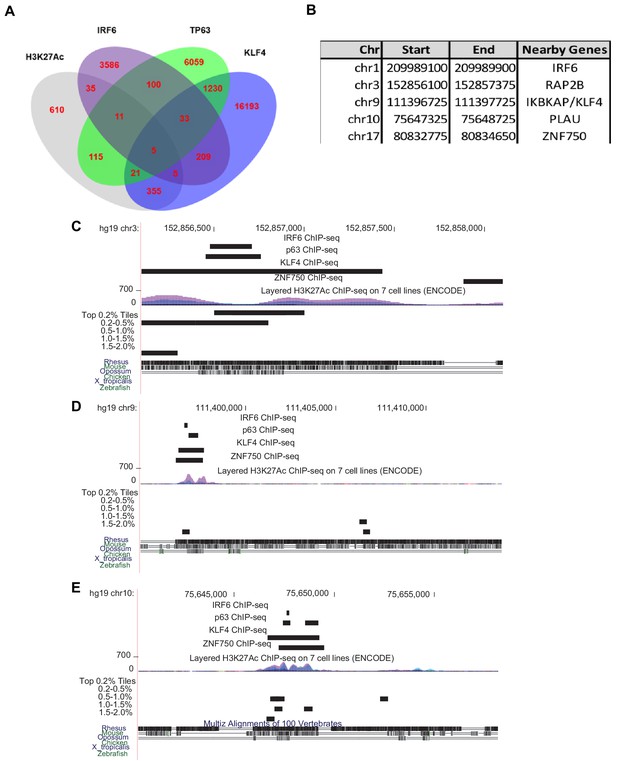
Browser views of all loci with mcs9.7 ChIP-seq features.
(A) Intersection of TP63, IRF6, KLF4 and H3K27Ac ChIP-seq peaks in human NHEK cells. (B) Coordinates for five genomic regions with overlapped TP63, IRF6, KLF4 and H3K27Ac ChIP-seq peaks. (C–E) Genome browser view for regions sharing this feature near RAP2B, KLF4, and PLAU.
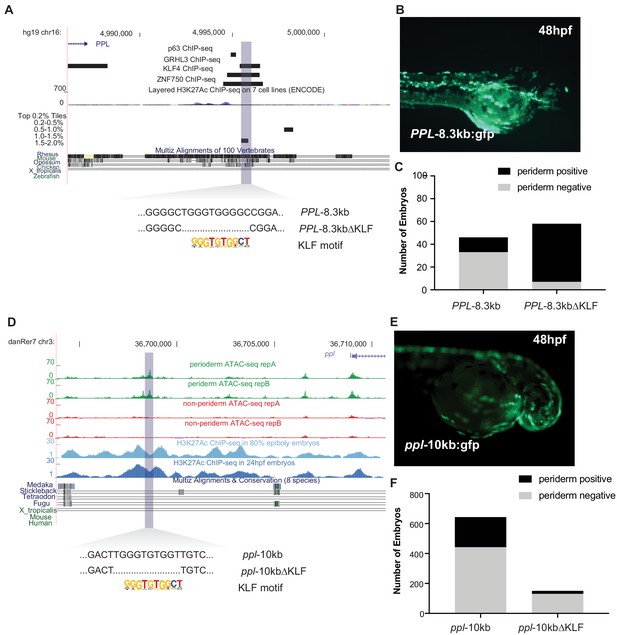
Reporter assay for human and zebrafish PPL elements predicted by zebrafish classifier.
Transient transgenic reporter assays for enhancer candidates near human PPL and zebrafish ppl. (A) Genome browser (hg19) view for PPL -8.3kb. (B) Lateral view, anterior to the right, of an embryo at 48 hpf injected at the one cell stage with the PPL-8.3kb:gfp reporter construct. (C) Bar chart showing number of embryos with 10 or more GFP-positive periderm cells injected with the indicated construct. PPL-8.3kb:DKLF: in the enhancer element, the motif matching the KLF4 binding site has been deleted. (D) Zebrafish genome (danRer 7) browser view for ppl-10kb (E) Lateral view, anterior to the right, of an embryo at 48 hpf injected at the one cell stage with the ppl-10kb:gfp reporter construct. (F) Bar chart showing number of embryos with 10 or more GFP-positive periderm cells injected with the indicated construct. ppl-810kb:DKLF: in the enhancer element, the motif matching the KLF4 binding site has been deleted.
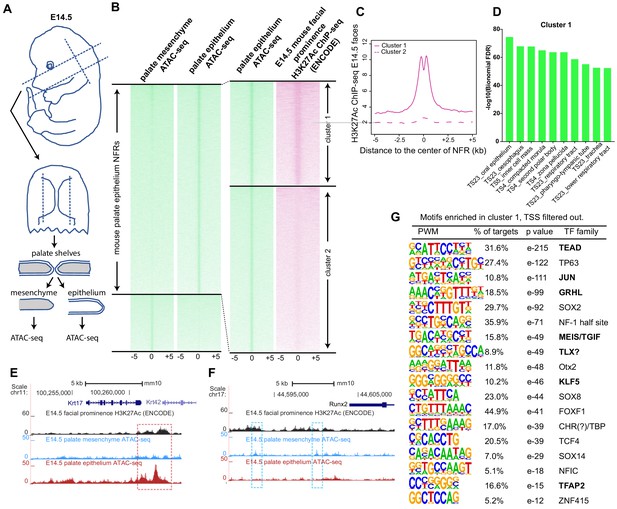
Identification of mouse embryonic palatal epithelium-specific active enhancers.
(A) Workflow of ATAC-seq in epithelium and mesenchyme cells isolated from palate shelves dissected from E14.5 embryos. (B) Heatmap plots of ATAC-seq and E14.5 mouse facial prominence H3K27Ac ChIP-seq (Klein and Andersen, 2015) in tissue-specific NFRs. (C) Plot of average density of H3K27Ac ChIP-seq signal, showing higher signal at cluster 1 elements than cluster 2 elements. (D) GO enrichment (MGI mouse gene expression pattern) of genes associated with cluster 1 elements. (E and F) UCSC Genome browser views of the mouse genome (mm10 build) showing the ATAC-seq and H3K27Ac ChIP-seq signals near the Krt17 and Runx2 genes. Red box, an example of a mouse palate-epithelium active enhancer (mPEAE). Blue boxes, examples of mouse palate mesenchyme active enhancers (mPMAEs). (G) Motifs enriched in cluster 1 of E14.5 palate-epithelium specific NFRs with elements overlying transcription start sites removed (i.e., mPEAEs). Motifs shared with zGPAEs are in bold.
-
Figure 5—source data 1
Density plot for H3K27Ac ChIP-seq in two clusters, as plotted in Figure 5C.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig5-data1-v2.csv
-
Figure 5—source data 2
Barchart for GO enrichment, as plotted in Figure 5D.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig5-data2-v2.xlsx
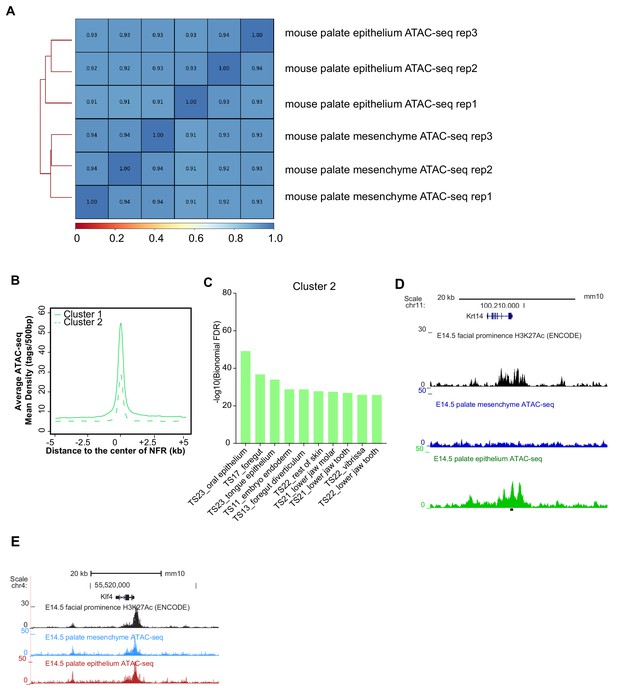
Concordance of replicates of mouse embryonic palatal epithelium ATAC-seq.
(A) Correlation of three biological replicates of E14.5 mouse palate epithelium and mesenchyme ATAC-seq results. (B) ATAC-seq density plot of different clusters of E14.5 mouse palate epithelium specific NFRs. (C) GO enrichment (MGI mouse gene expression pattern) of genes associated with cluster 2 elements. (D and E) UCSC Genome browser view showing the ATAC-seq and H3K27Ac ChIP-seq signals in Krt14 and Klf4 locus.
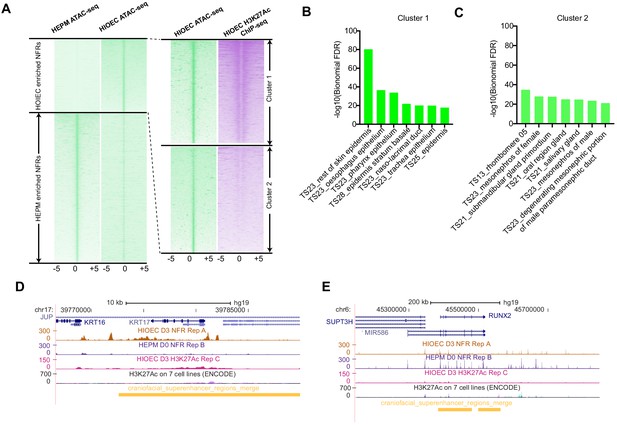
Summary of ATAC-seq in HIOEC and HEPM cells.
(A) Heatmap plots of ATAC-seq of HIOEC- and HEPM-specific NFRs. (B). GO enrichment for the genes near cluster 1 of HIOEC-specific NFRs. (C). GO enrichment for the genes near cluster 2 of HIOEC-specific NFRs. (D and E) UCSC Genome browser tracks showing the HIOEC and HEPM ATAC-seq and NHEK H3K27Ac ChIP-seq signals in IRF6 (D) and RUNX2 (E) locus.
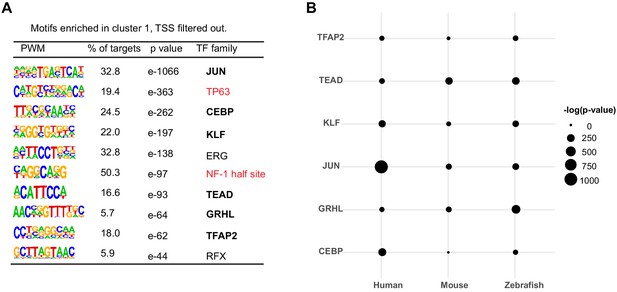
Motifs enriched in hOEAEs and shared among zGPAEs, mPEAEs and hOEAEs.
(A) Motifs enriched in hOEAEs. Bold, shared motifs enriched in all three epithelial tissues. (B) The significance of enrichment of each of the shared motifs among hOEAEs, mPEAEs and zPEAEs.
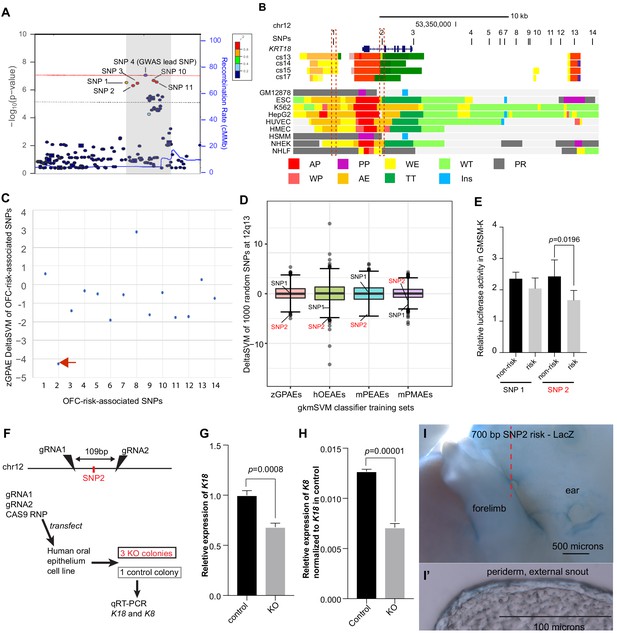
Use of a classifier trained on zGPAEs to prioritize orofacial clefting (OFC)-associated SNPs near KRT18 for functional tests .
(A) Regional plot showing OFC-risk-associated single nucleotide polymorphism (SNPs) near KRT18 from this study. SNP4 is the lead SNP from our meta-analysis of OFC GWAS (Leslie et al., 2017). (B) Browser view of the human genome, hg19, focused on this locus. Tracks: SNPs: OFC-risk-associated SNPs. SNP1: rs11170342, SNP2: rs2070875, SNP3: rs3741442, SNP4: rs11170344, SNP5: rs7299694, SNP6: rs6580920, SNP7: rs4503623, SNP8: rs2363635, SNP9: rs2682339, SNP10: rs111680692, SNP11: rs2363632, SNP12: rs4919749, SNP13: rs2638522, SNP14: rs9634243. Color coded bars: Chromatin status (color code explained in key), revealed by ChIP-seq to various chromatin marks. Cs13-cs17, facial explants from human embryos at Carnegie stage (cs) 13–17, encompassing the time when palate shelves fuse (Wilderman et al., 2018). Roadmap Epigenomics Project cell lines (Visel et al., 2008): GM12878, B-cell derived cell line; ESC, Embryonic stem cells; K562, myelogenous leukemia; HepG2, liver cancer; HUVEC, Human umbilical vein endothelial cells; HMEC, human mammary epithelial cells; HSMM, human skeletal muscle myoblasts; NHEK, normal human epidermal keratinocytes; NHLF, normal human lung fibroblasts. AP, active promoter; WP, weak promoter; PP, poised promoter; AE, active enhancer; WE, weak enhancer; TT, transcriptional transition; WT, weakly transcribed; Ins, insulator; PR, polycomb-repressed. (C) deltaSVM scores predicted by zGPAEs-derived classifier for the 14 OFC associated SNPs near KRT18. (D) Box and whisker plots of deltaSVM scores of 1000 randomly-selected SNPs near KRT18, scored by classifiers trained by zGPAEs (zebrafish periderm active enhancers), hOEAEs (human oral epithelium active enhancers), mPEAEs (mouse palatal epithelium active enhancers) and mPMAEs (mouse palatal mesenchyme active enhancers). The line is the median scoring SNP, the box contains the middle-scoring two quartiles, and the whisker represent the top and lower quartiles. Dots are outliers. deltaSVM scores for SNP1 and SNP2 are indicated. Number out of 1000 randomly selected SNPs with a lower deltaSVM than SNP2 with classifier trained on zGPAEs, 2; on mPEAEs, 9; on hOEAEs, 17; on mPMAEs, 186. (E) Dual luciferase assay for non-risk and risk alleles of rs11170342 (SNP1) and rs2070875 (SNP2) in GMSM-K cells. (F) Schematic diagram showing the workflow of generating GMSM-K cell colonies with 109 bp flanking SNP2 deleted by CRISPR-Cas9. (G,H) qRT-PCR showing relative RNA expression of KRT18 (G) and KRT8 (H) in three homozygous knockout colonies (KO) and one isolated wild-type colony (Control) of GMSM-K cell lines. (I) Lateral view of transgenic mice LacZ reporter assay for the 700 bp DNA fragment overlapping SNP2. (I’) Section of the facial prominence from I (red circled region).
-
Figure 6—source data 1
Barchart for relative dual luciferase activity in GMSM-K cells, as plotted in Figure 6E.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Barchart for relative gene expression of K18 and K8 in GMSM-K cells, as plotted in Figure 6G and H.
- https://cdn.elifesciences.org/articles/51325/elife-51325-fig6-data2-v2.xlsx

Dot plot of deltaSVM scores for each SNP calculated with classifiers trained on the indicated set of enhancer candidates.
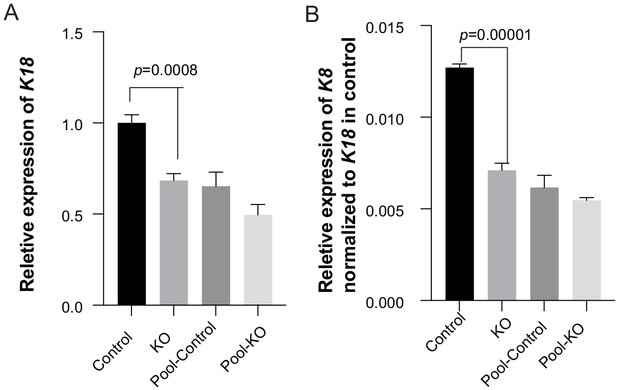
Bargraphs showing relative RNA expression of K18 (A) and K8 (B) in GMSM-K cells.
KO: three homozygous knockout colonies; Control: one isolated wildtype colony; Pool-control: pool of GMSM-K cells transfected with two gRNAs only; Pool-KO: Pool of GMSM-K cells transfected with two gRNA along with Cas9 RNP.

Lateral views of all wild-type mouse embryos for LacZ reporter assay.
(A) Embryos injected with a reporter construct built from a 701 bp element centered on SNP1, harboring the risk or non-risk allele as indicated. The large majority of embryos with SNP1 constructs, of either allele, were not blue, and no two blue embryos showed the same pattern. R-random integration, see below. No further copy number analysis was carried out. (B) Embryos injected with a reporter construct built from a 700 bp element centered on SNP2, harboring the risk or non-risk allele as indicated. Using the genomic DNA isolated from each embryo, PCR was carried out to determine if the reporter construct was present at all, and whether it was (S - single) present at the safe harbor locus in a single copy, (T - tandem), present at the safe harbor locus in more than one copy, or (R-random) was detectable but absent from the safe harbor locus, suggesting it integrated randomly into the genome (Kvon et al., 2020). One embryo (number 1, boxed) injected with a SNP2 construct (risk-allele) showed reporter activity in the periderm, as predicted. Quantitative PCR indicated this embryo had 8–10 copies of the reporter construct while the other T embryos had 2 copies.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | One Shot TOP10 | Life technologies | Cat# C4040-10 | Chemically competent cells |
| Cell line (Homo-sapiens) | GMSM-K (human embryonic oral epithelial cell line) | (Gilchrist et al., 2000) | RRID:CVCL_6A82 | a kind gift from Dr. Daniel Grenier |
| Cell line (Homo-sapiens) | HIOEC (human immortalized oral epithelial cells) | (Sdek et al., 2006) | RRID:CVCL_6E43 | |
| Cell line (Homo-sapiens) | HEPM (human embryonic palatal mesenchyme cells) | ATCC | ATCC Cat# CRL-1486, RRID:CVCL_2486 | |
| Antibody | anti-Histone H3, Acetylated Lysine 27 (Rabbit polyclonal) | Abcam | Abcam Cat# ab4729, RRID:AB_2118291; lot NO. GR3211959-1; | ChIP (4 ug per 500,000 HIOEC cells) |
| Recombinant DNA reagent | pXX330 (plasmid) | Addgene;{Cong, 2013 #2;Ran, 2013 #1} | RRID :Addgene_ 42230 | |
| Recombinant DNA reagent | cFos-GFP | (Fisher et al., 2006b) | a gift from Shannon Fisher | |
| Recombinant DNA reagent | cFos-tdTomato | This paper | Modified from cFos-GFP | |
| Recombinant DNA reagent | pENTR/D-TOPO | Life technologies | InvitrogenCat# K240020 | |
| Recombinant DNA reagent | cFos-FFLuc | (Liu et al., 2017a) | ||
| Sequence-based reagent | cFos-RLuc | (Liu et al., 2017a) | ||
| Sequence-based reagent | Klf17_+1.8_F | This paper | PCR primers | ATGCTGACTCCA CCATCCTC |
| Sequence-based reagent | Klf17_+1.8_R | This paper | PCR primers | CACCTACCCCTTGGC TAATCGTTG |
| Sequence-based reagent | Cavin2b_+18_F | This paper | PCR primers | TTCTGTTTTTGC CATCAGCA |
| Sequence-based reagent | Cavin2b_+18_R | This paper | PCR primers | CACCTTTTAATCAC CGCCTTTCCA |
| Sequence-based reagent | Gadd45ba_−0.7_F | This paper | PCR primers | TGGTTGGGTTC AGAGGTAGG |
| Sequence-based reagent | Gadd45ba_−0.7_R | This paper | PCR primers | CACCATGACTCGAC GAAAGCAAA |
| Sequence-based reagent | SNP2_gRNA_left | This paper | gRNA target | CTAAGAAGGATC TGCTCCCC |
| Commercial assay or kit | SNP2_gRNA_right | This paper | gRNA target | GAGGACAGTATTC TTAAACG |
| Commercial assay or kit | RNAqueous Total RNA Isolation Kit | Ambion | Cat# AM1912 | |
| Commercial assay or kit | RNA Clean and Concentrator-5 Kit | Zymo Research | Cat# R1013 | |
| Commercial assay or kit | SMART-Seq v4 Ultra Low Input RNA Kit | TAKARA | Cat# 634888 | |
| Commercial assay or kit | Agilent RNA 6000 Pico | Agilent Technologies | Cat# 5067–1513 | |
| Commercial assay or kit | Nextera XT DNA Sample Preparation Kit | Illumina | Cat# FC-131–1002 | |
| Commercial assay or kit | Nextera DNA Sample Preparation Kit | Illumina | Cat# FC-121–1030 | |
| Commercial assay or kit | VAHTS Universal DNA Library Prep Kit for Illumina | Vanzyme | Cat# ND606-01 | |
| Commercial assay or kit | KAPA Library Quantification Kit | Roche | Cat# KK4824 | |
| Commercial assay or kit | NEBNext High-Fidelity 2x PCR Master Mix | New England Biolabs | Cat# M0541S | |
| Chemical compound, drug | Ampure XP beads | Beckman Coutler | Cat# A63881 | |
| Chemical compound, drug | 0.25% trypsin-EDTA | Life Technologies | Cat# 25200056 | |
| Chemical compound, drug | Defined trypsin inhibitor | Life Technologies | Cat# R007100 | |
| Software, algorithm | Turbo DNase I | Ambion | Cat# AM2238 | |
| Software, algorithm | R | R | RRID:SCR_001905 | v 3.5.1 v 3.3.2 |
| Software, algorithm | Bowtie2 | (Langmead and Salzberg, 2012) | RRID:SCR_005476 | v 2.3.4.1 |
| Software, algorithm | Trimmomatic | (Bolger et al., 2014) | RRID:SCR_011848 | v.0.38 |
| Software, algorithm | DiffBind | (Ross-Innes et al., 2012) | RRID:SCR_012918 | |
| Software, algorithm | seqMINER | (Ye et al., 2011) | RRID:SCR_013020 | v 1.2.1 |
| Software, algorithm | HOMER | (Heinz et al., 2010) | RRID:SCR_010881 | v 3.0 |
| Software, algorithm | Gapped k-mer support vector machine | (Ghandi et al., 2016) | https://rdrr.io/cran/gkmSVM/ | v 0.79.0 |
| Software, algorithm | BEDTools | (Quinlan and Hall, 2010) | RRID:SCR_006646 | v 2.24.0 |
| Software, algorithm | Picard Tools | http://broadinstitute.github.io/picard/ | RRID:SCR_006525 | v 0.35 |
| Software, algorithm | SAMtools | (Li et al., 2009) | RRID:SCR_002105 | v 1.7 |
| Software, algorithm | MACS2 | (Zhang et al., 2008) | RRID:SCR_013291 | v 2.1.1 |
| Software, algorithm | DeepTools | (Ramírez et al., 2016) | RRID:SCR_016366 | v 2.0 |
Additional files
-
Supplementary file 1
Coordinates of ATAC-seq and ChIP-seq peaks identified in this study.
(a) Summary of peak numbers for all ATAC-seq and H3K27Ac ChIP-seq generated in this study (b) Coordinates of GFP-positive NFRs flanked by H3K27AcHigh (zGPAEs) (c) Coordinates of GFP-positive NFRs flanked low in H3K27Ac signals (d) Coordinates of GFP-negative NFRs flanked by H3K27AcHigh (GNAEs) (e). Coordinates of GFP-negative NFRs flanked low in H3K27Ac signals (f) Coordinates of fish zGPAEs training set (zv9) (g) Coordinates of mouse palate mesenchyme enriched NFR (h) Coordinates of mouse palate epithelium enriched NFR (i) Coordinates of mouse palate epithelium specific active enhancers (j) Coordinates of HIOEC-specific NFRs (k) Coordinates of HIOEC-specific active NFRs (flanked or overlapped with H3K27Ac ChIP-seq in HIOEC)
- https://cdn.elifesciences.org/articles/51325/elife-51325-supp1-v2.xlsx
-
Supplementary file 2
Zebrafish ppl and human PPL enhancer alignments using ClustalO.
(a) Alignments summary for enhancer homology test between ppl-10 and PPL-8.3. (b) Alignments details for enhancer homology test between ppl-10 and PPL-8.3. All alignments were conducted using the CLUSTALW algorithm with default parameters via the Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/). Alignments were then annotated to highlight identical blocks of length 5 to 6 bp long (cyan) or longer (yellow). See Materials and methods for further details on the choice of enhancer fragments used in these alignments.
- https://cdn.elifesciences.org/articles/51325/elife-51325-supp2-v2.docx
-
Supplementary file 3
deltaSVM score and JASPAR predicted TF binding changes in the KRT18 locus.
(a) List of OFC-associated SNPs near KRT18 locus (b) deltaSVM scores for 14 OFC-associated SNPs near KRT18 locus and 1000 random SNPs using classifiers trained by zGPAEs (c) deltaSVM scores for 14 OFC-associated SNPs near KRT18 locus and 1000 random SNPs using classifiers trained by mPEAEs (d) deltaSVM scores for 14 OFC-associated SNPs near KRT18 locus and 1000 random SNPs using classifiers trained by hOEAEs (e) deltaSVM scores for 14 OFC-associated SNPs near KRT18 locus and 1000 random SNPs using classifiers trained by mPMAEs (f) Effects of different alleles of SNP1 and SNP2 on transcription factor binding sites, predicted by JASPAR
- https://cdn.elifesciences.org/articles/51325/elife-51325-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51325/elife-51325-transrepform-v2.docx