Tissue-specific shaping of the TCR repertoire and antigen specificity of iNKT cells
Figures
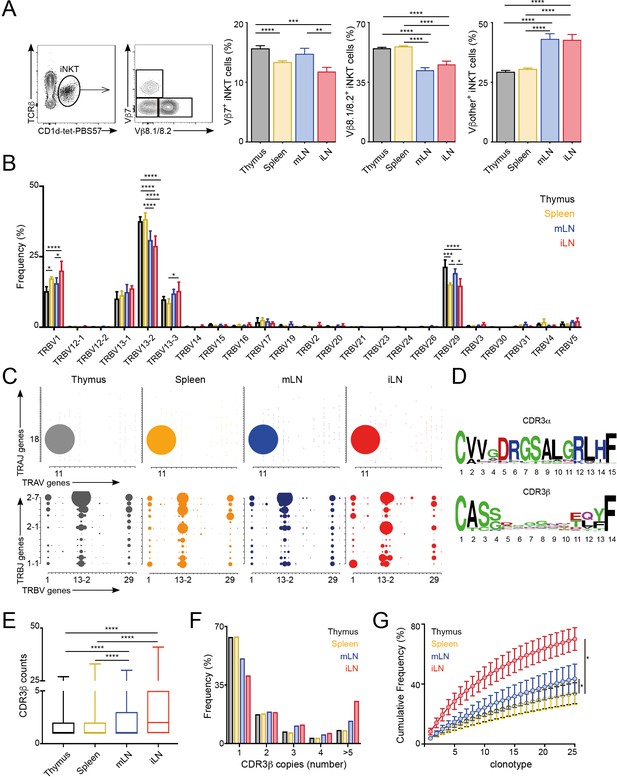
Different TCRVβ usage and clonal expansion in iNKT cells from several lymphoid tissues.
(A) Flow cytometry plots showing gating strategy (left) and quantification (right) for iNKT cells expressing Vβ7, Vβ8.1/8.2 or Vβother (non-Vβ7 or Vβ8.1/8.2) in the depicted tissues of WT C57BL/6. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 paired t-test. n = 15 mice from five independent experiments. (B) Frequency of TRBV gene usage in iNKT cell TCR sequences. Frequencies are calculated from RNAseq data from four samples per tissue. Bars represent mean ± SEM. *p<0.05, ***p<0.001, ****p<0.0001, ANOVA with Tukey’s multiple comparison test. (C) Gene usage plot (2D) showing TRAV-TRAJ (top) and TRBV-TRBJ (bottom) pairing for total TCR sequences obtained from iNKT cells isolated from the depicted tissues of WT mice. The circle size represents the percentage of sequences with each specific V-J pairing from the total TCR sequences for each tissue. (D) Visual representation for aa enrichment at each position for CDR3α (top) and CDR3β (bottom) sequences for iNKT cells (pooled from all tissues). Analyses were performed with sequences of 15 or 14 aa for CDR3α and CDR3β respectively. Graphics were generated with Weblogo. (E) Median value for counts for CDR3β sequences identified in the depicted tissues. Data obtained from RNAseq and pooled from four samples per tissue. Data have been calculated using the counts for each CDR3β sequence and expressed as box-and-whisker diagrams depicting the median ± lower quartile, upper quartile, sample minimum and maximum. ****p<0.0001 Mann-Whitney test. (F) Frequency of CDR3β clonotype usage in relation to the repertoire size for iNKT cells isolated from the depicted tissues (data obtained from RNAseq and pooled from four samples per tissue). Frequency of CDR3β sequences present once, twice, 3, 4 or five or more times are shown. (G) Cumulative frequencies occupied by the 25 most prevalent CDR3β clonotypes for iNKT cells isolated from the depicted tissues. Data has been calculated for sequences from four samples per tissue and represented as mean ± SEM. *p<0.05, paired t-test.
-
Figure 1—source data 1
Different TCRVβ usage and clonal expansion in iNKT cells from several lymphoid tissues.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig1-data1-v3.xlsx
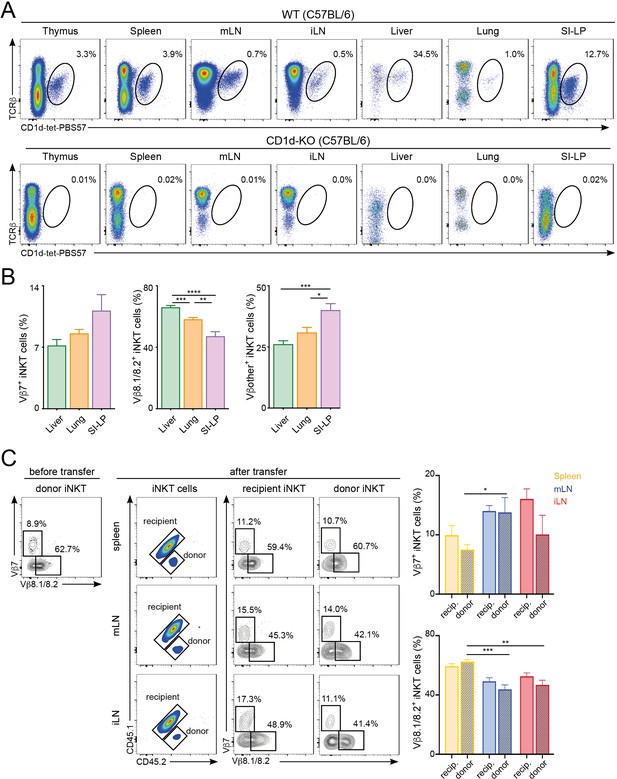
Tissue-dependent bias for TCRVβ usage in iNKT cells.
(A) Flow cytometry profiles showing iNKT cells (TCRβ+CD1d-tet-PBS57+) in the depicted tissues of WT and CD1d-KO mice. Numbers indicate percentage of iNKT cells within TCRβ+ cells. (B) Quantification of iNKT cells expressing Vβ7, Vβ8.1/8.2 or Vβother (non-Vβ7 or Vβ8.1/8.2) in the depicted tissues of WT C57BL/6 mice. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001 two-tailed unpaired t-test. n = 7 mice from three independent experiments. (C) iNKT cells enriched from spleen and thymus were adoptively transferred into congenic mice and TCRVβ usage in iNKT cells homing to various tissues was analysed 12 days later. Flow-cytometry plots show expression of Vβ7 and Vβ8.1/8.2 in iNKT cells from donor (CD45.2+) and recipient (CD45.1+CD45.2+) mice before or 12 days after transfer. Bar plots show frequency of donor and recipient iNKT cells expressing Vβ7 or Vβ8.1/8.2 in the depicted tissues. Bars represent mean ± SEM. n = 5 recipient mice from two independent experiments. *p<0.05, **p<0.01, ***p<0.001 two-tailed unpaired t-test.
-
Figure 1—figure supplement 1—source data 1
Tissue-dependent bias for TCRVβ usage in iNKT cells.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig1-figsupp1-data1-v3.xlsx
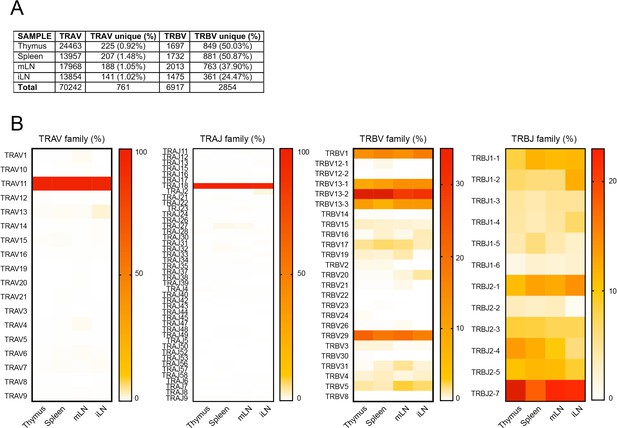
Frequency of TRBV and TRAV gene usage in iNKT cell TCR sequences.
(A) Total number of TRAV and TRBV sequences and percentages of unique sequences obtained from cells on the indicated tissues. (B) Heat map representation of the frequency of TRAV, TRAJ, TRBV and TRBJ sequences for iNKT cells isolated from the depicted tissues. Data obtained from RNAseq and pooled from four samples per tissue.
-
Figure 1—figure supplement 2—source data 1
Frequency of TRBV and TRAV gene usage in iNKT cell TCR sequences.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig1-figsupp2-data1-v3.xlsx
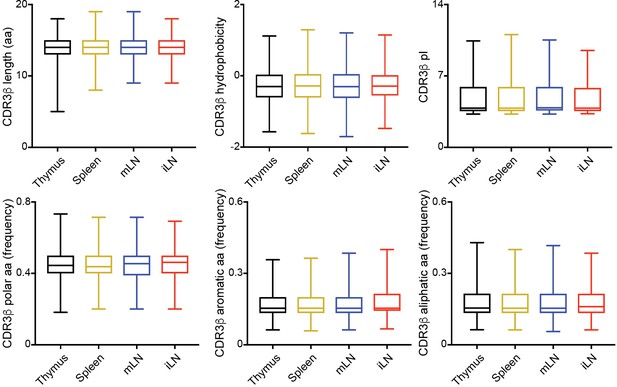
Physicochemical properties of CDR3β sequences.
Median values for length, hydrophobicity, pl, and frequency of polar, aromatic or aliphatic aa in CDR3β sequences identified in the depicted tissues. Data are expressed as box-and-whisker diagrams depicting the median ± lower quartile, upper quartile, sample minimum and maximum. Data obtained from RNAseq and pooled from four samples per tissue.
-
Figure 1—figure supplement 3—source data 1
Physicochemical properties of CDR3β sequences.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig1-figsupp3-data1-v3.xlsx
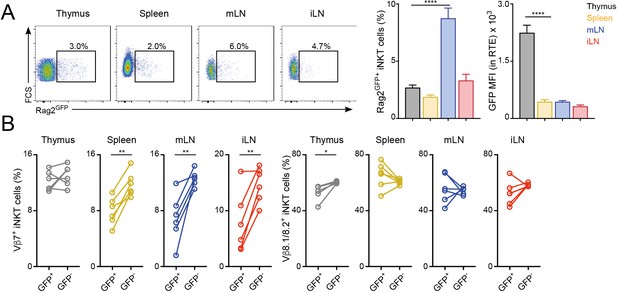
Distinct TCRVβ usage for iNKT RTE.
(A–B) iNKT RTE were identified as GFP+ cells in the tissues of RAG2GFP mice (6–9 weeks/old). (A) Flow-cytometry (left) and quantification (right) showing the percentage of RAG2GFP+ iNKT cells and the MFI for the GFP expression in RTE in the depicted tissues. (B) Frequency of Vβ7 or Vβ8.1/8.2-expressing GFP+ and GFP- iNKT cells from the tissues of RAG2GFP mice. Bars represent mean ± SEM. *p<0.05, **p<0.01, ****p<0.0001 two-tailed unpaired (A) or paired t-test (B). n = 6 mice from two independent experiments.
-
Figure 2—source data 1
Distinct TCRVβ usage for iNKT RTE.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig2-data1-v3.xlsx
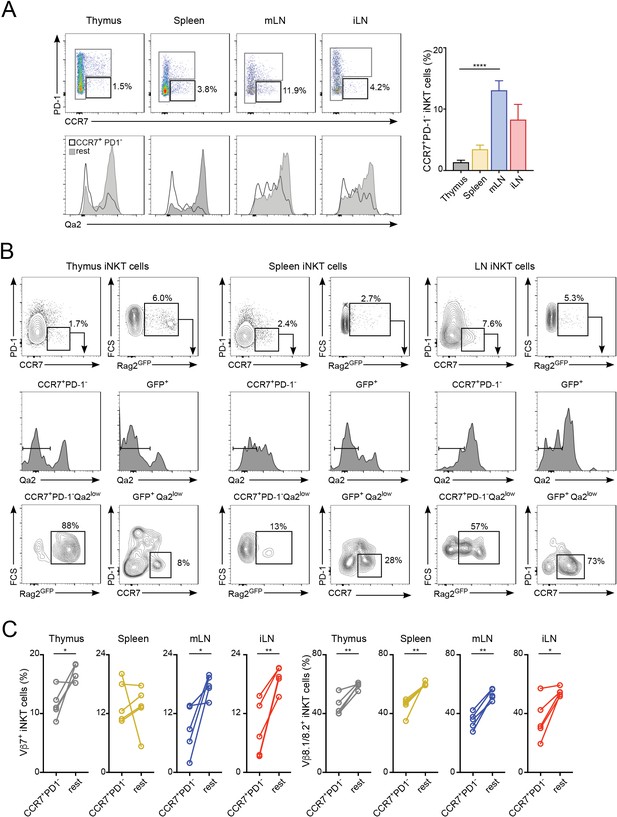
TCRVβ usage for iNKT cell precursors.
(A) iNKT cell precursors were identified by flow-cytometry (left top) as CCR7+PD-1- cells. Q2a expression in iNKT cell precursors (empty profile) and non-precursors (grey, rest) is shown (left bottom). Bar plot shows quantification of CCR7+PD-1- iNKT cells in the depicted tissues (right). ****p<0.0001, two-tailed unpaired paired t-test. n = 5 mice from three independent experiments. (B) Flow-cytometry plots showing expression of CCR7, PD1, Qa2 and GFP in iNKT cells from Rag2-GFP mice. (C) Frequency of Vβ7 or Vβ8.1/8.2-expressing iNKT cell precursors (CCR7+PD-1-) and non-precursors (rest) from the tissues of WT mice. *p<0.05, **p<0.01, two-tailed paired t-test. n = 5 mice from three independent experiments.
-
Figure 2—figure supplement 1—source data 1
TCRVβ usage for iNKT cell precursors.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig2-figsupp1-data1-v3.xlsx
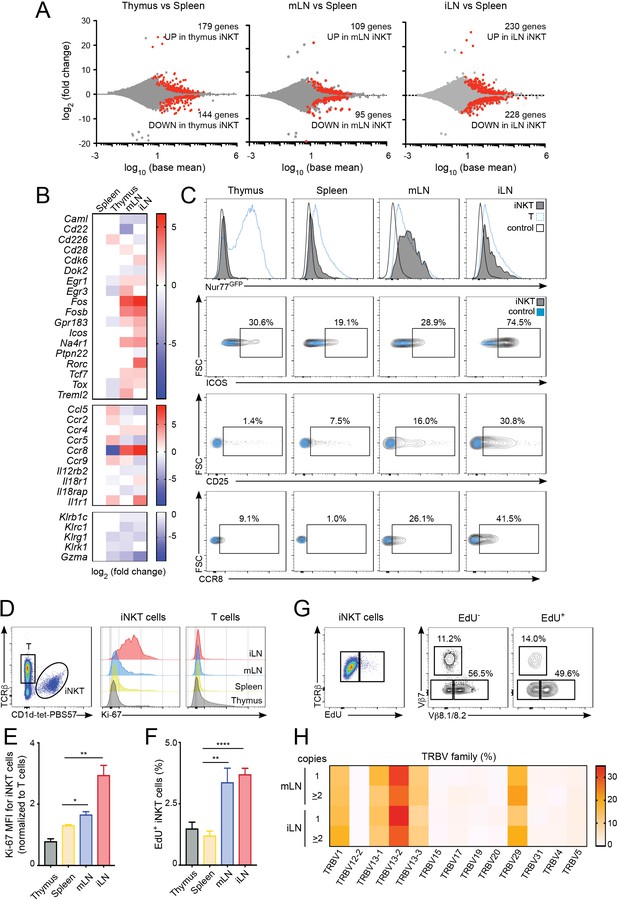
Increased activation and proliferation of LN iNKT cells.
(A) Plots show differentially expressed genes for pairwise comparisons from iNKT cells from the depicted tissues. A fold change cut-off of 1.5 and adjusted p-value cut off of 0.01 were applied to colour code differentially expressed genes on the plot (red). The numbers of differentially expressed genes are indicated in the graphs. (B) Heat map showing RNAseq analyses of selected transcripts significantly changed in iNKT cells from thymus, mLN or iLN versus spleen. n = 4 samples. (C) Top, Representative flow-cytometry histograms showing GFP expression in iNKT cells (grey) and T cells (blue) from the depicted tissues of Nur77GFP mice. iNKT cells from the tissues of Nur77GFP- mice are shown as control (empty profile). Bottom, representative flow-cytometry plots showing CD25, ICOS and CCR8 expression in iNKT cells from the tissues of WT mice (grey) and control (blue). (D–E) Ki-67 expression in iNKT cells and T cells from the depicted tissues of WT mice. Ki-67 MFI for iNKT cells (E) is related to T cells from each tissue. Bars represent mean ± SEM. *p<0.05, **p<0.01, two-tailed unpaired t-test. n = 5 mice from two independent experiments. (F) Quantification of EdU incorporation for iNKT cells from the depicted tissues after 48 hr of EdU administration in vivo. Bars represent mean ± SEM. **p<0.01, ****p<0.0001 two-tailed unpaired t-test. n = 6 mice from three independent experiments. (G) Representative flow-cytometry plot showing EdU incorporation in iNKT cells (left) and frequency of Vβ7 or Vβ8.1/8.2-expressing EdU+ and EdU- iNKT cells (right). n = 4 mice. (H) Heat map representation of the frequency of TRBV amongst low-abundance (one copy) or more abundant (>2 copies) CDR3β sequences obtained from iLN and mLN as indicated. Data obtained from RNAseq and pooled from four samples per tissue.
-
Figure 3—source data 1
Increased activation and proliferation of LN iNKT cells.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig3-data1-v3.xlsx
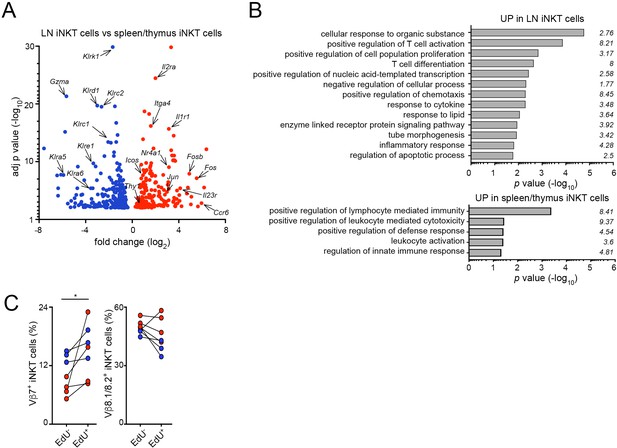
Gene expression analyses for iNKT cells from various tissues.
(A) Differentially expressed genes upregulated (red) or downregulated (blue) in iNKT cells from LNs vs. iNKT cells from spleen/thymus. A fold change cut-off of 1.5 and adjusted p-value cut off of 0.01 were applied to colour code differentially expressed genes on the plot. (B) Functional enrichment analysis of genes upregulated in LN iNKT cells (vs spleen/thymus iNKT; top) or in spleen/thymus iNKT cells (vs LN iNKT; bottom). The GO terms are shown for both sets of genes ranked by p values and fold-enrichments are depicted on the graph. Enrichment and p values (from a Fisher’s exact test with Bonferroni correction) were calculated with PANTHER tools. (C) Frequency of Vβ7 or Vβ8.1/8.2-expressing EdU+ or EdU- iNKT cells from mLN (blue) or iLN (red). n = 4 mice from two experiments. *p<0.05, two-tailed paired t-test.
-
Figure 3—figure supplement 1—source data 1
Gene expression analyses for iNKT cells from various tissues.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig3-figsupp1-data1-v3.xlsx
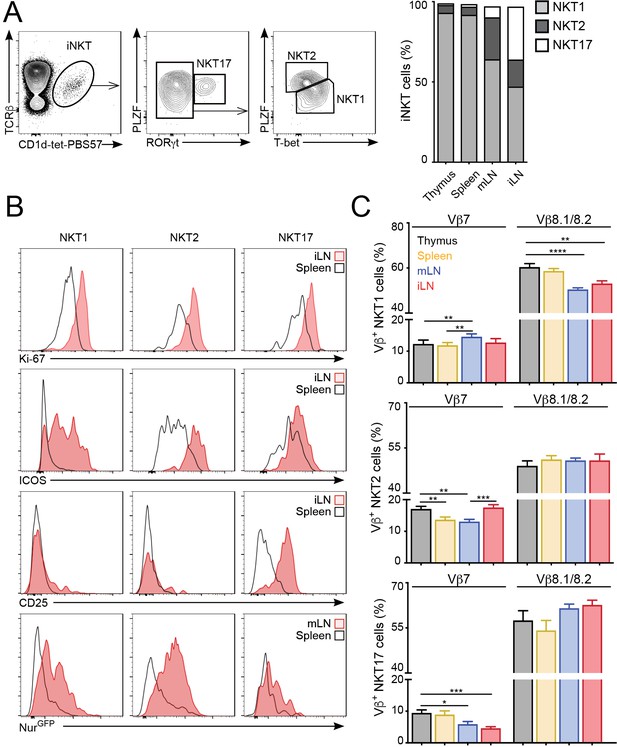
The tissue of origin dictates the basal activation and TCRβ repertoire of all iNKT subsets.
(A) Analysis of iNKT cell populations in the tissues of WT C57BL/6 mice, showing flow-cytometry plots (A, left) and frequency (A, right) of NKT1 (RORγt-PLZFloT-bet+), NKT2 (RORγt-PLZFhiT-bet-) and NKT17 (PLZFintRORγt+) cells. n = 10 mice from four independent experiments. (B) Top, Representative flow-cytometry plots showing Ki-67, CD25 and ICOS expression in NKT1, NKT2 and NKT17 cells from the depicted tissues. Subpopulations were identified as in (A). Bottom, GFP expression in iNKT cell subsets from the depicted tissues from Nur77GFP mice. iNKT cell subsets were identified as described (Engel et al., 2016): NKT1 (CD27+NK1.1+), NKT2 (CD27+, NK1.1-, CD1d-Tethi, CD4+), NKT17 (CD27-, CD4-, CCR6+). (C) Frequency of Vβ7- or Vβ8.1/8.2-expressing iNKT cells within the NKT1 (top), NKT2 (middle) or NKT17 (bottom) populations in the depicted tissues of WT mice. n = 10 mice from four independent experiments. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 paired t-test.
-
Figure 4—source data 1
The tissue of origin dictates the basal activation and TCRβ repertoire of all iNKT subsets.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig4-data1-v3.xlsx
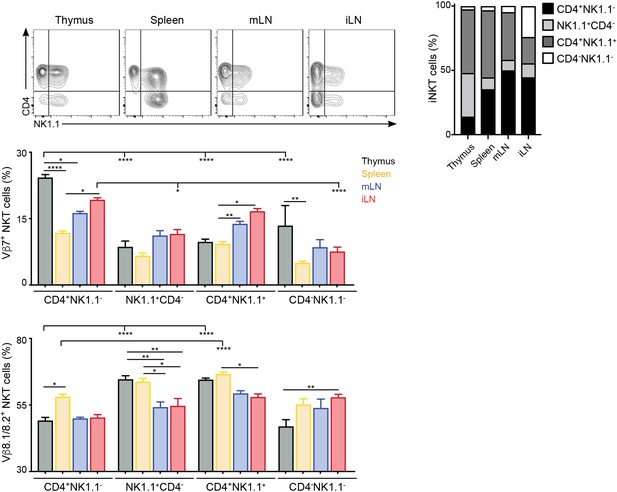
TCRVβ usage for iNKT cell subpopulations.
(Top) Analysis of iNKT cell populations in the tissues of WT C57BL/6 mice, showing flow-cytometry plots (left) and frequency (right) of CD4+NK1.1-, NK1.1+CD4-, CD4+NK1.1+ or CD4-NK1.1- iNKT cells. (Bottom) Frequency of Vβ7- or Vβ8.1/8.2-expressing iNKT cells within the CD4+NK1.1-, NK1.1+CD4-, CD4+NK1.1+ or CD4-NK1.1- populations in the depicted tissues of WT mice. n = 5 mice from two independent experiments. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ANOVA with Tukey’s multiple comparisons test.
-
Figure 4—figure supplement 1—source data 1
TCRVβ usage for iNKT cell subpopulations.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig4-figsupp1-data1-v3.xlsx
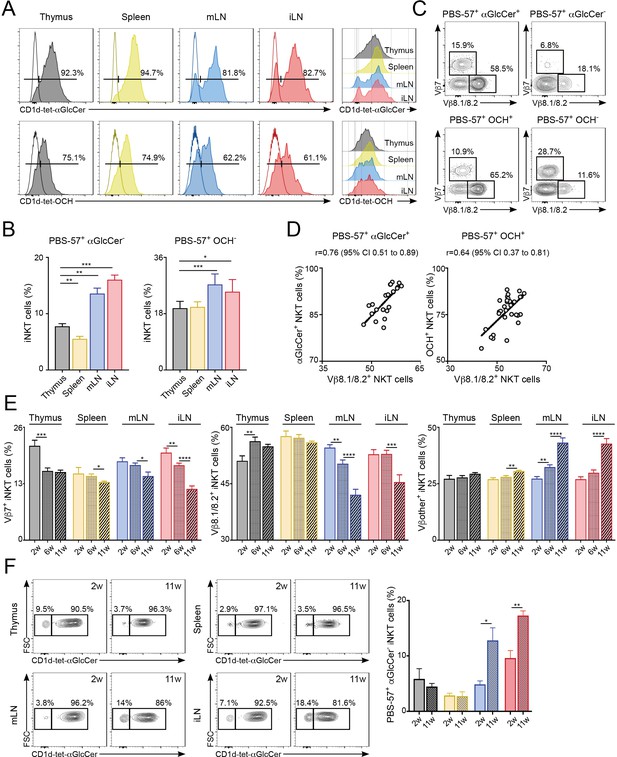
Differential lipid antigen recognition for iNKT cells from various lymphoid tissues.
(A–D) iNKT cells from the depicted tissues were co-stained with CD1d-tet-PBS57 and CD1d-tet-αGlcCer or CD1d-tet-OCH. (A) Flow-cytometry profiles. (B) Quantification of αGlcCer- (left) or OCH- (right) iNKT cells. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, two-tailed paired t-test. (C) Vβ usage for the depicted iNKT cell populations from the spleen. (D) Frequency of αGlcCer+ (left) or OCH+ (right) iNKT cells was related to the frequency of Vβ8.1/8.2 usage for each sample. Pearson correlation analyses are shown for each graph. n = 6–8 mice from 3 to 4 independent experiments. (E–F) TCR Vβ repertoire and lipid antigen recognition of iNKT cells from the depicted tissues were analysed at indicated time points (weeks of age). (E) Frequency of iNKT cells expressing Vβ7, Vβ8.1/8.2 or Vβother (no Vβ7 or Vβ8.1/8.2) in the tissues of WT C57BL/6 mice of 2, 6 or 11 weeks of age. n = 10–15 mice from 10 independent experiments. (F) Flow-cytometry profiles (left) and quantification of αGlcCer- iNKT cells (right) for iNKT cells from the tissues of WT mice of 2 or 11 weeks of age as depicted. n = 3 mice from two independent experiments. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 two-tailed unpaired t-test.
-
Figure 5—source data 1
Differential lipid antigen recognition for iNKT cells from various lymphoid tissues.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig5-data1-v3.xlsx
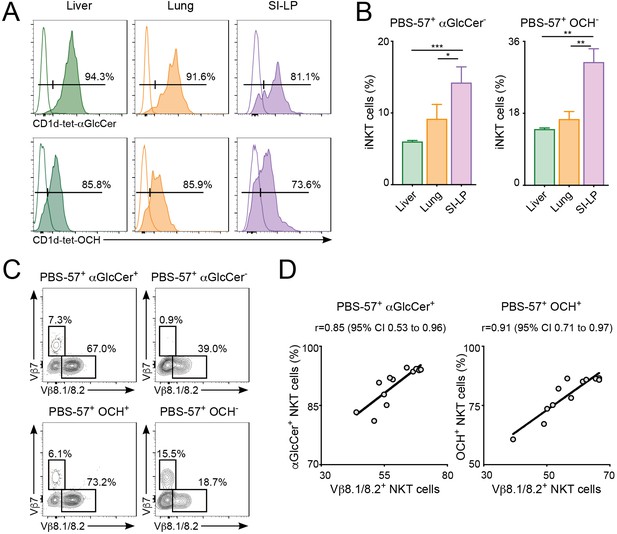
Differential lipid antigen recognition for iNKT cells from non-lymphoid tissues .
(A–D) iNKT cells from liver, lung or SI-LP were co-stained with CD1d-tet-PBS57 and CD1d-tet-αGlcCer or CD1d-tet-OCH. (A) Flow-cytometry profiles. (B) Quantification of αGlcCer- (left) or OCH- (right) iNKT cells. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, two-tailed paired t-test. n = 4 mice from two independent experiments. (C) Vβ usage for the depicted iNKT cell populations from the liver. (D) Frequency of αGlcCer+ (left) or OCH+ (right) iNKT cells was related to the frequency of Vβ8.1/8.2 usage for each sample. Pearson correlation analyses are shown for each graph.
-
Figure 5—figure supplement 1—source data 1
Differential lipid antigen recognition for iNKT cells from non-lymphoid tissues.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig5-figsupp1-data1-v3.xlsx
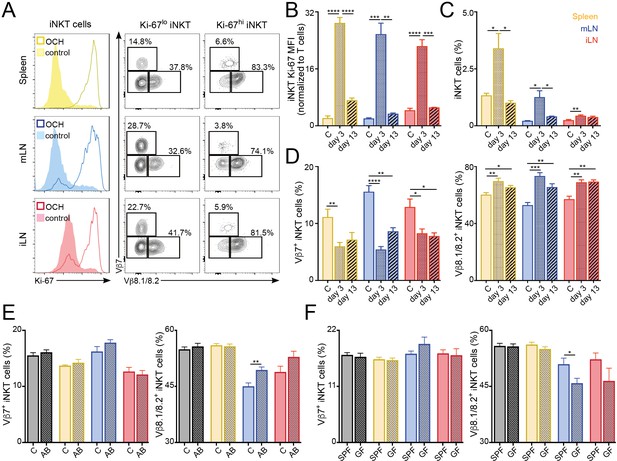
TCR repertoire of iNKT cells changes following immunisation and environmental challenges.
(A–D) Mice were injected with OCH (or PBS as control, c) and iNKT cells from spleen and lymph nodes were analysed 3 or 13 days later. (A) Flow-cytometry profiles showing Ki-67 expression in all iNKT cells (left) and Vβ7, Vβ8.1/8.2 expression for iNKT cells expressing high (Ki-67hi, right) or low (Ki-67lo, middle) Ki-67 3 days after OCH administration. (B–D) Bar plots showing expression of Ki-67 (B), frequency of iNKT cells (C) and percentage of iNKT cells expressing Vβ7 or Vβ8.1/8.2 (D) at the depicted time points in spleen (yellow), mLN (blue) or iLN (red). n = 3–5 mice from 2 to 3 independent experiments. (E–F) Frequency of iNKT cells expressing Vβ7 or Vβ8.1/8.2 in the tissues of mice (Thymus = grey; Spleen = yellow; mLN = blue; iLN = red) treated with antibiotics in the drinking water vs control mice (E) or SPF vs GF mice (F). n = 10 mice (E) and n = 6 mice (F) from two independent experiments. (A–F) Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 two-tailed unpaired t-test.
-
Figure 6—source data 1
TCR repertoire of iNKT cells changes following immunisation and environmental challenges.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig6-data1-v3.xlsx
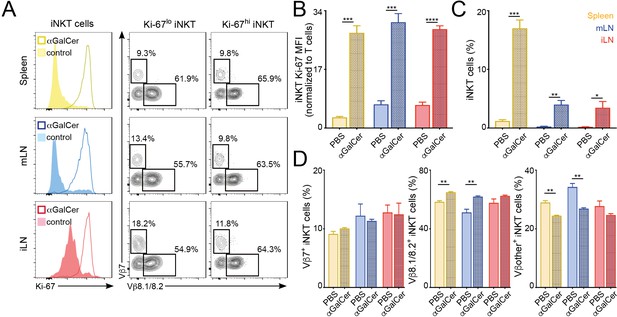
Changes in the iNKT cell TCR repertoire following immunisation with αGalCer.
Mice were injected with αGalCer (or PBS as control) and iNKT cells from spleen and lymph nodes were analysed 3 days later. (A) Flow-cytometry profiles showing Ki-67 expression in all iNKT cells (left) and Vβ7, Vβ8.1/8.2 expression for iNKT cells expressing high (Ki-67hi, right) or low (Ki-67lo, middle) Ki-67 3 days after αGalCer administration. (B–D) Bar plots showing expression of Ki-67 (B), frequency of iNKT cells (C) and percentage of iNKT cells expressing Vβ7, Vβ8.1/8.2 or Vβother (D) at the depicted time points in spleen (yellow), mLN (blue) or iLN (red). n = 3 mice from two independent experiments. Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 two-tailed unpaired t-test.
-
Figure 6—figure supplement 1—source data 1
Changes in the iNKT cell TCR repertoire following immunisation with αGalCer.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig6-figsupp1-data1-v3.xlsx
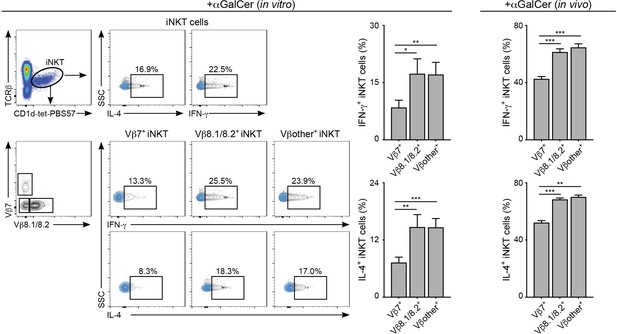
Cytokine secretion by iNKT cells relates to TCRVβ usage.
Splenocytes were stimulated in vitro with αGalCer (left, in vitro) or WT mice were injected in vivo with αGalCer (right, in vivo) and cytokine production was measured by intracellular staining 2 hr later. Flow cytometry graphs and quantification (bar plots) showing the frequency of IL-4 or IFN-γ-producing iNKT cells within the Vβ7+, Vβ8+ or Vβother+ populations. n = 7 mice (in vitro) and n = 4 mice (in vivo). Bars represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ANOVA with Tukey’s multiple comparisons test.
-
Figure 6—figure supplement 2—source data 1
Cytokine secretion by iNKT cells relates to TCRVβ usage.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig6-figsupp2-data1-v3.xlsx
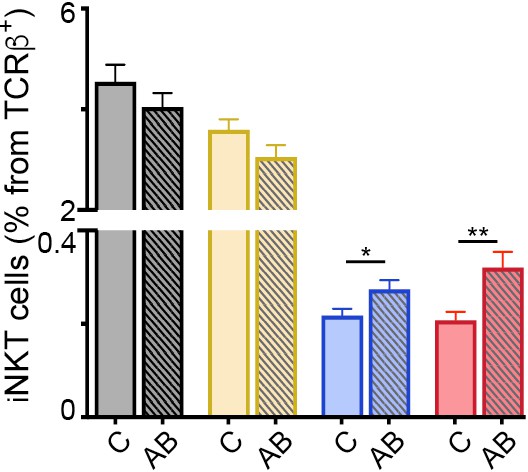
Frequency of iNKT cells after antibiotic treatment.
Frequency of iNKT cells in the tissues of mice (Thymus = grey; Spleen = yellow; mLN = blue; iLN = red) treated with antibiotics in the drinking water vs control mice. n = 10 mice from two independent experiments. Bars represent mean ± SEM. **p<0.01 two-tailed unpaired t-test.
-
Figure 6—figure supplement 3—source data 1
Frequency of iNKT cells after antibiotic treatment.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig6-figsupp3-data1-v3.xlsx
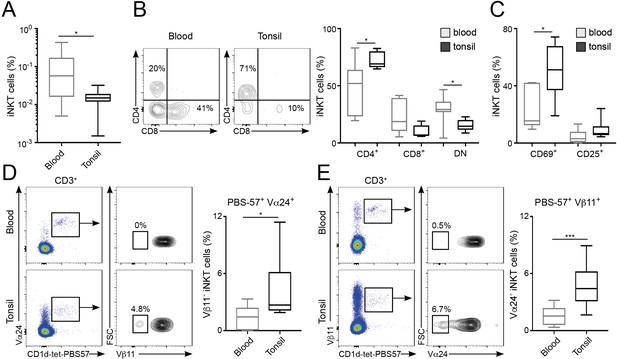
Distinct phenotype and TCR repertoire for human iNKT cells from different anatomical locations.
(A) Mean percentage of iNKT cells (CD1d-tet-PBS57+CD3+) from total CD3+B220-CD14- cells in blood or tonsils. (B–C) Flow-cytometry (left) and quantification (right) showing the percentage of CD4+, CD8+ and DN cells (B) or CD69+ and CD25+ cells (C) within the iNKT cell population in the depicted tissues. (D–E) Flow-cytometry (left) and quantification (right) showing the percentage of Vβ11- iNKT cells within CD1d-tet-PBS57+Vα24+ cells (D); or the percentage of Vα24- cells within CD1d-tet-PBS57+Vβ11+ cells (E). (A–E) Data are expressed as box-and-whisker diagrams depicting the median ± lower quartile, upper quartile, sample minimum and maximum. *p<0.05, ***p<0.001, two-tailed unpaired t-test. n = 10 samples per tissue from four independent experiments.
-
Figure 7—source data 1
Distinct phenotype and TCR repertoire for human iNKT cells from different anatomical locations.
- https://cdn.elifesciences.org/articles/51663/elife-51663-fig7-data1-v3.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Rag2-GFP: Tg(Rag2-EGFP)1Mnz | PMID: 10458165 | MGI:3784416 | |
| Strain, strain background (Mus musculus) | Nur77-GFP: C57BL/6-Tg(Nr4a1-EGFP/cre)820Khog/J | PMID: 21606508 | MGI:5007644 | |
| Strain, strain background (Mus musculus) | CD1d-KO: Del(3Cd1d2-Cd1d1)1SbpJ | PMID: 14632651 | MGI:5582477 | |
| Antibody | PBS57-loaded CD1d-tetramer (mouse) | NIH Tetramer Core Facility | https://tetramer.yerkes.emory.edu | (1:1000) |
| Antibody | anti-mouse B220 (rat monoclonal) | BioLegend | 103224 | (1:200) |
| Antibody | anti-mouse CD8α (rat monoclonal) | BioLegend | 100714 | (1:200) |
| Antibody | anti-mouse CD11b (rat monoclonal) | BioLegend | 101226 | (1:200) |
| Antibody | anti-mouse CD11c (armenian hamster monoclonal) | BioLegend | 117323 | (1:200) |
| Antibody | anti-mouse PLZF (armenian hamster monoclonal) | BioLegend | 145807 | (1:200) |
| Antibody | anti-mouse RORγt (mouse monoclonal) | BD Biosciences | 564722 | (1:200) |
| Antibody | anti-mouse T-bet (mouse monoclonal) | BioLegend | 644823 | (1:200) |
| Antibody | anti-mouse TCRβ (armenian hamster monoclonal) | BioLegend | 109233 | (1:200) |
| Antibody | anti-mouse Vβ7 (rat monoclonal) | BioLegend | 118306 | (1:200) |
| Antibody | anti-mouse Vβ8.1/8.2 (rat monoclonal) | eBiosciences | 46-5813-80 | (1:200) |
| Antibody | anti-mouse CCR7 (rat monoclonal) | BioLegend | 120105 | (1:100) |
| Antibody | anti-mouse PD1 (rat monoclonal) | BioLegend | 135219 | (1:200) |
| Antibody | anti-mouse Qa2 (mouse monoclonal) | BD Biosciences | 743312 | (1:200) |
| Antibody | anti-mouse CD25 (rat monoclonal) | BioLegend | 102015 | (1:200) |
| Antibody | anti-mouse Ki-67 (rat monoclonal) | BioLegend | 652425 | (1:200) |
| Antibody | anti-mouse ICOS (rat monoclonal) | BioLegend | 117405 | (1:200) |
| Antibody | anti-mouse CD4 (rat monoclonal) | BioLegend | 100433 | (1:200) |
| Antibody | anti-mouse NK1.1 (mouse monoclonal) | eBiosciences | 11-5941-85 | (1:200) |
| Antibody | anti-mouse CD27 (armenian hamster monoclonal) | BioLegend | 124215 | (1:200) |
| Antibody | anti-mouse CCR6 (armenian hamster monoclonal) | BioLegend | 129809 | (1:200) |
| Antibody | anti-mouse CD45.1 (mouse monoclonal) | BioLegend | 110731 | (1:200) |
| Antibody | anti-mouse CD45.2 (mouse monoclonal) | BioLegend | 109805 | (1:200) |
| Antibody | anti-mouse CCR8 (rat monoclonal) | BioLegend | 150320 | (1:200) |
| Antibody | anti-mouse IL-4 (rat monoclonal) | BioLegend | 504111 | (1:200) |
| Antibody | anti-mouse IFN-γ (rat monoclonal) | BioLegend | 505810 | (1:200) |
| Antibody | PBS57-loaded CD1d-tetramer (human) | NIH Tetramer Core Facility | https://tetramer.yerkes.emory.edu | (1:500) |
| Antibody | anti-human CD3 (mouse monoclonal) | BioLegend | 300418 | (1:200) |
| Antibody | anti-human CD4 (rat monoclonal) | BioLegend | 357415 | (1:200) |
| Antibody | anti-human CD8a (mouse monoclonal) | BioLegend | 300913 | (1:200) |
| Antibody | anti-human CD25 (mouse monoclonal) | BioLegend | 302613 | (1:200) |
| Antibody | anti-human CD69 (mouse monoclonal) | BioLegend | 310921 | (1:200) |
| Antibody | anti-human Vα24 (mouse monoclonal) | BioLegend | 360003 | (1:200) |
| Antibody | anti-human Vβ11 (human monoclonal) | Miltenyi Biotech | 130-108-799 | (1:200) |
| Antibody | anti-human CD19 (mouse monoclonal) | BioLegend | 302223 | (1:200) |
| Antibody | anti-human CD14 (mouse monoclonal) | BioLegend | 325615 | (1:200) |
| Commercial assay or kit | Zombie (fixable viability dye) | BioLegend | 423105 | |
| Commercial assay or kit | Dynabeads Biotin binder | Invitrogen | 11047 | |
| Commercial assay or kit | Click-iT Plus EdU Flow-cytometry assay kit | Invitrogen | C10418 | |
| Chemical compound, drug | αGalCer (α-Galactosylceramide, KRN7000) | Enzo Life Sciences | ALX-306–027 | |
| Chemical compound, drug | OCH | Enzo Life Sciences | ALX-306–029 | |
| Software, algorithm | MiXCR | Bolotin et al. (2013) | https://mixcr.readthedocs.io/en/master/ | |
| Software, algorithm | Weblogo | Crooks et al. (2004) | https://weblogo.berkeley.edu | |
| Software, algorithm | Brepertoire | Margreitter et al. (2018) | http://mabra.biomed.kcl.ac.uk/BRepertoire_5/? |





