Etv transcription factors functionally diverge from their upstream FGF signaling in lens development
Figures
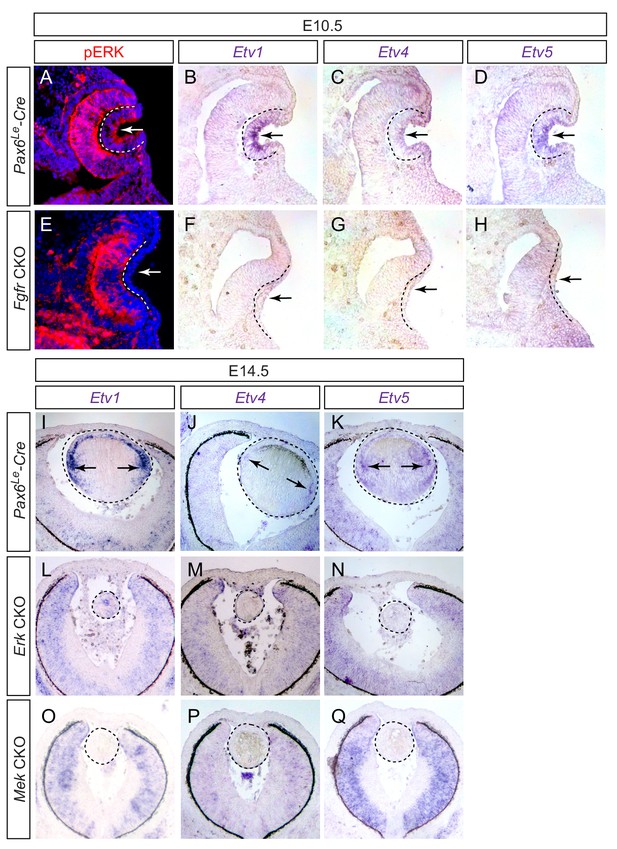
Etv transcription factors are controlled by FGF-ERK signaling in the lens.
(A–D) At E10.5, the invaginating lens ectoderm displays ERK phosphorylation and expression of Etv1, 4 and 5 (arrows). (E–H) Genetic ablation of Fgfr1/2 in Fgfr CKO mutants prevented lens vesicle formation and abrogated phospho-ERK and Etv expression (arrows). The lens ectoderms are marked by dotted lines. (I–K) At E14.5, Etv1, 4 and 5 are predominantly expressed in the transitional zone of the lens (arrows). (L–Q) Deletion of either Erk1/2 (Erk CKO) or Mek1/2 (Mek CKO) abolished expression of Etv genes. The lenses are circled in dotted lines.
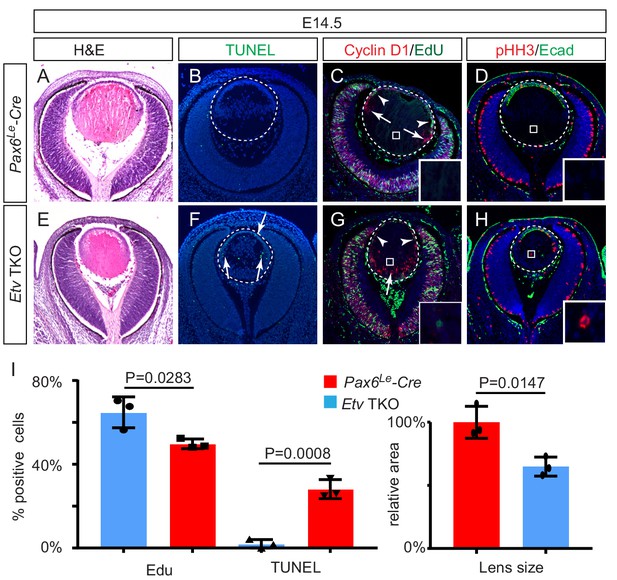
Lens development requires Etv transcription factors.
(A–H) Hematoxylin and eosin (H and E) staining reveal reduced lens size in Etv1/4/5 deletion (Etv TKO) mutants (A and E). Etv null lens exhibited increased cell apoptosis shown by TUNEL staining (B and F, arrows) and reduced cell proliferation as indicated by EdU staining (C and G, arrowheads). In contrast, ectopic expression of Cyclin D1 (G, arrows) and proliferation markers EdU and pHH3 (C, D, G and H, inserts) were detected in the posterior lens. (I) Quantification of EdU and TUNEL staining.
-
Figure 2—source data 1
Source data for Figure 2I.
- https://cdn.elifesciences.org/articles/51915/elife-51915-fig2-data1-v2.xlsx
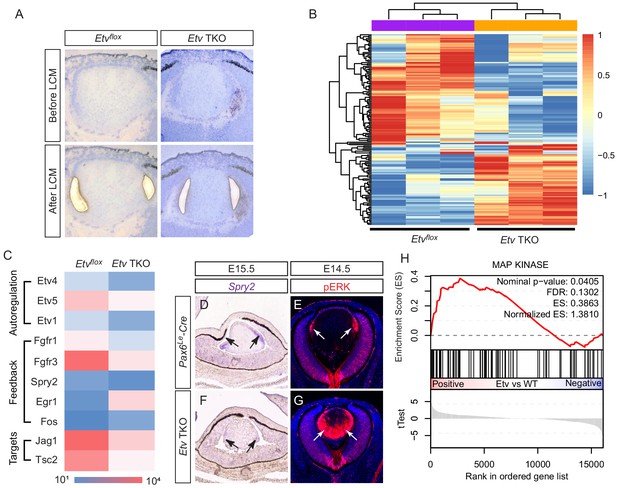
Transcriptomic analysis shows ERK signaling dysregulation in Etv mutant lens.
(A) The transitional zone of the lens was isolated by laser capture microscope (LCM) for RNA sequencing analysis. (B) Cluster analysis of the top differentially expressed genes in the RNA sequencing data. (C) Heat map of the Etv regulated genes. (D and F) Spry2 is significantly down-regulated in Etv TKO mutants. (H) Gene set enrichment analysis (GSEA) indicates the MAPK pathway is elevated.
-
Figure 3—source data 1
Source data for Figure 3C.
- https://cdn.elifesciences.org/articles/51915/elife-51915-fig3-data1-v2.xlsx
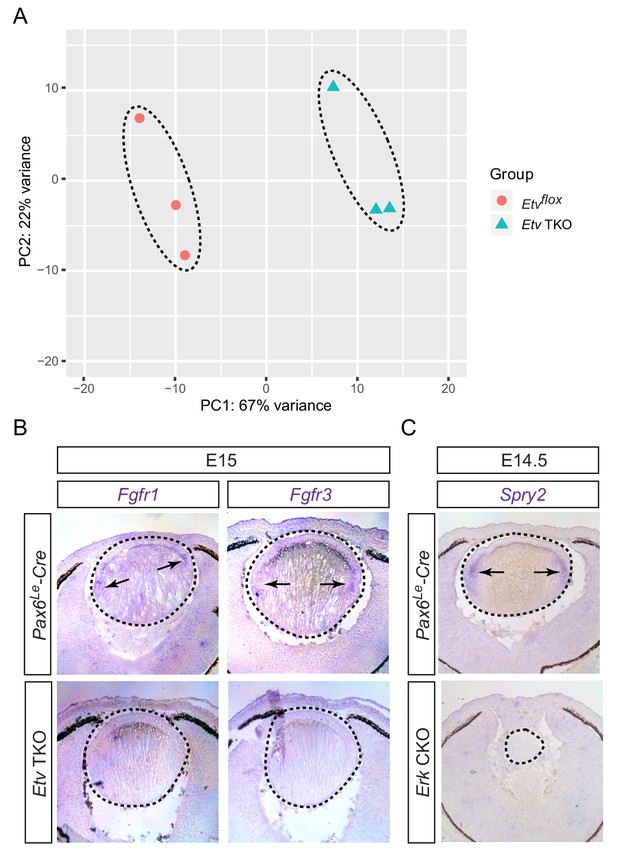
RNA sequencing analysis identified Etv-regulated genes.
(A) Principle component analysis (PCA) showed that control and Etv TKO mutant lenses were well separated in their transcriptomes. (B) Fgfr1 and Fgfr3 were strongly expressed in the transitional zones (arrows) of the control lenses, but down regulated in Etv TKO mutant lenses. (C) Genetic deletion of Erk1/2 abolished Spry2 expression in Erk CKO mutants.
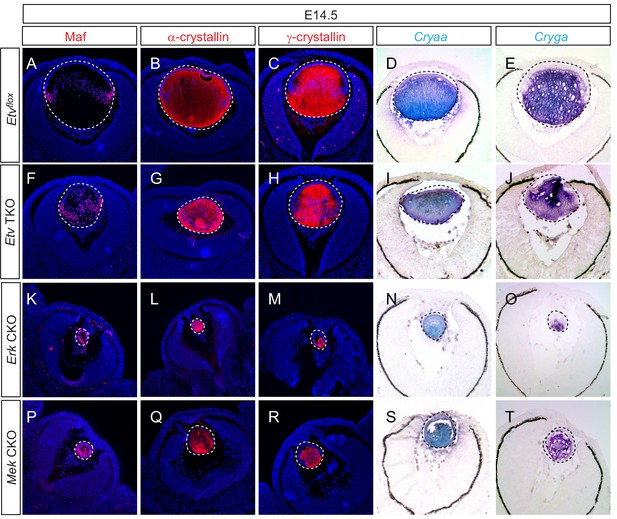
Lens fiber differentiation proceeds in the absence of Etv and ERK.
(A–J) Expression of fiber cell markers Maf, α- and γ-crystallin is unaffected in Etv TKO lenses. (K–T) Maf, α- and γ-crystallin are still expressed in Erk and Mek CKO lens despite the severely reduced lens size.
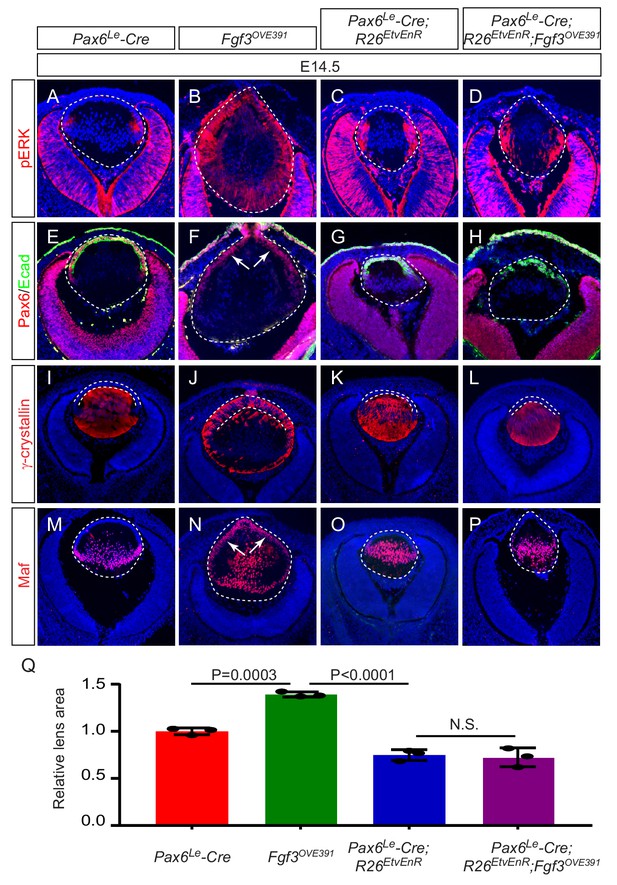
Etv deletion prevents FGF from inducing aberrant differentiation of the lens epithelium.
(A–P) Overexpression of Fgf3 in Fgf3OVE391 lens induced expansion of ERK phosphorylation into the anterior lens epithelium (B), which lost the epithelial marker E-cadherin (F, arrows) but expressed the fiber cell markers Maf and γ-crystallin (J and N). The expression of dominant negative EtvEnR restored normal lens differentiation pattern in Pax6Le-Cre; Fgf3OVE391;R26EtvEnR lenses (H, L and P). (Q) The lens size was enlarged in Fgf3OVE391 embryos, but reduced in both Pax6Le-Cre;R26EtvEnR and Pax6Le-Cre; Fgf3OVE391;R26EtvEnR embryos.
-
Figure 5—source data 1
Source data for Figure 5Q.
- https://cdn.elifesciences.org/articles/51915/elife-51915-fig5-data1-v2.xlsx
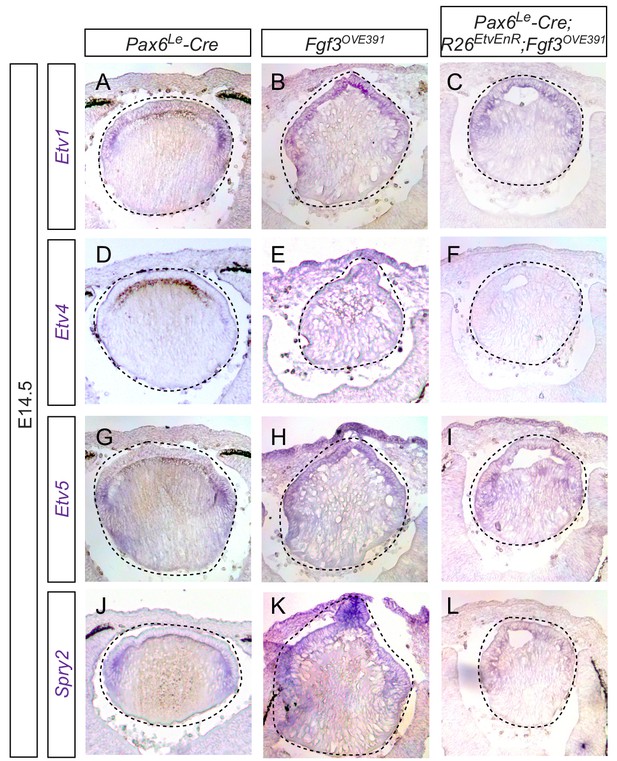
Fgf3 overexpression stimulates expressions of Etv and Spry in the lens.
(A–L) Overexpression of Fgf3 in Fgf3OVE391 led to upregulation of Etv1, 4, five and Spry2 in the anterior lens epithelium, which were greatly reduced by expression of dominant negative EtvEnR.
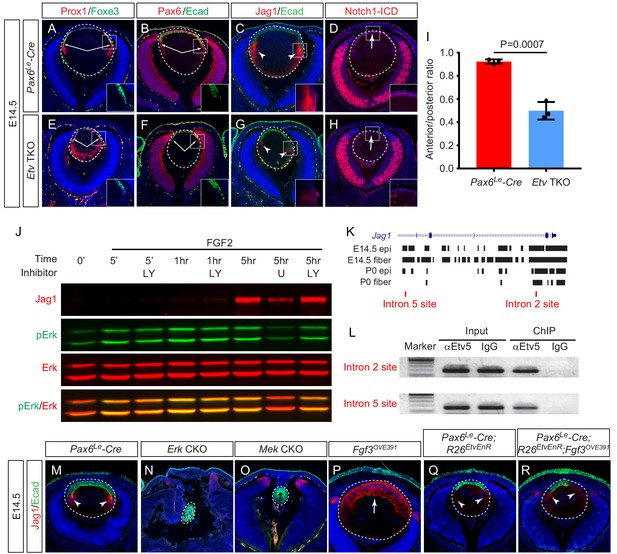
Etv induces Jag1 expression to control Notch signaling.
(A–H) The transitional zone marked by the boundaries of Prox1, Foxe3, Pax6 and E-cadherin expressions are shifted anteriorly in Etv TKO mutant lenses (A, B, E and F, arrows). This was caused by reduced expression of Jag1 in the nascent lens fibers (C and G, arrowheads) and down regulation of Notch signaling as indicated by Notch1-ICD staining in the lens epithelium (D and H, arrows). (I) Quantification of the anterior and posterior perimeters of the lens shows the relative shortening of the lens epithelium. (J) FGF2 induced Jag1 expression in the lens culture after 5 hr, which was blocked by Mek inhibitor U1206, but not by PI3K inhibitor LY294002. (K) Bioinformatic analysis identifies two Etv binding sites within the intron 2 and 5 of the Jag1 locus. The bars indicate the open chromatin regions obtained from the ATAC-seq analysis of lens epithelium and fibers. (L) Chromatin immunoprecipitation experiment in lens cultures showed that both the introns 2 and 5 sites were pulled down by Etv5 antibody but not IgG control. (M–R) The endogenous Jag1 expression in lens fiber cells (M, arrowheads) is abolished by deletion of either Erk or Mek (N and O). Overexpression of Fgf3 induces ectopic Jag1 expression in the lens epithelium (P, arrow), which is suppressed by dominant negative EtvEnR (Q and R, arrowheads).
-
Figure 6—source data 1
Source data for Figure 6I.
- https://cdn.elifesciences.org/articles/51915/elife-51915-fig6-data1-v2.xlsx
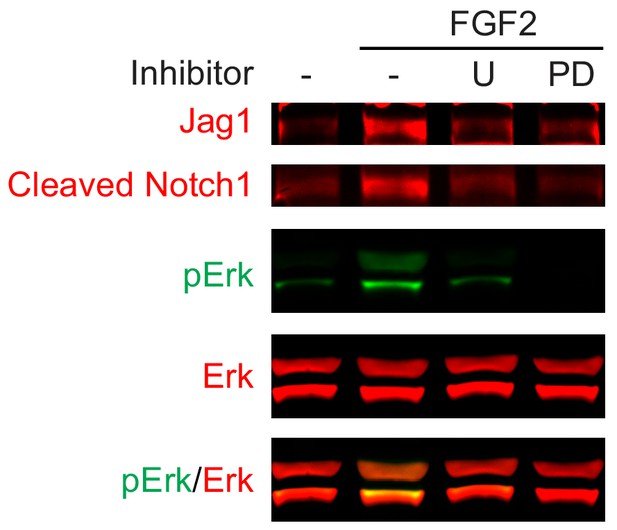
FGF induces Jag1-Notch pathway in an ERK-dependent manner.
FGF2 induced expression of Jag1 and Notch1 cleavage in lens cells, which were blocked by Mek inhibitors U0126 (U) and PD0325901 (PD).
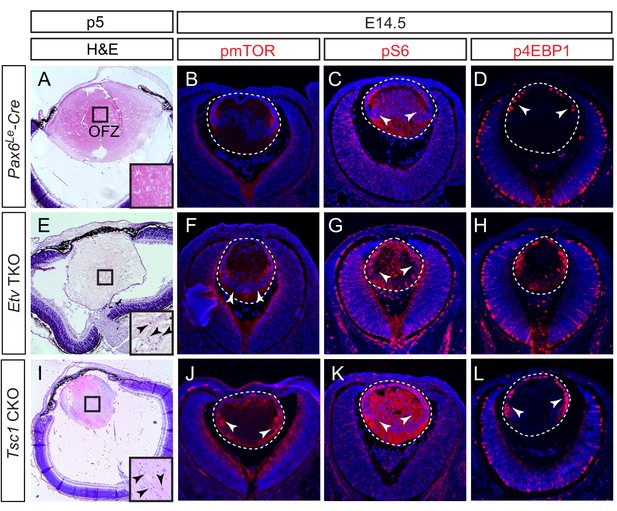
Activation of mTOR signaling disrupts nuclei degradation in the Etv mutant.
(A–J) The center of the wild type lens was an organelle free zone (OFZ) at P5 (F), but it still contains nuclei in Etv mutants (F, arrowheads). This is associated with increased mTOR signaling as indicated by the elevated phosphorylation of mTOR, S6 and 4EBP1 (B-D, F-H, arrowheads). (I–L) The aberrant retention of nuclei and activation of mTOR signaling are reproduced in Tsc1 knockout lens.
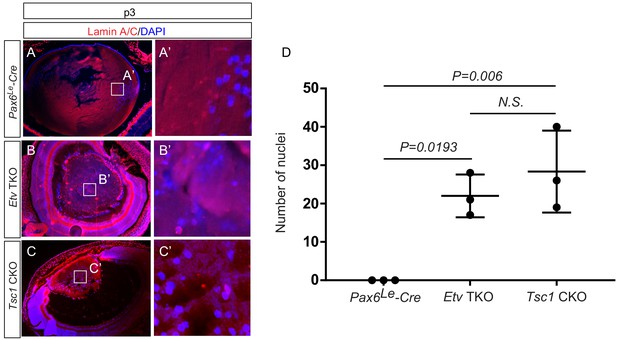
Persistent nuclei in Etv and Tsc1 mutant lenses.
(A–C, A–C’) At postnatal day 3 (P3), the lens nuclei can be visualized with strong staining of DAPI and weak staining of Lamin A/C. Whereas the nuclei in control lens were restricted to the periphery (A and A’), both Etv and Tsc1 mutants displayed persistent nuclei in the lens center (B and B’, C and C’). (D) The number of DAPI-positive nuclei within the central half of lenses were quantified.
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1D.
- https://cdn.elifesciences.org/articles/51915/elife-51915-fig7-figsupp1-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit monoclonal anti-cyclin D1 | Cell Signaling | Cat.# 55506, RRID:AB_2827374 | IHC-1/200 |
| Antibody | Rabbit polyclonal anti-Maf | Santa Cruz Biotechnology | Cat.#sc-7866, RRID:AB_638562 | IHC-1/200 |
| Antibody | Mouse monoclonal anti-Ecadherin | BD | Cat.#610181, RRID:AB_397580 | IHC-1/500 |
| Antibody | Rabbit monoclonal anti-Erk | Cell Signaling | Cat.# 4695, RRID:AB_390779 | WB-1/2000 |
| Antibody | Mouse monoclonal anti-Foxe3 | Santa Cruz Biotechnology | Cat.#sc-377465 | IHC-1/200 |
| Antibody | Rabbit polyclonal anti-Jag1 | Santa Cruz Biotechnology | Cat.#sc-8303,RRID:AB_649685 | IHC-1/200, WB-1/500 |
| Antibody | Mouse monoclonal anti-Ki67 | BD | Cat.#550609, RRID:AB_393778 | IHC-1/200 |
| Antibody | Rabbit monoclonal Lamin A/C | Abcam | Cat.# ab133256, RRID:AB_2813767 | IHC-1/1000 |
| Antibody | Rabbit monoclonal Notch1 | Cell Signalling | Cat.#4380, RRID:AB_10691684 | WB-1/1000 |
| Antibody | Rabbit monoclonal Notch1-ICD | Cell Signaling | Cat.#4147, RRID:AB_2153348 | IHC-1/200 |
| Antibody | Rabbit polyclonal anti-Prox1 | Covance | Cat.#PRB-238C, RRID:AB_291595 | IHC-1/200 |
| Antibody | Rabbit polyclonal anti-pHH3 | Millipore | Cat.#06-570, RRID:AB_310177 | IHC-1/200 |
| Antibody | Rabbit anti-pS6 | Cell Signaling | Cat.#5364, RRID:AB_10694233 | IHC-1/200 |
| Antibody | Rabbit monoclonal anti-pmTOR | Cell Signaling | Cat.#5536, RRID:AB_10691552 | IHC-1/200 |
| Antibody | Rabbit polyclonal anti-Pax6 | Covance | Cat.#PRB-278P, RRID:AB_291612 | IHC-1/200 |
| Antibody | Rabbit anti-p4EBP1 | Cell Signaling | Cat.#2855, RRID:AB_560835 | IHC-1/200 |
| Antibody | Rabbit monoclonal anti-pERK | Cell Signaling | Cat.#9101, RRID:AB_331646 | IHC-1/200 |
| Antibody | Mouse monoclonal anti-pERK | Santa Cruz Biotechnology | Cat# sc-7383, RRID:AB_627545 | WB-1/1000 |
| Antibody | Rabbit polyclonal anti-α-Crystallin | Sam Zigler (National Eye Institute) | IHC-1/5000 | |
| Antibody | Rabbit polyclonal anti-γ-Crystallin | Sam Zigler (National Eye Institute) | IHC-1/5000 | |
| EdU | DNA synthesis monitoring probe | Abcam | cat. # ab14618 | |
| Commercial assay Kit | Click IT EdU Cell proliferation kit | Invitrogen | cat. # C10337 | |
| Commercial assay Kit | In situ cell death detection kit | Roche | cat.# 1168479510 | |
| Peptide, recombinant protein | recombinant murine FGF2 | ScienCell | cat.# 124-02 | |
| Genetic reagent (M. musculus) | Etv1flox | PMID:12741988 | MGI:2663693 | Dr. Silvia Arber (University of Basel, Basel, Switzerland) |
| Genetic reagent (M. musculus) | Etv4-/- | PMID:11094084 | MGI:2445834 | Dr. Xin Sun (University of California at San Diego, San Diego, CA) |
| Genetic reagent (M. musculus) | Etv5flox | PMID:19386269 | MGI:3849047 | Dr. Xin Sun (University of California at San Diego, San Diego, CA) |
| Genetic reagent (M. musculus) | Tsc1flox | PMID:12205640 | MGI:2656240 | Dr. Stephen Tsang (Columbia University, New York, NY) |
| Genetic reagent (M. musculus) | R26EtvEnR | PMID:19386268 | MGI:3848910 | Drs. Andrew McMahon (University of Southern California, Los Angeles, CA) and James Li (University of Connecticut Health Center, Farmington, CT) |
| Genetic reagent (M. musculus) | Map2k1flox | PMID:16887817 | MGI:3714918 | |
| Genetic reagent (M. musculus) | Map2k2KO | PMID:12832465 | MGI:2668345 | |
| Genetic reagent (M. musculus) | Mapk3-/- | PMID:11160759 | MGI:3042006 | |
| Genetic reagent (M. musculus) | Mapk1flox | PMID:18596172 | MGI:3803954 | |
| Genetic reagent (M. musculus) | Fgf3OVE391 | PMID:7539358 | MGI:6393977 | Dr. Michael Robinson, Miami University. |
| Genetic reagent (M. musculus) | Pax6Le-Cre | PMID:11069887 | MGI:3045795 | |
| Genetic reagent (M. musculus) | Fgfr1flox | PMID:16421190 | Stock #: 007671 MGI:3713779 | |
| Genetic reagent (M. musculus) | Fgfr2flox | PMID:12756187 | MGI:3044679 | Dr. David Ornitz, Washington University Medical School, St Louis, MO |





