Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon–anticodon pairing
Figures
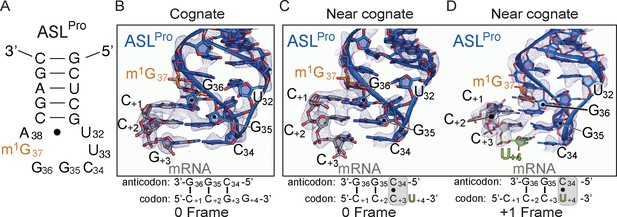
The ability of ASLPro to +1 frameshift in the peptidyl (P) site is dependent on the near-cognate mRNA codon.
(A) Secondary structure of the anticodon stem-loop (ASL) of tRNAPro. The m1G37 modified nucleotide is shown in orange and the anticodon nucleotides (C34, G35, and G36) and the U32-A38 pairing are labeled. (B) Structure of 70S-ASLPro bound to a cognate CCG codon in the P site with the codon in the 0 or canonical frame. All 2Fo-Fc electron density maps shown in panels B-D are contoured at 1.0σ. (C,D) Structure of 70S-ASLPro bound to a +1 slippery CCC-U codon shows the mRNA position is either in the 0 (panel C) or +1 frame (panel D) in the two 70S molecules in the crystallographic asymmetric unit. In either the 0 or +1 frame, a cis Watson–Crick interaction at the third base-pair forms (C+3•C34 or U+4•C34). mRNA numbering starts at +1 according to the first position in the P site.
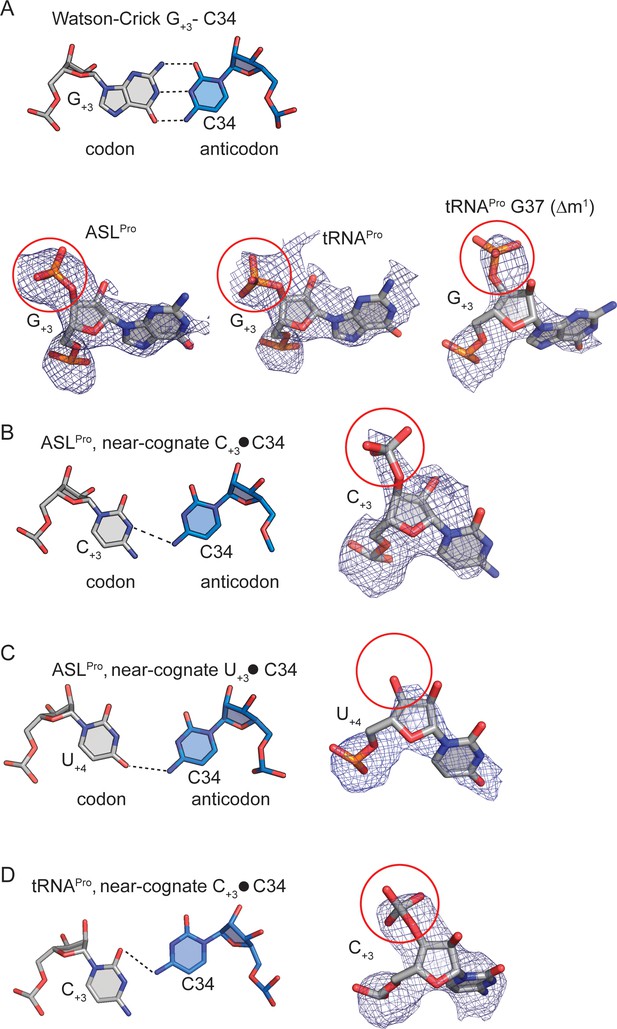
Determining the mRNA frame by visualizing the +4 nucleotide mRNA phosphate density.
(A) A cognate G+3-C34 base-pair (mRNA nucleotide:anticodon nucleotide) is shown above. Representative 2Fo-Fc density is shown for the same cognate base-pairingbetween mRNA and ASLPro/tRNAPro(below). The phosphate of G+4is circled in red and its presence signifies that the mRNA is in the 0 frame (B) A near-cognate ASLPro C+3-C34 base-pair is shown with 2Fo-Fc density. The phosphate of U+4 is circled in red and its presence signifies that the mRNA is in the 0 frame. (C) A near-cognate ASLPro U+3-C34 base-pairi is shown with 2Fo-Fc density surrounding U+4.There is a lack of density for an addition phosphate group (circled in red) indicating that another nucleotide is not present and the mRNA is in the +1 frame. (D) A near-cognate tRNAPro C+3-C34 base-pair is shown with 2Fo-Fc density surrounding C+3.The phosphate of U+4 is circled in red and its presence signifies that the mRNA is in the 0 frame. 2Fo-Fc density are set at 1σ for all structures. The cutoff for a base-pair hydrogen bond is 3.5 Å.
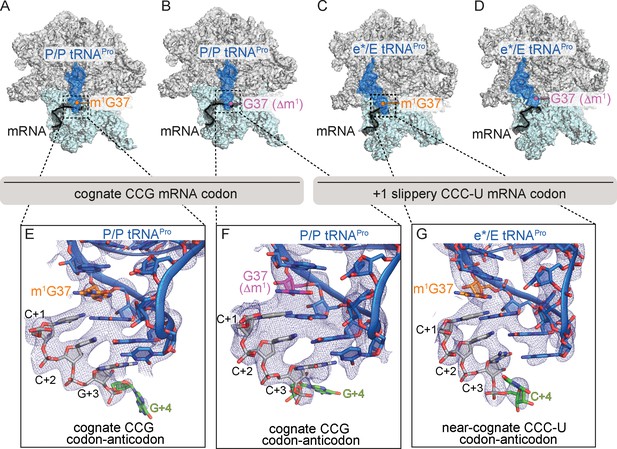
Identity of the mRNA proline codon regulates 30S head domain swivel and tilting.
Overview of 70S ribosome-tRNAPro complex structures: (A) tRNAPro m1G37 on a cognate CCG codon adopts a P/P orientation (located on the P site on the 30S and 50S subunits); (B) tRNAPro lacking the m1G37 modification (G37 Δm1) on a cognate CCG codon also adopts a P/P orientation; (C) tRNAPro on a +1 slippery CCC-U codon adopts an e*/E orientation (e* denotes the location between the E and P sites on the 30S while "E" is the E site of the 50S); and (D) tRNAPro lacking the m1G37 (G37 Δm1) on a +1 slippery CCC-U codon adopts an e*/E orientation. In this complex (panel D), the 30S head domain and anticodon-codon interaction are disordered. In panels A-D, the 16S rRNA of the 30S head domain is removed for clarity. Zoomed-in view of 2Fo-Fc density of (E) the codon–anticodon for tRNAPro on a cognate CCG codon, (F) tRNAPro G37 (Δm1) on a cognate CCG codon in the P site, and (G) tRNAPro G37 (Δm1) on a near-cognate CCC-U codon in position between the E and the P sites (e*). All 2Fo-Fc electron density maps shown in panels E-G are contoured at 1.0σ.

mRNA and full-length tRNAProelectron density.
(A) 2Fo-Fc electron density for full-length tRNAPro on a cognate codon in the P site. (B) Zoomed-in view of codon–anticodon 2Fo-Fc density for full-length tRNAPro on a cognate CCG codon. (C) 2Fo-Fc density for full-length tRNAPro G37 (Δm1) on a cognate codon in the P site. (D) Zoomed-in view of codon–anticodon 2Fo-Fc density for full-length tRNAPro G37 (Δm1) on a cognate CCG codon. (E) 2Fo-Fc density for full-length tRNAPro on a near-cognate codon in the e* site. (F) Zoomed-in view of codon–anticodon 2Fo-Fc density for tRNAPro on a near-cognate CCC-U codon. (G) 2Fo-Fc density for 16S rRNA and full-length tRNAPro G37 (Δm1) on a near-cognate CCC-U codon in the e* site. The portion of the ASL that lacks density is represented as a hollow blue dashed line. 2Fo-Fc density is is contoured at 1σ for all structures. In the zoomed-in views of panels B, D, and F, the +four position on the mRNA is highlighted in green to indicate the structures are in the 0 frame.
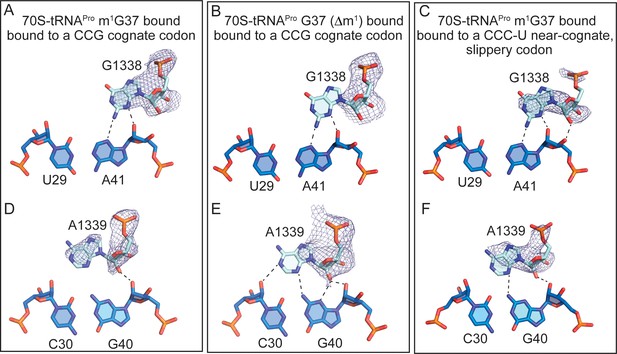
Full-length tRNAPro interactions with 16S nucleotides G1338 and A1339.
Comparison of the A-minor motif interactions of G1338 with tRNAPro U29-A41 base-pairs (A) and A1339 with tRNAPro C30-G40 base-pairs in (D) 70S-tRNAPro bound to a cognate CCG codon. (B,E) 70S-tRNAPro G37 (Δm1) bound to a cognate CCG codon. (C,F) 70S-tRNAPro bound to a near-cognate CCC-U codon. 2Fo-Fc density for G1338 and A1339 is contoured at 1σ.
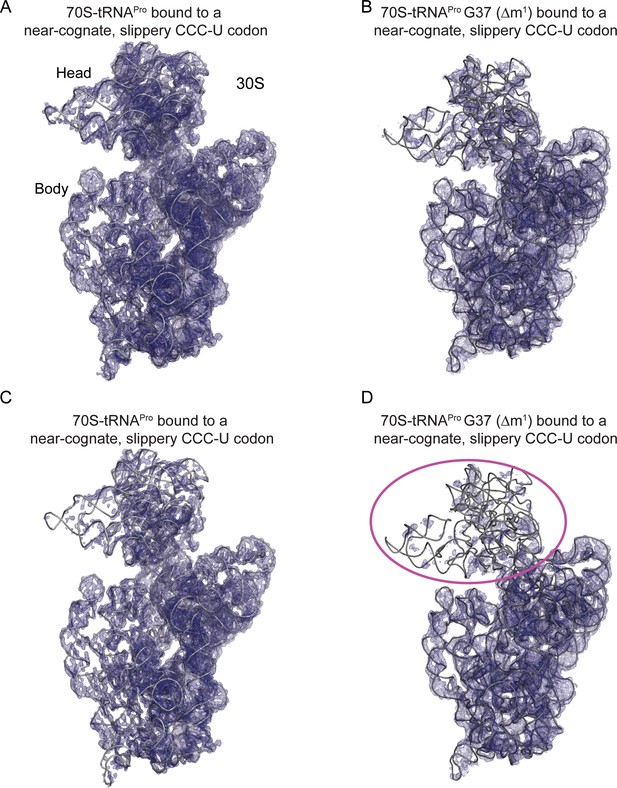
tRNAPro G37 (Δm1) coupled with a slippery codon capable of +1 frameshifting induces disorder in the 30S head domain.
(A) 2Fo-Fc density at 1.0σ of 16S rRNA in the 70S-tRNAPro bound to a cognate CCG codon. (B) 2Fo-Fc density at 1.0σ of 16S rRNA in the 70S-tRNAPro G37 (Δm1) bound to a near-cognate CCC-U codon. (C,D) The same ribosome complexes as in panels A and B but with 2Fo-Fc electron density increased to 1.5σ.
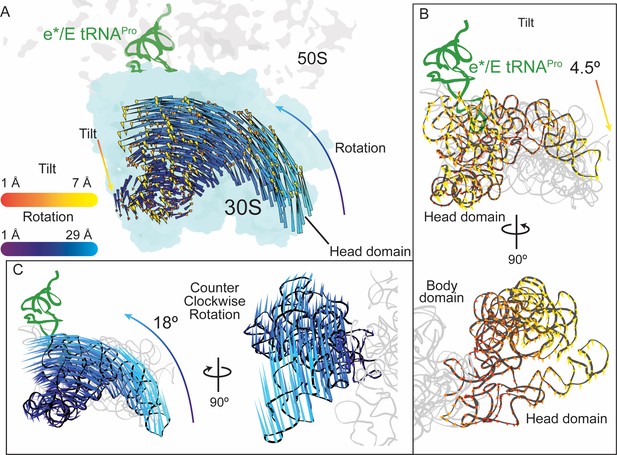
30S head domain movement in the presence of a +1 frameshift-prone tRNA.
(A) Overview of the 70S ribosome containing an e*/E tRNAPro bound to a +1 slippery CCC-U codon. Shifts in phosphate atom positions of the 30S head domain (16S rRNA nucleotides 930–1380) in this structure as compared to the unrotated 70S (PDB code 4V5C) are shown as two vectors corresponding to the two directions of rotation/swivel (blue) and tilt (orange and yellow). (B) Top, same view as in panel A but showing only the tilt of the head domain. Bottom, a 90° rotated view showing the tilt is downward resulting in movement of the head domain away from the body domain. (C) Left, the same view as in panel A with only the counterclockwise swivel/rotation of the head domain indicated (left). Right, a 90° horizontal rotated view shows that the swivel is greatest toward the subunit interface, close to e*/E tRNAPro and on the surface of the ribosome.
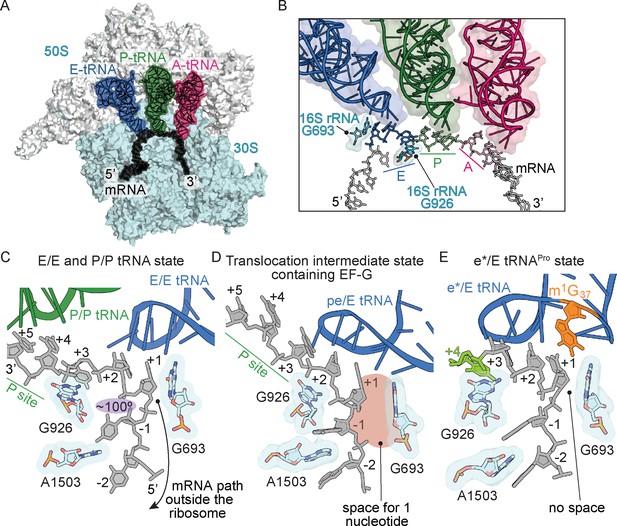
Frame-dependent conformations of the mRNA in the E and P sites.
(A) Overview of the 70S ribosome with (B) a zoomed-in view of the mRNA-tRNA interaction in the A, P, and E sites (PDB code 4V6F). 16S rRNA nucleotides G693 and G926 interact with the E-site codon–anticodon. (C) The normal path of the mRNA (black) in a ribosome structure containing P/P and E/E tRNAs demonstrates only a three-nucleotide codon (nucleotides +1, +2 and +3) is accommodated in the E site (PDB code 4V5F). 16S rRNA G693 defines the starts of the E-site codon and interacts with the first nucleotide. As the mRNA leaves the E site, there is a 100° kink between the first nucleotide of the E-site codon (+1) and the −1 nucleotide (shaded in purple). Panel C is rotated ~180° relative to the view in panel B. (D) A translocation intermediate structure induced by EF-G contains a tRNA positioned between the P and the E sites on the 30S (denoted ‘pe’). The pe/E tRNA has not undergone full translocation to the E site and thus only two nucleotides (+1 and +2) are located in the E site (PDB code 4W29). In this translocation intermediate state, there is space available to accommodate an additional nucleotide of the codon (shaded in red) that would occur upon full translocation. (E) tRNAPro bound to a +1 slippery CCC-U codon reveals that although the codon–anticodon pair has not been fully translocated, this placement of the mRNA is different as compared to normal translocation intermediates structures as shown in panel D. The additional nucleotide (+4) of the four-nucleotide codon is shown in green.
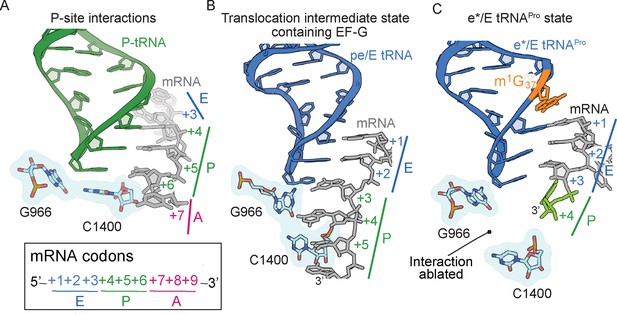
The 16S rRNA G966-C1400 gate between the 30S head and body domains is disrupted during a frameshift event.
(A) In a post-translocation state containing E/E, P/P and A/A tRNAs, 16S rRNA nucleotides G966 and C1400 are located beneath the P-site tRNA (PDB code 4V5F). (B) The G966 and C1400 interaction remains intact during EF-G-mediated translocation as the position of 30S head domain nucleotide G966 shifts while 30S body nucleotide C1400 remains constant (PDB code 4W29). (C) In a ribosome undergoing a +1 frameshift induced by tRNAPro and a slippery CCC-U codon, the G966-C1400 interaction is broken. The additional nucleotide (+4) of the four-nucleotide codon is shown in green.

30S interactions with the P-site ASLPro.
(A) 70S P-site ASLPro interacting with a cognate CCG codon in the 0 frame. (B) 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the 0 frame. (C) 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the +1 frame. 16S rRNA nucleotides G1338, A1339, C1400, and G966 are shown in panels A-C. The following interactions for each of these three structures are shown beneath. G1338 contacts the minor groove of ASLPro at nucleotides U29-A41 in the structrue of 70S P-site ASLPro interacting with a cognate CCG codon in the 0 frame (D), in the structure of 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the 0 frame (E) and in the structure of 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the +1 frame (F). A1339 contacts ASLPro at nucleotides C30-G40 in the structrue of 70S P-site ASLPro interacting with a cognate CCG codon in the 0 frame (G), in the structure of 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the 0 frame (H) and in the structure of 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the +1 frame (I). 2Fo-Fc density for G1338 and A1339 is contoured at 1σ for all structures. (J) C1400 stacks with G+3 mRNA and ASLPro C34 in the structrue of 70S P-site ASLPro interacting with a cognate CCG codon in the 0 frame (J), in the structure of 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the 0 frame (K) and in the structure of 70S P-site ASLPro interacting with a near-cognate CCC-U codon in the +1 frame (L). The distance cutoff for a hydrogen bondt between 16S and ASLPro is 3.5 Å.
Videos
mRNA +1 frameshifting of ASLPro in the peptidyl (P) site is dependent on the slippery mRNA codon.
An overview of the 70S ribosome (50S is light cyan and the 30S in gray) showing how the anticodon stem-loop (ASL) of tRNAPro (blue) interacts with either a cognate CCG codon or a slippery CCC-U codon (black). The m1G37 modified nucleotide is shown in orange. A zoomed-in view of the anticodon nucleotides (G36, G35, and C34) interacting with the cognate C+1, C+2 and G+3 mRNA respectively, in the 0 frame. All 2Fo-Fc electron density maps shown are contoured at 1.0 σ. A morph from the 0 frame structure to the structure of ASLPro interacting with a slippery CCC-U codon is shown to demonstrate the movement of the mRNA into the +1 frame. The fourth nucleotide of the mRNA codon is shown in green. In either the 0 or +1 frame, a cis Watson–Crick interaction at the third base-pair forms (C+3•C34 or U+4•C34). mRNA numbering starts at +1 according to the first position of the mRNA in the P site.
Influence of m1G37 and the slippery codon on +1 frameshifting and conformational changes of the 30S head domain.
An overview of the 70S ribosome (50S is light cyan and the 30S in gray) showing how tRNAPro (blue) (+/- m1G37) interacts with either a cognate CCG codon or a slippery CCC-U codon (black). A zoomed-in view of the anticodon nucleotides (G36, G35, and C34) interacting with the cognate C+1, C+2, and G+3 mRNA respectively, in the 0 frame. All 2Fo-Fc electron density maps shown are contoured at 1.0σ. The m1G37 modified nucleotide is shown in orange and the U32-A38 pairing is indicated. A morph of the changes of the U32-A38 pairing when the tRNA interacts with either a cognate CCG or a slippery CCC-U codon is shown. An overview and a morph of P/P tRNAPro bound to a cognate CCG codon to the e*/E site along with movement of the 30S head domain is shown. Lastly, a zoomed-in view shows a morph from a 0 frame codon-anticodon interaction that forms in the P/P site to the pairing in the e*/E site.
Tables
RNAs used in this study.
| tRNAPro 5’ half | 5’-CGGUGAUUGGCGCAGCCUGGUAGCGCACUUCGUUCGGm1GA-3’ |
| tRNAPro 3’ half | 5’-CGAAGGGGUCGGAGGUUCGAAUCCUCUAUCACCGACCA-3’ |
| mRNA_cognate | 5’- GGCAAGGAGGUAAAA CCGG-3’ |
| mRNA_slippery | 5’- GGCAAGGAGGUAAAA CCCU-3’ |
-
The underlined nucleotides indicates the Shine-Dalgarno region while the bold nucleotides are the P-site codons.
Data collection and refinement statistics for 70S-ASLPro structures.
| 70S-ASLPro-CCG codon | 70S-ASLPro-CCC-U codon | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Wavelength (Å) | 0.9791 | 0.9791 |
| Cell dimensions | ||
| a, b, c (Å) | 209.79,451.91,621.58 | 210.12,451.80,622.96 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 49.70–3.10 (3.21–3.10) | 49.70–3.40 (3.52–3.40) |
| Rpim (%) | 21.2 (80.5) | 14.5 (65.5) |
| I/σI | 4.7 (1.1) | 5.9 (1.3) |
| Completeness (%) | 97.99 (87.41) | 98.85 (94.87) |
| Redundancy | 3.7 (3.1) | 4.2 (3.4) |
| CC1/2 | 0.968 (0.250) | 0.987 (0.359) |
| Refinement | ||
| Reflections | 1034968 (91757) | 797600 (75973) |
| Rwork/Rfree (%) | 24.1/27.2 | 19.8/23.7 |
| No. atoms | 289313 | 290047 |
| B-factors (Å2) | ||
| Overall | 90.37 | 103.79 |
| Macromolecule | 90.6 | 104.05 |
| Ligand/ion | 39.43 | 43.84 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.004 | 0.010 |
| Bond angles (°) | 0.83 | 0.98 |
| PDB ID | 6NTA | 6NSH |
-
Highest resolution shell is shown in parentheses.
Data collection and refinement statistics for 70S-tRNAPro structures.
Highest resolution shell is shown in parentheses.
| tRNAPro m1G37-CCG codon | tRNAPro m1G37-CCC-U codon | tRNAPro G37(Δm1)-CCG codon | tRNAPro G37(Δm1)-CCC-U codon | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 | 0.9792 |
| Cell dimensions | ||||
| a, b, c (Å) | 210.20,451.47,620.21 | 210.74,450.26,626.11 | 209.97,450.71,619.40 | 210.09,450.32,622.89 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 49.20–3.20 (3.31–3.20) | 49.93–3.50 (3.63–3.50) | 49.82–3.97 (4.11–3.97) | 49.83–4.14 (4.29–4.14) |
| Rpim (%) | 11.5 (51.7) | 9.00 (44.1) | 8.60 (86.0) | 9.807 (95.5) |
| I/σI | 6.7 (1.5) | 7.8 (1.7) | 6.3 (1.0) | 5.1 (1.0) |
| Completeness (%) | 99.11 (98.45) | 97.55 (89.72) | 98.60 (95.72) | 98.39 (95.63) |
| Redundancy | 5.9 (4.4) | 4.2 (2.2) | 14.1 (10.0) | 6.5 (3.6) |
| CC1/2 | 0.991 (0.377) | 0.996 (0.464) | 0.998 (0.313) | 0.998 (0.37) |
| Refinement | ||||
| Reflections | 951115 (93932) | 723555 (66052) | 4959167 (47742) | 439195 (42360) |
| Rwork/Rfree (%) | 22.8/25.6 | 23.4/25.6 | 22.8/25.5 | 24.8/29.4 |
| No. atoms | 291966 | 292039 | 291793 | 291185 |
| B-factors (Å2) | ||||
| Overall | 103.87 | 113.6 | 177.39 | 247.09 |
| Macromolecule | 104.12 | 113.9 | 177.77 | 247.54 |
| Ligand/ion | 42.04 | 37.71 | 72.42 | 123.12 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.011 | 0.010 | 0.007 | 0.007 |
| Bond angles (°) | 1.00 | 1.23 | 0.94 | 1.38 |
| PDB ID | 6NUO | 6NWY | 6O3M | 6OSI |





