Genetic inactivation of mTORC1 or mTORC2 in neurons reveals distinct functions in glutamatergic synaptic transmission
Figures
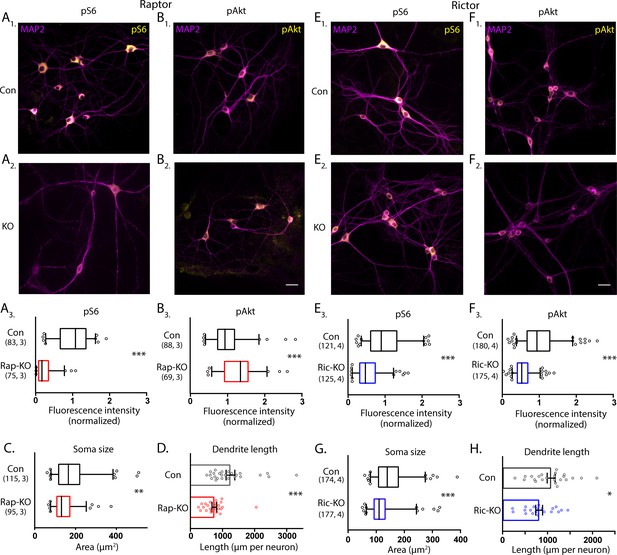
Loss of Raptor or Rictor in primary hippocampal neurons alters mTOR signaling and decreases neuron growth.
(A) Representative images of Raptor-Con (A1) and Raptor-KO neurons (A2) showing the structure of the neurons revealed by MAP2 immunofluorescence (purple) and the immunofluorescence signal from phospho-S6 (pS6, yellow) (A3) Box plot (median and 95%) showing the fluorescence intensity measurements from the pS6 signal in Raptor-Con and Raptor-KO neurons. (B) Representative images of Raptor-Con (B1) and Raptor-KO neurons (B2) showing the structure of the neurons revealed by MAP2 immunofluorescence (purple) and the immunofluorescence signal from phospho-AKT473 (pAkt, yellow) Scale bar is 25 µm. (B3) Box plot (median and 95%) showing the fluorescence intensity measurements from the pAkt signal in Raptor-Con and Raptor-KO neurons. (C) Box plot (median and 95%) showing the area measurements of the somatic compartment in Raptor-Con and Raptor-KO neurons. (D) Dot plot showing the measurements of the total dendritic length in Raptor-Con and Raptor-KO neurons and mean ± s.e.m. Each dot is one neuron sampled from three independent cultures. (E) Representative images of Rictor-Con (E1) and Rictor-KO neurons (E2) showing the structure of the neurons revealed by MAP2 immunofluorescence (purple) and the immunofluorescence signal from phospho-S6 (pS6, yellow) (E3) Box plot (median and 95%) showing the fluorescence intensity measurements from the pS6 signal in Rictor-Con and Rictor-KO neurons. (F) Representative images of Rictor-Con (F1) and Rictor-KO neurons (F2) showing the structure of the neurons revealed by MAP2 immunofluorescence (purple) and the immunofluorescence signal from phospho-AKT473 (pAkt, yellow) Scale bar is 25 µm. (F3) Box plot (median and 95%) showing the fluorescence intensity measurements from the pAkt signal in Rictor-Con and Rictor-KO neurons. (G) Box plot (median and 95%) showing the area measurements of the somatic compartment in Rictor-Con and Rictor-KO neurons. (H) Dot plot showing the measurements of the total dendritic length in Rictor-Con and Rictor-KO neurons and mean ± s.e.m. Each dot is one neuron sampled from three independent cultures. The numbers underneath the groups indicate the number of neurons analyzed and the number of cultures. *=p < 0.05, **=p < 0.01, ***=p < 0.001, as tested with Generalized Estimating Equations.
-
Figure 1—source data 1
Values and statistical analysis of band intensities for Western blot analysis of RAPTOR and RICTOR protein levels.
- https://cdn.elifesciences.org/articles/51440/elife-51440-fig1-data1-v2.xlsx
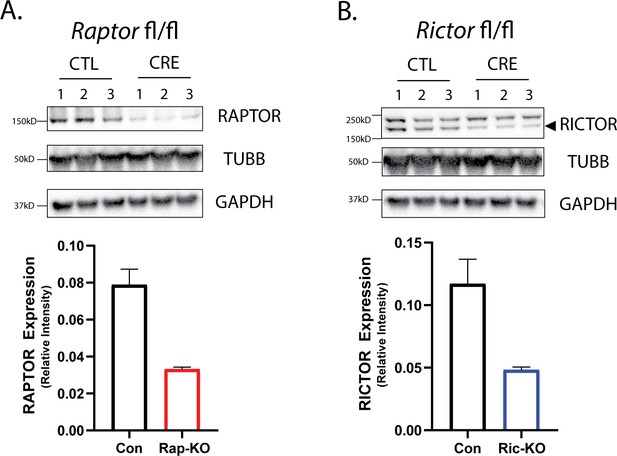
Western blot analysis of Raptor and Rictor levels after Cre-mediated gene deletion.
A control (CTL) or Cre recombinase containing virus (CRE) was applied to hippopcampal neurons from mice harboring floxed alleles at either the Raptor (A) or Rictor locus (B). Western blots were subsequently performed on protein lysates from wells (n = 3) containing neurons from two mice each. Visual inspection of the western blot (top) reveals lower expression of RAPTOR and RICTOR in the CRE condition relative to the associated control. Normalized expression of RAPTOR* and RICTOR** confirmed a significant decrease of protein expression in their respective CRE condition (bottom – graphs). *t(4)=5.4, p=0.0005; **t(4)=3.5, p=0.03.
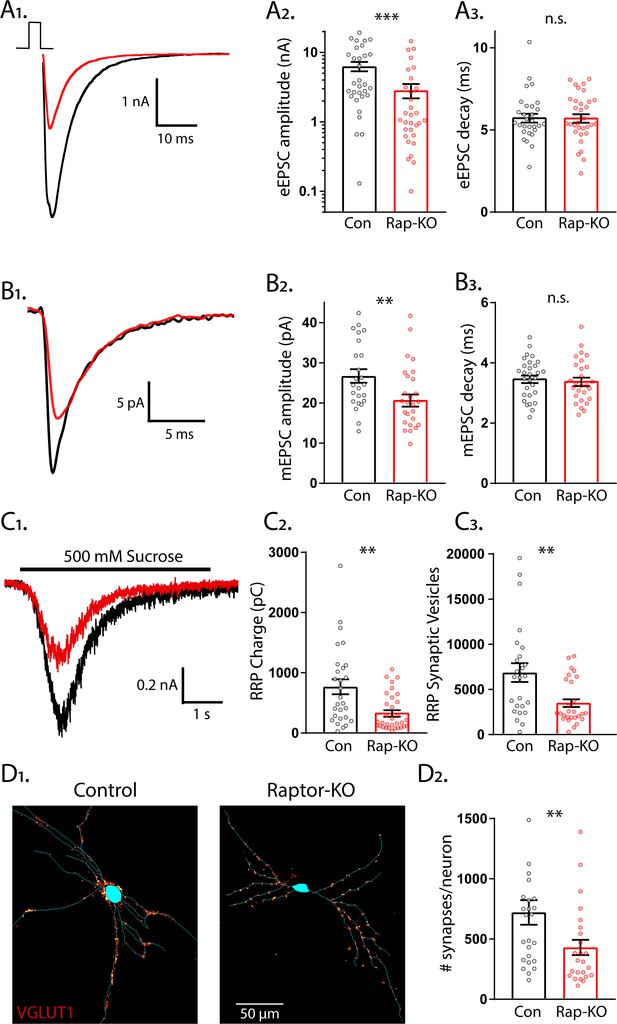
Loss of Raptor decreases the strength of evoked excitatory synaptic transmission via changes in quantal size and synapse number.
(A1) Example traces of evoked EPSCs (eEPSCs) recorded from single-neuron primary hippocampal cultures of Raptor-Con (black) and Raptor-KO (red) neurons. (A2) Plot showing the values of peak eEPSC amplitudes recorded from Raptor-Con (black) and Raptor-KO (red) neurons on a logarithmic scale. (A3) Plot showing the values of single exponential fits to the fast component of the eEPSC decay recorded from Raptor-Con (black) and Raptor-KO (red) neurons on a linear scale. (B1) Example traces of average miniature EPSCs (mEPSCs) recorded from single-neuron primary hippocampal cultures of Raptor-Con (black) and Raptor-KO (red) neurons. (B2) Plot showing the values of mEPSC peak amplitudes recorded from Raptor-Con (black) and Raptor-KO (red) neurons. (B3) Plot showing the distributions of mEPSC decay time constants recorded from Raptor-Con (black) and Raptor-KO (red) neurons. (C1) Example traces of the current response to 500 mM sucrose application recorded from single-neuron primary hippocampal cultures of Raptor-Con (black) and Raptor-KO (red) neurons. The black line indicates the time of sucrose application. (C2) Plot showing the values of the charge contained in the readily releasable pool (RRP) of Raptor-Con (black) and Raptor-KO (red) neurons, as determined by integrating the sucrose response after subtracting the steady state component. (C3) Plot showing the number of vesicles contained in the RRP of Raptor-Con (black) and Raptor-KO (red) neurons, as determined by dividing the RRP charge by the mean mEPSC charge for each neuron. (D1) Representative images showing fluorescence intensity in a red color look up table (LUT) from VGLUT1 immunostaining superimposed on a tracing of the cell body and dendrites from a Raptor-Con (left) and a Raptor-KO (right) neuron. (D2) Plot showing the values of synapse number per neuron for Raptor-Con (black) and Raptor-KO (red) neurons. For all dot plots, each dot represents the mean response from one neuron sampled from three independent cultures and the bars show the estimated marginal means and standard errors. ** indicates a p value of < 0.01, *** indicates p<0.001 and n.s. indicates p>0.05, as tested with Generalized Estimating Equations.
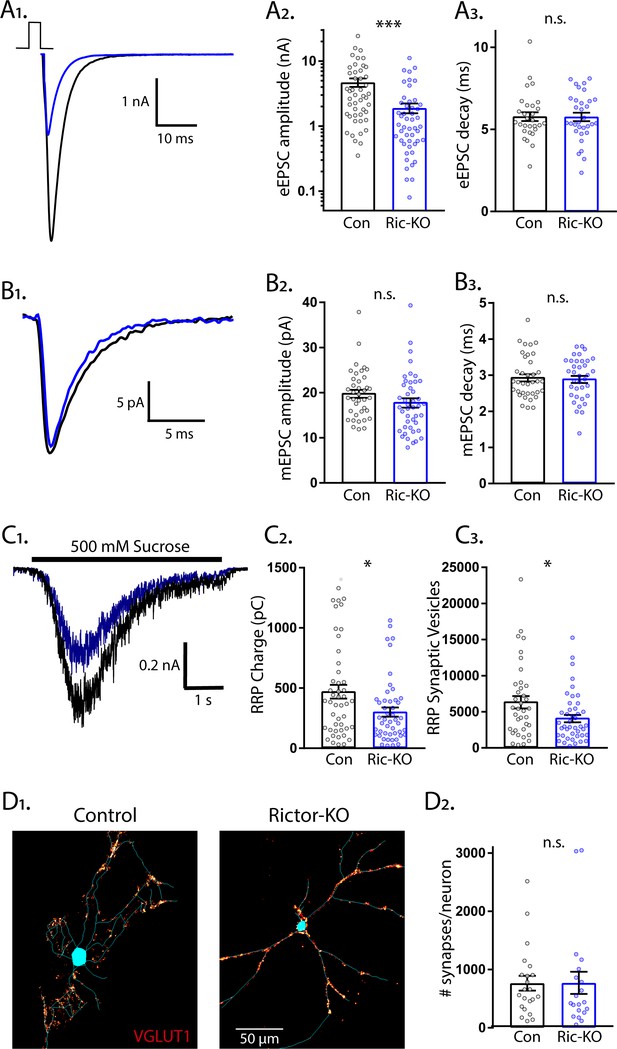
Loss of Rictor decreases the strength of evoked excitatory synaptic transmission without altering quantal size or synapse number.
(A1) Example traces of eEPSCs recorded from single-neuron primary hippocampal cultures of Rictor-Con (black) and Rictor-KO (blue) neurons. (A2) Plot showing the values of peak eEPSC amplitudes recorded from Rictor-Con (black) and Rictor-KO (blue) neurons on a logarithmic scale. (A3) Plot showing the values of single exponential fits to the fast component of the eEPSC decay recorded from Rictor-Con (black) and Rictor-KO (blue) neurons on a linear scale. (B1) Example traces of average mEPSCs recorded from single-neuron primary hippocampal cultures of Rictor-Con (black) and Rictor-KO (blue) neurons. (B2) Plot showing the values of mEPSC peak amplitudes recorded from Rictor-Con (black) and Rictor-KO (blue) neurons. (B3) Plot showing the distributions of mEPSC decay time constants recorded from Rictor-Con (black) and Rictor-KO (blue) neurons. (C1) Example traces of the current response to 500 mM sucrose application recorded from single-neuron primary hippocampal cultures of Rictor-Con (black) and Rictor-KO (blue) neurons. The black line indicates the time of sucrose application. (C2) Plot showing the values of the charge contained in the readily releasable pool (RRP) of Rictor-Con (black) and Rictor-KO (blue) neurons, as determined by integrating the sucrose response after subtracting the steady state component. (C3) Plot showing the number of vesicles contained in the RRP of Rictor-Con (black) and Rictor-KO (blue) neurons, as determined by dividing the RRP charge by the mean mEPSC charge for each neuron. (D1) Representative images showing fluorescence intensity in a red color look up table (LUT) from VGLUT1 immunostaining superimposed on a tracing of the cell body and dendrites from a Rictor-Con (left) and a Rictor-KO (right) neuron. (D2) Plot showing the values of synapse number per neuron neuron for Rictor-Con (black) and Rictor-KO (blue) neurons. For all dot plots, each dot represents the mean response from one neuron sampled from four independent cultures, except the synapse counts, which are from three cultures. The bars show the estimated marginal means and standard errors of the mean. ** indicates a p value of < 0.01, *** indicates p<0.001 and n.s. indicates p>0.05, as tested with Generalized Estimating Equations.
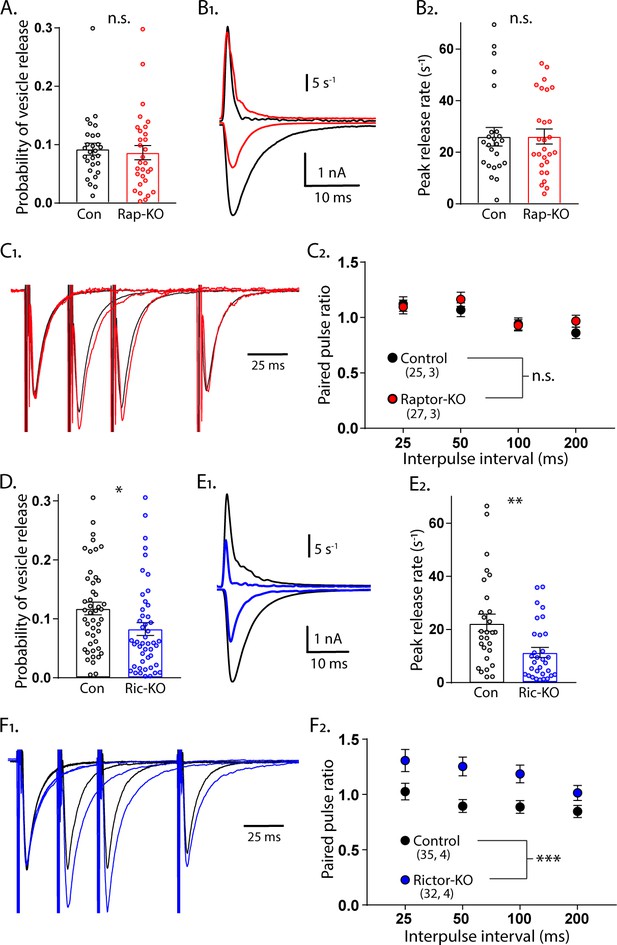
Loss of Rictor reduces evoked SV release efficiency but Raptor loss does not.
(A) Plot showing no difference in the vesicular release probability of Raptor-Con (black) and Raptor-KO (red) neurons. (B1) Example traces showing the rate of SV release (top traces) over their corresponding EPSCs from Raptor-Con (black) and Raptor-KO (red) neurons. (B2) Plot of the peak vesicle release rates of Raptor-Con (black) and Raptor-KO (red) neurons. (C1) Example traces of EPSCs evoked in response to 2 ms depolarizations spaced at 25, 50, and 100 ms. The three sweeps at different intervals are overlayed, as are the responses from Raptor-Con (black) and Raptor-KO (red) neurons. The values are normalized to the peak amplitude of the first EPSC in each sweep. (C2) Summary data showing the estimated marginal means and standard errors for Raptor-Con (black) and Raptor-KO (red) groups at different interpulse intervals. All data summarized in panels (A-C) were obtained from three independent cultures. (D) Plot showing the decrease in vesicular release probability between Rictor-Con (black) and Rictor-KO (blue) neurons. (E1) Example traces showing the rate of SV release (top traces) over their corresponding EPSCs from Rictor-Con (black) and Rictor-KO (blue) neurons. (E2) Plot of the peak vesicle release rates of Raptor-Con (black) and Raptor-KO (red) neurons. (F1) Example traces of EPSCs evoked in response to 2 ms depolarizations spaced at 25, 50, and 100 ms. The three sweeps at different intervals are overlayed, as are the responses from Rictor-Con (black) and Rictor-KO (blue) neurons. The values are normalized to the peak amplitude of the first EPSC in each sweep. (F2) Summary data showing the estimated marginal means and standard errors for Rictor-Con (black) and Raptor-KO (red) groups at different interpulse intervals. All data in panels D-F were obtained from four independent cultures. * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.001, and n.s. = p > 0.05, effect of group tested with Generalized Estimating Equations.
-
Figure 4—source data 1
Source data for Figure 4C2.
- https://cdn.elifesciences.org/articles/51440/elife-51440-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Source data for Figure 4F2.
- https://cdn.elifesciences.org/articles/51440/elife-51440-fig4-data2-v2.xlsx
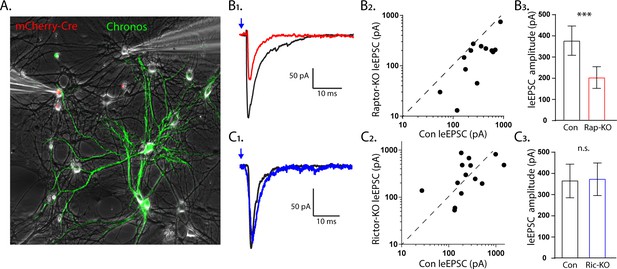
The effect of Raptor loss on evoked EPSCs, but not Rictor, is due to postynaptic impairments.
(A1) Representative image illustrating the experimental setup. The red fluorescence from the mCherry-Cre and the green fluorescence from the optogenetic protein Chronos fused to GFP are shown overlayed on a phase contrast image showing the patch pipettes attached to one Cre-positive (left pipette) and one Cre-negative (right pipette) neuron. (B1) Example light-evoked (le)EPSCs obtained simultaneously from a control (black) and a Raptor-KO (red) neuron held at −55 mV in response to a 2 ms flash of blue light. (B2) Scatter plot showing the leEPSC responses of neuron pairs to blue light stimulation. The peak amplitude from the control neuron in each pair is represented by the symbol’s value on the x-axis, and the peak amplitude of the Raptor-KO neuron is represented by the symbol’s value on the y-axis. Pairs in which the KO response is smaller than the control response will be below the dashed line. (B3) Bar graph showing the leEPSC amplitudes (mean ± s.e.m.) of control (black) and Raptor-KO (red) neurons. Data were obtained from three independent cultures. (C1) Example leEPSCs obtained simultaneously from a control (black) and a Rictor-KO (blue) neuron held at −55 mV in response to a 2 ms flash of blue light. (C2) Scatter plot showing the leEPSC responses of neuron pairs to blue light stimulation. The peak amplitude from the control neuron in each pair is represented by the symbol’s value on the x-axis, and the peak amplitude of the Rictor-KO neuron is represented by the symbol’s value on the y-axis. (C3) Bar graph showing the leEPSC amplitudes (mean ± s.e.m.) of control (black) and Rictor-KO (blue) neurons. Data were obtained from three independent cultures. *** indicates p<0.001 and n.s. = p > 0.05, effect of group tested with Generalized Estimating Equations.
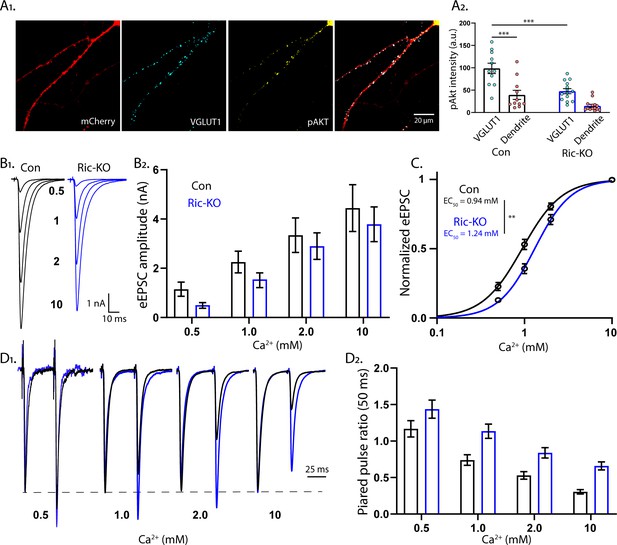
mTORC2 acts presynaptically to alter the calcium sensitivity of SV release.
(A1) Representative images showing the colocalization of VGLUT1 (cyan) and pAkt (yellow) immunofluorescent signals on a Rictor-Con neuron (red). (A2) The pAkt signal colocalized with VGLUT1 is more intense than the signal in neighboring dendrites and is significantly reduced by Rictor loss. Each dot represents the estimated marginal mean value of measurements from one neuron and bars are means ± s.e.m. (B1) Example traces of eEPSCs in indicated [Ca2+]ext from Rictor-Con (black) and Rictor-KO (blue) neurons. (B2) Estimated marginal means ± s.e.m values for eEPSC amplitudes in indicated [Ca2+]ext from Rictor-Con (black) and Rictor-KO (blue) neurons. (C) Linear-log plot of the normalized eEPSC amplitude versus [Ca2+]ext for Rictor-Con (black) and Rictor-KO (blue) neurons. Each point is the mean amplitude ± s.e.m. The solid lines are Hill equation fits to the data. (D1) Example traces of eEPSCs evoked in response to 2 ms depolarizations spaced at 50 ms from Rictor-Con (black) and Rictor-KO (blue) neurons in indicated [Ca2+]ext. The traces are normalized to the peak amplitude of the first eEPSC in each [Ca2+]ext. (D2) Summary data showing the paired pulse ratios (estimated marginal means ± s.e.m) for Rictor-Con (black) and Rictor-KO (blue) groups at different [Ca2+]ext. All data summarized in panels A-D were obtained from three independent cultures. *** indicates p<0.001 as tested with Generalized Estimating Equations.
-
Figure 6—source data 1
Source data for Figure 6B2 and C.
- https://cdn.elifesciences.org/articles/51440/elife-51440-fig6-data1-v2.xlsx
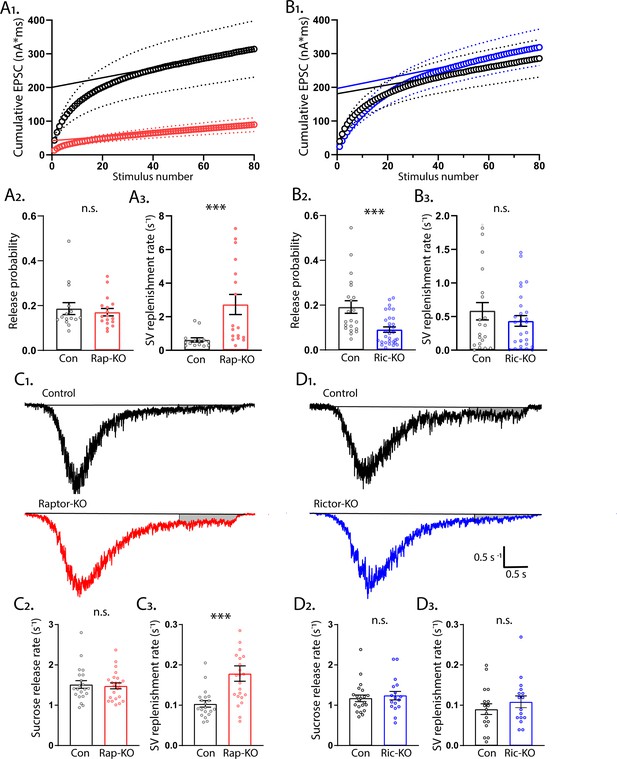
mTORC1 inactivation increases SV replenishment rate.
(A1) Cumulative EPSC charge in Raptor-Con (black) and Raptor-KO (red) neurons. Circles are the population means and the dotted lines are the s.e.m. The solid lines are linear fits to the final 20 responses and the y-intercept indicates the RRPtrain. (A2) Release probability calculated using RRPtrain is unchanged in Raptor-KO neurons. (A3) The SV replenishment rate is significantly increased by Raptor loss. (B1) Cumulative EPSC charge in Rictor-Con (black) and Rictor-KO (blue) neurons. (B2) Release probability calculated using RRPtrain is significantly decreased in Rictor-KO neurons. (B3) The SV replenishment rate is unaffected by Rictor loss. (C1) Example traces of normalized current responses to 500 mM sucrose application in Raptor-Con (black) and Raptor-KO (red) neurons. The black line shows the pre-sucrose baseline, and the gray shaded area shows the area used to calculate the replenishment rate. (C2) Plot showing the rate constants for sucrose-induced synaptic vesicle (SV) release in Raptor-Con (black) and Raptor-KO (red) neurons. (C3) Plot showing the replenishment rate constants after sucrose-induced SV release in Raptor-Con (black) and Raptor-KO (red) neurons. (D1) Example traces of normalized current responses to 500 mM sucrose application in Rictor-Con (black) and Rictor-KO (blue) neurons. The black line shows the pre-sucrose baseline, and the gray shaded area shows the area used to calculate the replenishment rate. (D2) Plot showing the rate constants for sucrose-induced SV release in Rictor-Con (black) and Rictor-KO (blue) neurons. (D3) Plot showing the replenishment rate constants after sucrose-induced SV release in Rictor-Con (black) and Rictor-KO (blue) neurons. In the dot plots, each dot represents the mean value from one neuron obtained from three independent cultures per genotype, and the bars show the estimated marginal means and s.e.m. ***=p < 0.001, and n.s. = p > 0.05, as tested with Generalized Estimating Equations.
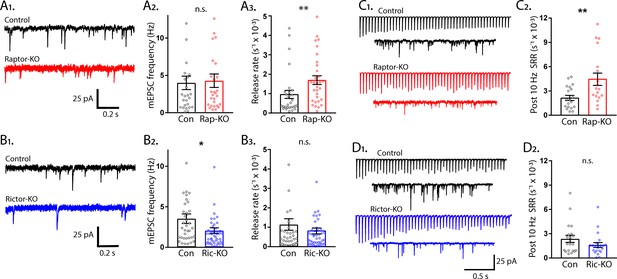
The rate constant for spontaneous vesicle fusion is regulated by mTORC1 activity.
(A1) Example traces of miniature synaptic currents recorded in single-neuron cultures of Raptor-Con (black) and Raptor-KO (red) neurons. (A2) Plot showing the mEPSC frequencies from Raptor-Con (black) and Raptor-KO (red) neurons. (A3) Plot showing the spontaneous release rate (SRR) of Raptor-Con (black) and Raptor-KO (red) neurons. (B1) Example traces of miniature synaptic currents recorded in single-neuron cultures of Rictor-Con (black) and Rictor-KO (blue) neurons. (B2) Plot showing the mEPSC frequencies from Rictor-Con (black) and Rictor-KO (blue) neurons. (B3) Plot showing the spontaneous release rate (SRR) of Rictor-Con (black) and Rictor-KO (blue) neurons. (C1) Example traces of EPSCs in response to 50 stimulations at 10 Hz (top traces) over traces of spontaneous SV release in the 10 s following the train. Example trace from a Raptor-Con neuron is in black, and a Raptor-KO neuron in red. (C2) Plot showing the rate constants for spontaneous SV release of Raptor-Con (black) and Raptor-KO (red) neurons in the 10 s following the 10 Hz stimulation. (D1) Example traces of EPSCs in response to 50 stimulations at 10 Hz (top traces) over traces of spontaneous SV release in the 10 s following the train. Example trace from a Rictor-Con neuron is in black, and a Rictor-KO neuron in blue. (D2) Plot showing the rate constants for spontaneous SV release of Rictor-Con (black) and Rictor-KO (blue) neurons in the 10 s following the 10 Hz stimulation. In the dot plots, each dot represents the value from one neuron, and the bars show the estimated marginal means and s.e.m. Raptor data was obtained from three independent cultures and Rictor data from four independent cultures. *=p < 0.05, **=p < 0.01, and n.s. = p > 0.05, as tested with Generalized Estimating Equations.
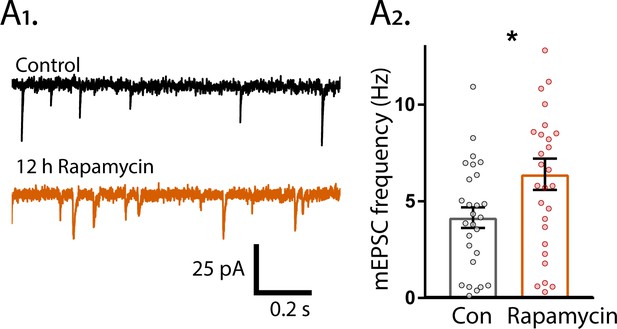
12 hr Rapamycin treatment increases mEPSC frequency.
(A1) Example traces of mEPSC activity recorded in mass cultures after 12 hr treatment with DMSO (black trace) or 20 nM rapamycin (orange trace). (A2) Summary data showing the increase in mEPSC frequency. Each dot represents one neuron obtained from two independent cultures.
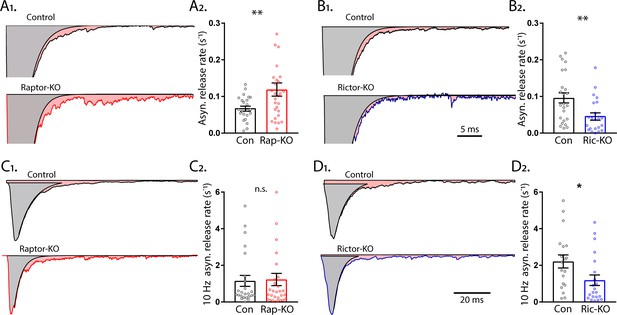
mTORC1 and mTORC2 inactivation have opposite effects on asynchronous synaptic vesicle release.
(A1) Example traces of normalized EPSCs evoked at 0.1 Hz from Raptor-Con (black) and Raptor-KO (red) neurons. The gray shaded area represents the area under the curve of the synchronous component of synaptic vesicle (SV) release, and the pink shaded area represents the asynchronous component. (A2) Plot showing the rate constants for asynchronous SV release of Raptor-Con (black) and Raptor-KO (red) neurons. (B1) Example traces of normalized EPSCs evoked at 0.1 Hz from Rictor-Con (black) and Rictor-KO (blue) neurons shaded to highlight the synchronous (gray) and asynchronous (pink) components of SV release. (B2) Plot showing the rate constants for asynchronous SV release of Rictor-Con (black) and Rictor-KO (blue) neurons. (C1) Example traces of normalized EPSCs at the end of a 10 Hz train from Raptor-Con (black) and Raptor-KO (red) neurons. The gray shaded area represents the area under the curve of the synchronous component of SV release and the pink shaded area represents the asynchronous component. (C2) Plot showing the rate constants for asynchronous SV release at the end of a 10 Hz train from Raptor-Con (black) and Raptor-KO (red) neurons. (D1) Example traces of normalized EPSCs at the end of a 10 Hz train from Rictor-Con (black) and Rictor-KO (blue) neurons shaded to highlight the synchronous (gray) and asynchronous (pink) components of SV release. (D2) Plot showing the rate constants for asynchronous SV release at the end of a 10 Hz train from Rictor-Con (black) and Rictor-KO (blue) neurons. In the dot plots, each dot represents the mean value from one neuron obtained from three independent cultures per genotype, and the bars show the estimated marginal means and s.e.m. *=p < 0.05, **=p < 0.01, and n.s. = p > 0.05, as tested with Generalized Estimating Equations.
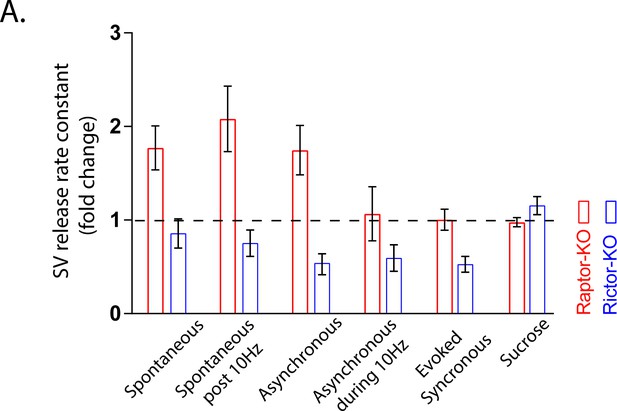
Summary of SV release rates in Raptor-KO and Rictor-KO neurons.
(A) Plot showing the release rate constants for both Raptor-KO (red) and Rictor-KO (blue) neurons over the range of conditions tested in the study. Each bar represents the mean log2 fold change for that genotype to illustrate the relatively stronger effect of mTORC1 inactivation in potentiating SV release when the rate is low, and of mTORC2 inactivation when the rate is high. Error bars are 95% Confidence Intervals.
Tables
Summary of the measurements of basic membrane properties.
Measurements are estimated marginal means ± s.e.m. Significance tested with Generalized Estimating Equations.
| Raptor-KO | ||||
|---|---|---|---|---|
| Con, n = 13 | Rap-KO, n = 15 | P value | 95% CI of difference | |
| Resting Potential, mV | 53.3 ± 2.2 | 55.9 ± 2.0 | 0.38 | −3.4–8.6 |
| Input Resistance, MΩ | 277 ± 37 | 442 ± 33 | 0.003 | 63–268 |
| Capacitance, pF | 138 ± 8.2 | 87 ± 7.4 | <0.001 | −73.2 - −27.8 |
| Time constant, ms | 36.8 ± 2.9 | 36.2 ± 2.6 | 0.89 | −8.6–7.4 |
| AP threshold, mV | 37.1 ± 2.2 | 38.0 ± 2.0 | 0.76 | −5.1–7.0 |
| AP amplitude, mV | 77.8 ± 3.5 | 67.7 ± 3.1 | 0.043 | −19.7 - −0.34 |
| Rictor-KO | ||||
| Con, n = 15 | Ric-KO, n = 17 | P value | 95% CI of difference | |
| Resting Potential, mV | 53.5 ± 1.1 | 52.2 ± 1.2 | 0.41 | −4.5–1.8 |
| Input Resistance, MΩ | 237 ± 28 | 329 ± 37 | 0.033 | 6–176 |
| Capacitance, pF | 173 ± 14 | 135 ± 10 | 0.065 | −78.6–2.7 |
| Time constant, ms | 37.4 ± 5.1 | 40.3 ± 5.2 | 0.65 | −9.6–15.4 |
| AP threshold, mV | 35.0 ± 1.3 | 32.3 ± 1.3 | 0.14 | −6.5–0.99 |
| AP amplitude, mV | 73.9 ± 3.2 | 69.0 ± 2.99 | 0.27 | −13.8–4.1 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent Mus musculus | Raptor-cKO mice | The Jackson Laboratories | Jackson Labs stock: 013188 | |
| Genetic reagent Mus musculus | Rictor-cKO mice | The Jackson Laboratories | Jackson Labs stock: 020649 | |
| Strain, strain background Mus musculus | C57BL/6J | The Jackson Laboratories | Jackson Labs stock: 000664 | |
| Recombinant DNA reagent | AAV8-SYN-mCherry-Cre | UNC Vector Core | ||
| Recombinant DNA reagent | AAV8-SYN-mCherry | UNC Vector Core | ||
| Recombinant DNA reagent | AAV9-Syn-Chronos-GFP | UNC Vector Core | ||
| Antibody | Rabbit monoclonal anti-Raptor | Cell Signaling | Cat #2280 RRID:AB_561245 | (1:1000) |
| Antibody | Rabbit monoclonal anti-Rictor | Cell Signaling | Cat #2114 RRID:AB_2179963 | (1:1000) |
| Antibody | Mouse monoclonal anti-MAP2 | Synaptic Systems | Cat #188 011 | (1:1000) |
| Antibody | rabbit polyclonal anti-VGLUT1 | Synaptic Systems | Cat #135 302 | (1:5000) |
| Antibody | guinea pig polyclonal anti-VGLUT1 | Synaptic Systems | Cat #135 304 | (1:5000) |
| Antibody | Rabbit polyclonal anti-pAkt (S473) | Cell Signaling | Cat # 9271 RRID:AB_2315049 | (1:1000) |
| Antibody | Rabbit monoclonal anti-pS6 (S240/244) | Cell Signaling | Cat # 5364 RRID:AB10694233 | (1:1000) |
| Chemical compound, drug | rapamycin | Cayman | Item # 13346 | |
| Chemical compound, drug | Kynurenic acid | Tocris | Cat. #0223 | |
| Chemical compound, drug | Bicuculline Methiodide | Tocris | Cat. #2503 | |
| Chemical compound, drug | Tetrodotoxin | Enzo | BML-NA120-0001 | |
| Software, algorithm | pClamp | Molecular devices | RRID:SCR_011323 | |
| Software, algorithm | Axograph X | Axograph | RRID:SCR_014284 | |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | |
| Software, algorithm | Prism | Graphpad | RRID:SCR_002798 | |
| Software, algorithm | FIJI | NIH | RRID:SCR_002285 | |
| Software, algorithm | Intellicount | PMID:29218324 | ||
| Software, algorithm | Matlab | Mathworks | RRID:SCR_001622 |





