Retromer subunit, VPS29, regulates synaptic transmission and is required for endolysosomal function in the aging brain
Figures
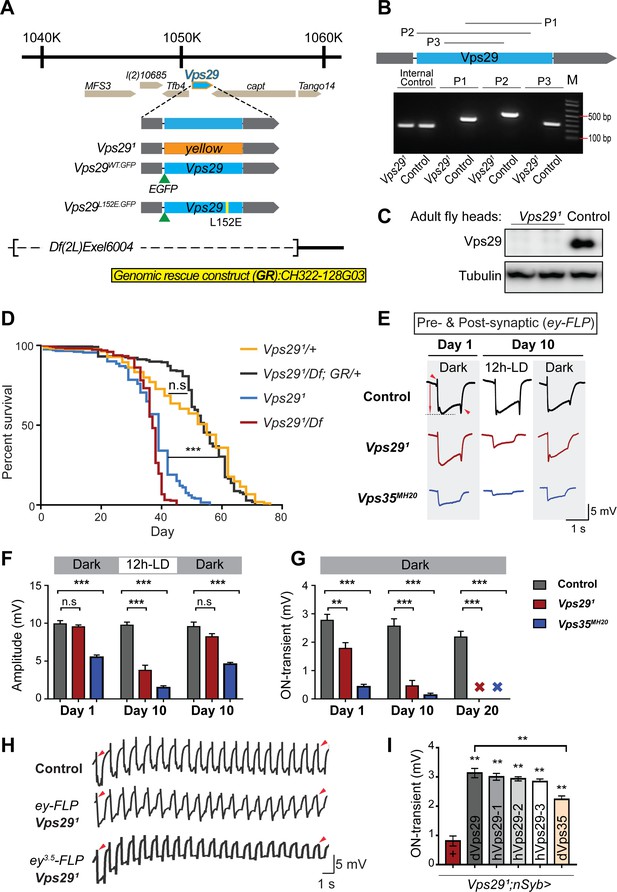
Vps29 is required for age-dependent retinal function.
(A) The Vps29 genomic locus is shown, highlighting reagents used in this study. In the null allele, Vps291, the gene coding sequence (blue, 2L: 1150852–1151420) is replaced by a ywing2+ marker gene. Vps29WT.GFP and Vps29L152E.GFP are identical N-terminal tagged-Vps29 alleles, except for the L152E variant. A chromosomal deficiency Df(2L)Exel6004 is shown, with the deleted regions indicated by dashed lines. A bacterial artificial chromosome (BAC) (yellow) was used for transgenic genomic rescue (GR). (B) Genomic polymerase chain reaction (PCR) showing loss of Vps29 coding sequence in Vps291 homozygotes versus control (w) flies. P1, P2, and P3 denote expected PCR products from primer pairs targeting Vps29 genomic sequence. As an additional control, PCR was also performed for Vps35 genomic sequence. We also performed PCRs using primer pairs that span both sides of the breakpoint junctions abutting the inserted ywing2+ marker gene cassette (not shown), and these products were Sanger sequenced to confirm the depicted molecular lesion. (C) Western blot from adult heads probed with anti-Vps29 antibody, confirming Vps291 is a protein null allele. (D) Vps291 homozygotes and Vps291/Df transheterozygotes (Vps291/Df(2L)Exel6004) show reduced survival that is rescued by the Vps29 genomic rescue strain (Vps291/Df; GR/+). Quantitation based on n = 200–235 per group. See also Figure 1—figure supplement 1A. (E) Representative ERG traces at 1- and 10 days after generation of ey-FLP clones from (i) control (FRT40A), (ii) Vps291, or (iii) Vps35MH20. Flies were raised using an alternating 12 hr light/dark cycle (12h-LD) or in complete darkness. Loss of either Vps29 or Vps35 disrupted light-induced depolarization (arrow) in 10-day-old flies. On- and off- transient ERG potentials (top and bottom arrowheads, respectively) were also lost. Raising flies in the dark restored ERG depolarization, but not the transients, indicating persistent defects in synaptic transmission. (F–G) Quantification (n = 6–8) of ERG depolarization and on-transient potentials. ‘X’ denotes undetectable on-transients in 20-day-old animals. See also Figure 1—figure supplement 1B. (H) Compared with controls (FRT40A clones), ERG transient potentials are extinguished by rapid stimulation in Vps291 clones. Consistent results were obtained using either ey-FLP or ey3.5-FLP, which targets presynaptic neurons only. Flies were raised in complete darkness and examined at 1 day. See also Figure 1—figure supplement 1C. (I) Rescue of Vps291 synaptic transmission defects by pan-neuronal expression (nSyb-GAL4 driver) of either Drosophila Vps29 or Vps35 (dVps29 and dVps35) or human Vps29 (hVps29-1,−2, or −3, representing three alternate isoforms). Quantification of ERG on-transients in 15-day-old flies (n = 9–12 per group) from the following genotypes: (1) Vps291;nSyb-Gal4/+; (2) Vps291;nSyb-Gal4/UAS-dVps29; (3-5) Vps291;nSyb-Gal4/UAS-hVps29; and (6) Vps291;nSyb-Gal4/UAS-dVps35. Statistical analysis was based on log-rank test with Bonferroni's correction (D) or one-way ANOVA (F, G, I). All error bars denote SEM. n.s., not significant; **, p<0.01; ***, p<0.001.
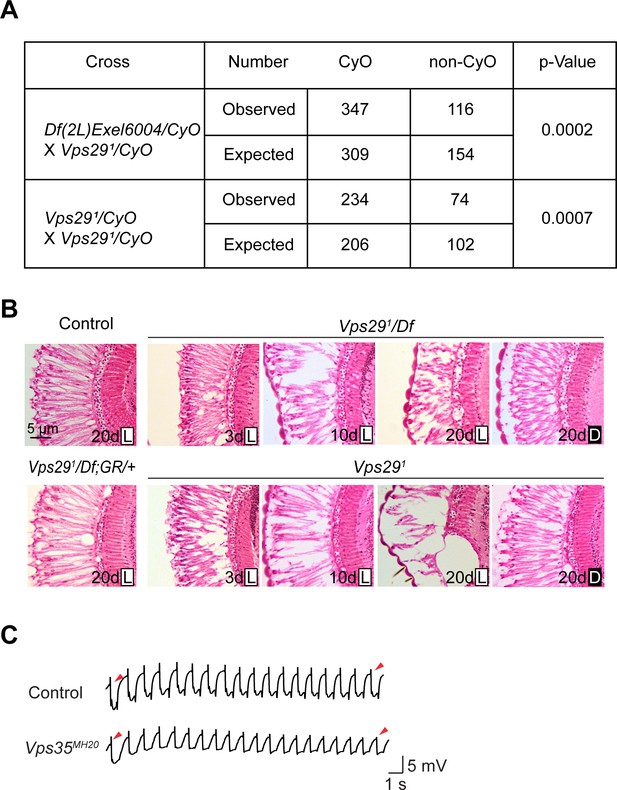
Additional studies of the adult retina.
(A) Adult flies lacking Vps29 (Vps291 homozygotes and Vps291/Df) are recovered at ratios below Mendelian expectations (~25% vs. 33%). For statistical analysis, a chi-square test was performed. (B) Loss of Vps29 causes light- and age-dependent retinal degeneration. Frontal sections through the retina are shown, stained with hematoxylin and eosin, revealing vacuolar changes, which was suppressed by either introducing the Vps29 genomic rescue construct (GR) or raising animals in dark conditions. GMR-w-RNAi was used to remove eye pigment. We examined the following genotypes: (1, control) GMR-w-RNAi/+; Vps291/+; (2) GMR-w-RNAi/+; Vps291/Df; (3) GMR-w-RNAi/+; Vps291; (4) GMR-w-RNAi/+; Vps291/Df; GR/+. Flies were raised in 12 hr light/12 hr dark cycle (L) or in the dark (D). (C) Compared with controls (FRT40A clones), ERG transient potentials are partially extinguished by rapid stimulation in Vps35MH20 clones. Clones were generated using ey-FLP. Flies were raised in complete darkness and examined at 1 day.
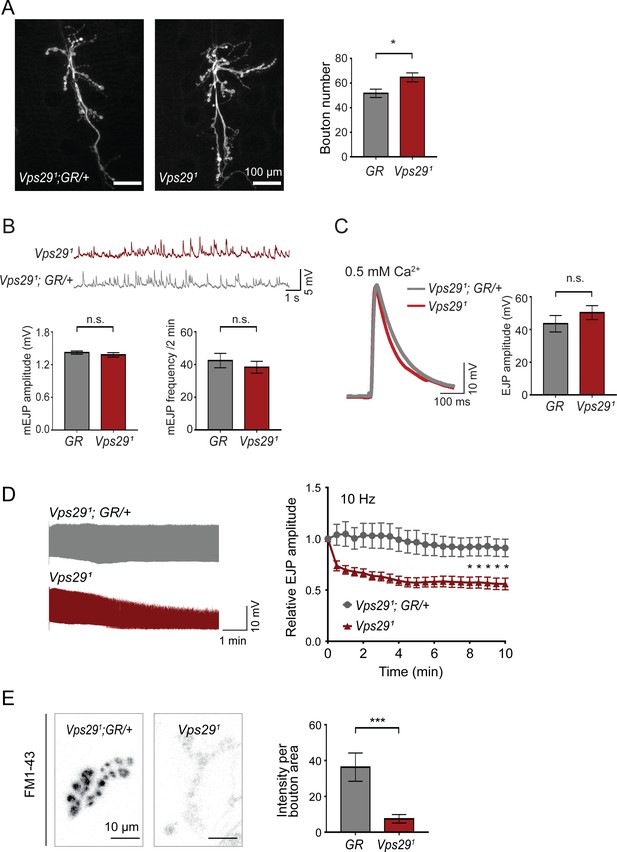
Retromer regulates synaptic vesicle endocytosis and recycling.
(A) Vps29 loss of function causes an increased number of synaptic terminal boutons at the larval neuromuscular junction (NMJ). NMJ preparations from Vps291 homozygotes or control larvae (GR = Vps291;GR/+) were stained with an antibody against horseradish peroxidase, and Type IIb boutons at abdominal segments A2 and A3 were quantified (n = 5 animals per group). The Vps29 transgenic BAC (GR) was heterozygous for all comparisons. (B) Larval NMJ electrophysiology in Vps291 reveals normal miniature excitatory junction potential (mEJP) amplitude and frequency (in 2 min) (n = 14–16). (C) Evoked excitatory junction potentials (EJPs) are normal in the absence of Vps29. Representative EJP traces from Vps291 and Vps291;GR/+ (control, GR) larvae and quantification (n = 13–15). 0.2 Hz stimulation was performed using 0.5 mM Ca2+. (D) Vps291 NMJs show synaptic depression following rapid stimulation (10 Hz, 0.5 mM extracellular Ca2+). Representative traces (10 min) are shown, with quantification (n = 13). Data was normalized to initial EJP amplitude. (E) Vps291 NMJs show reduced FM 1-43 dye uptake following KCl stimulation, consistent with impaired synaptic vesicle endocytosis. FM1-43 signal intensity (per bouton area) was quantified (n = 8–11). See also Figure 2—figure supplement 1. Statistical analysis (A–E) based on Student’s t-test. All error bars denote SEM. n.s., not significant; *, p<0.05; ***, p<0.001.
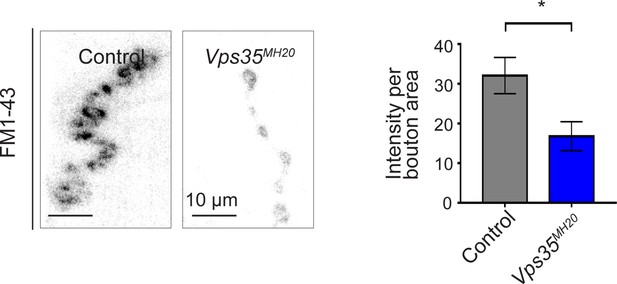
Additional studies of the neuromuscular junction.
Vps35MH20 larval neuromuscular junctions (NMJs) show reduced FM 1-43 dye uptake following KCl stimulation, consistent with impaired synaptic vesicle endocytosis. FM 1-43 signal intensity (per bouton area) was quantified (n = 9–11) in Vps35MH20 homozygotes or controls (w). Statistical analysis based on Student’s t-test. All error bars denote SEM. *p<0.05.
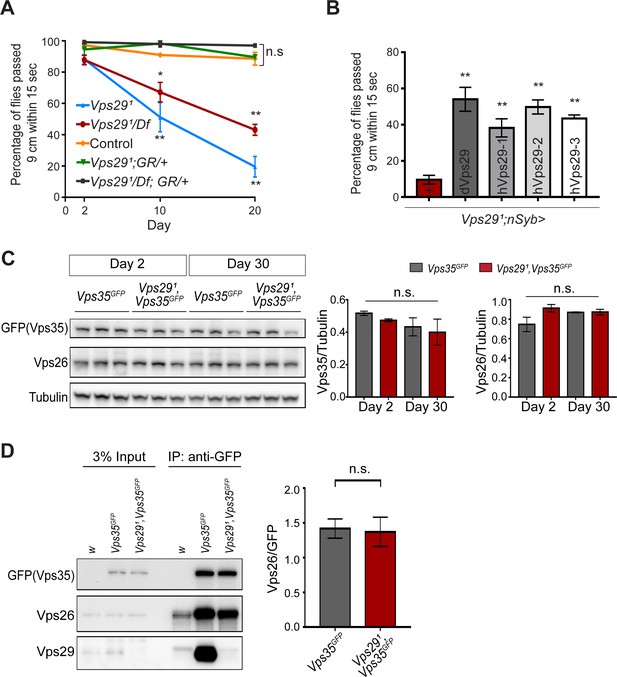
Progressive locomotor impairment in Vps29 mutants but preserved Vps35 and Vps26 protein levels.
(A) Vps291/Df and Vps291 adults demonstrate age-dependent locomotor impairment, based on startle-induced negative geotaxis, and this phenotype is fully rescued by a single, heterozygous copy of the Vps29 BAC transgenic (Vps291/Df; GR/+ and Vps291; GR/+). Control: yw/w. The Vps291 homozygote genotype manifests a stronger locomotor phenotype than Vps291/Df (p<0.01 in comparisons at 20 days). Quantification based on n = 4 groups, each consisting 14–16 flies. See also Figure 3—figure supplement 1A. (B) Pan-neuronal expression of either Drosophila or human Vps29 (dVps29 or hVps29, respectively), using the nsyb-GAL4 driver rescues the Vps291 locomotor phenotype. Quantification based on n = 4 groups of 20-day-old flies from the following genotypes: (1) Vps291;nSyb-Gal4/+; (2) Vps291;nSyb-Gal4/UAS-dVps29; and (3-5) Vps291;nSyb-Gal4/UAS-hVps29. (C) Vps35 and Vps26 protein levels are normal in the absence of Vps29. Western blots of adult fly head homogenates from either Vps291, Vps35GFP homozygotes or controls (Vps35GFP), probed for Vps35GFP (anti-GFP), Vps26, or Tubulin (loading control). Quantification based on n = 3 replicate experiments. See also Figure 3—figure supplement 1B,C. (D) The association of Vps35 and Vps26 proteins is preserved in the absence of Vps29. Vps35GFP was immunoprecipitated from either Vps291, Vps35GFP homozygotes or control animals (Vps35GFP) (2 day old), and western blots were probed for Vps35GFP (anti-GFP), Vps26, and Vps29. Quantification based on n = 4 replicate experiments. Statistical analysis based on one-way ANOVA (A–C) or Student’s t- test (D). All error bars denote SEM. n.s., not significant; **, p<0.01; ***, p<0.001.
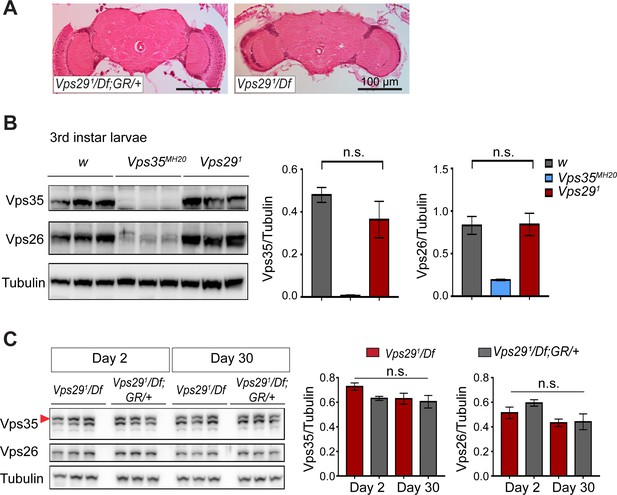
Loss of Vps29 does not affect Vps35 or Vps26 protein expression.
(A) Preserved adult brain histology in aged animals lacking Vps29. Hematoxylin and eosin stained frontal sections from 45-day-old Vps291/Df or control animals (Vps291/Df; GR/+) demonstrate no overt neuropathology in the central brain complex. Note that the retina and lamina are absent from frontal sections prepared from Vps29 mutants due to severe degeneration of these structures. See also Figure 1—figure supplement 1B. (B) Vps35 and Vps26 protein levels are normally expressed in Vps291 homozygous third instar larvae but absent or reduced, respectively, in Vps35MH20 homozygotes. Western blots were probed for Vps35, Vps26, and Tubulin (loading control), and quantified based on n = 3 replicate experiments. For statistical analysis, a Student’s t-test was performed. (C) Vps35 and Vps26 proteins are normally expressed in Vps291/Df animals. Adult fly head homogenates were prepared from 2-day-old or 30-day-old Vps291/Df or control animals (Vps291/Df; GR/+). Western blots were probed for Vps35, Vps26, and Tubulin (loading control), and quantified based on n = 3 replicate experiments. For statistical analysis, one-way ANOVA was performed. Red arrowhead denotes the Vps35-specific band. All error bars denote SEM. n.s., not significant.
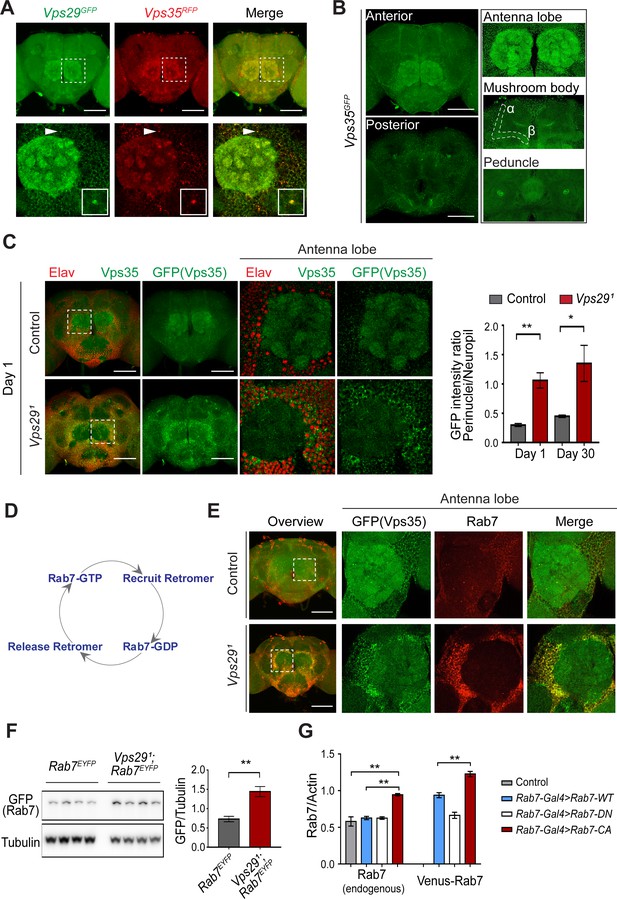
Vps29 regulates Vps35 localization in the adult brain.
(A) Vps29GFP (anti-GFP, green) and Vps35RFP (red) are expressed throughout the adult brain and co-localize. Boxed region of interest including antennal lobe (top row) is shown magnified (bottom row). Representative puncta (arrowhead) co-staining for Vps35 and Vps29 is further magnified in inset. Scale bars, 100 µm. (B) Vps35GFP (anti-GFP, green) is modestly enriched in neuropil regions, including the antennal lobes and the mushroom body (α-/β- lobes and peduncles). Scale bars, 100 µm. See also Figure 4—figure supplement 1A. (C) In the absence of Vps29, Vps35GFP (green) protein is redistributed in the adult brain, shifting from neuropil to soma and forming large perinuclear puncta. Neuronal nuclei are labeled with anti-Elav (red). Within the boxed region of interest, including antennal lobe and surrounding nuclei, the GFP intensity ratio (perinuclear to neuropil) was quantified (n = 3) in Vps291, Vps35GFP homozygotes or Vps35GFP controls. Scale bars, 100 µm. See also Figure 4—figure supplement 1B and Figure 4—figure supplement 2. (D) Schematic highlighting the Rab7 cycle including GTP-bound (active) and GDP-bound (inactive) forms and putative interactions regulating retromer recruitment and release. (E) In Vps29 mutants, Vps35GFP (green) colocalizes with Rab7 (anti-Rab7, red), which appears increased and similarly redistributed from neuropil to soma. The region of interest centered on the antenna lobe is indicated in the boxed region of the lower power image (maximal intensity projection); the magnified images are representative anterior single sections taken through the antennal lobe. Scale bars, 100 µm. See also Figure 4—figure supplement 3. (F) Rab7 protein levels are increased in the absence of Vps29. Western blots of adult head homogenates, including Vps291, Rab7EYFP homozygotes or Rab7EYFP controls (2-day-old), were probed for Rab7-YFP (anti-GFP) and Tubulin (loading control). Quantification based on n = 4 replicate experiments. See also Figure 4—figure supplement 4A. (G) Rab7 protein levels are increased in the constitutive active, ‘GTP-locked’ Rab7Q67L mutant. Western blots of adult head homogenates from 5-day-old animals were probed for Rab7 or Actin, including the following genotypes: (1, Rab7-Gal4 > Rab7 CA) UAS-Venus-Rab7 Q67L/+; Rab7-Gal4/+; (2, Rab7-Gal4 > Rab7 WT) UAS-Venus-Rab7 WT/+; Rab7-Gal4/+; (3, Rab7-Gal4 > Rab7 DN) UAS-Venus-Rab7 T22N/+; Rab7-Gal4/+; (4, control) w. Venus-tagged or endogenous Rab7 protein levels were separately quantified, based on n = 4 replicate experiments. See also Figure 4—figure supplement 4B. Statistical analysis was based on Student’s t-test (C, F) or one-way ANOVA (G). To address the possibility of a modestly skewed distribution, the data in C were log2-transformed, and the results of t-tests were unchanged. All error bars denote SEM. *, p<0.05; **, p<0.01.
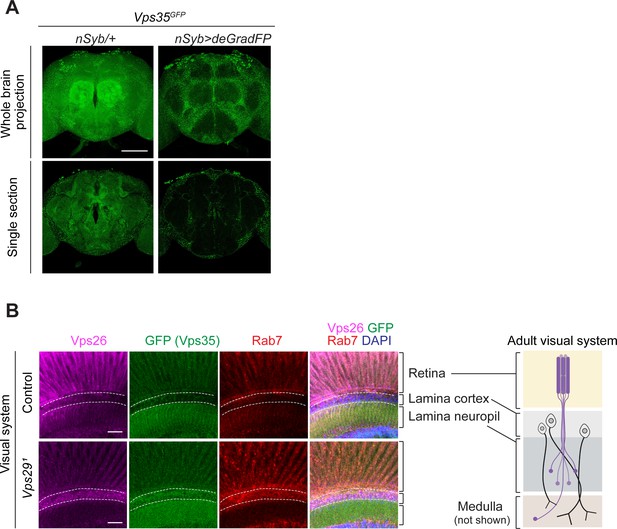
Loss of Vps29 causes mislocalization of retromer in adult brain.
(A) Neuronal expression of Vps35 (anti-GFP) in the adult fly brain was examined by selective depletion using deGradFP. The following genotypes were examined: Vps35GFP; nSyb-Gal4/UAS-deGradFP (also known as Nslmb-vhhGFP4) and Vps35GFP; nSyb-Gal4/+. Crosses were established and maintained at 18°C, and adult animals were shifted to 29°C for 7 days post-eclosion, prior to brain dissection. Scale bars, 100 µm. (B) In the adult retina of Vps29 mutants, Vps35 (anti-GFP, green), Vps26 (anti-Vps26, magenta), and Rab7 (anti-Rab7, red) are increased in the cortex of the lamina where neuronal cell bodies (marked by DAPI) are concentrated. The following genotypes were examined: Vps291, Vps35GFP homozygotes and Vps35GFP (control). Scale bars, 20 µm.
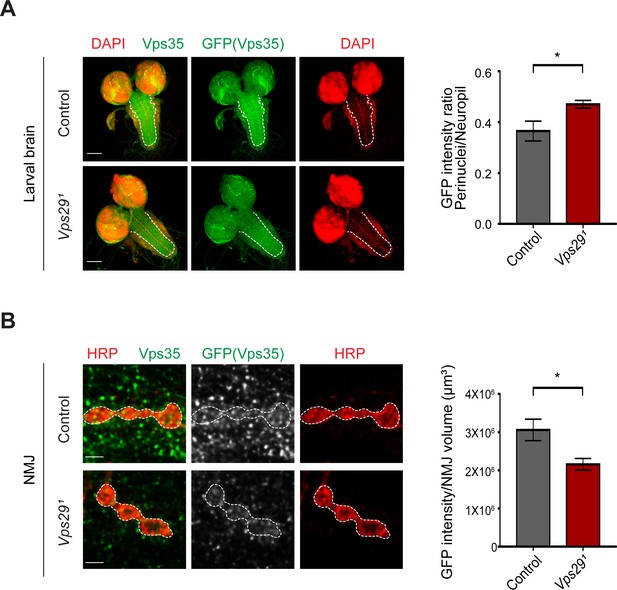
Loss of Vps29 causes mislocalization of Vps35 in larvae.
(A) In Vps291 larval brain, Vps35GFP (green) protein is reduced in the neuropil (dashed line). Nuclei are labeled with DAPI (red). The GFP intensity ratio (perinuclear to neuropil) was quantified (n = 7–8) in Vps291, Vps35GFP homozygotes or Vps35GFP controls. Scale bars, 100 µm. (B) Vps35GFP is reduced at the larval NMJ in Vps291 mutant animals. NMJ preparations from Vps291, Vps35GFP homozygotes or Vps35GFP controls were co-stained with anti-HRP and anti-GFP, and GFP intensity within Type IIb boutons at abdominal segments A2 and A3 were calculated relative to HRP volume (n = 7 animals per group). Scale bars, 2 µm. Statistical analysis based on Student’s t test. All error bars denote SEM. *, p<0.05.
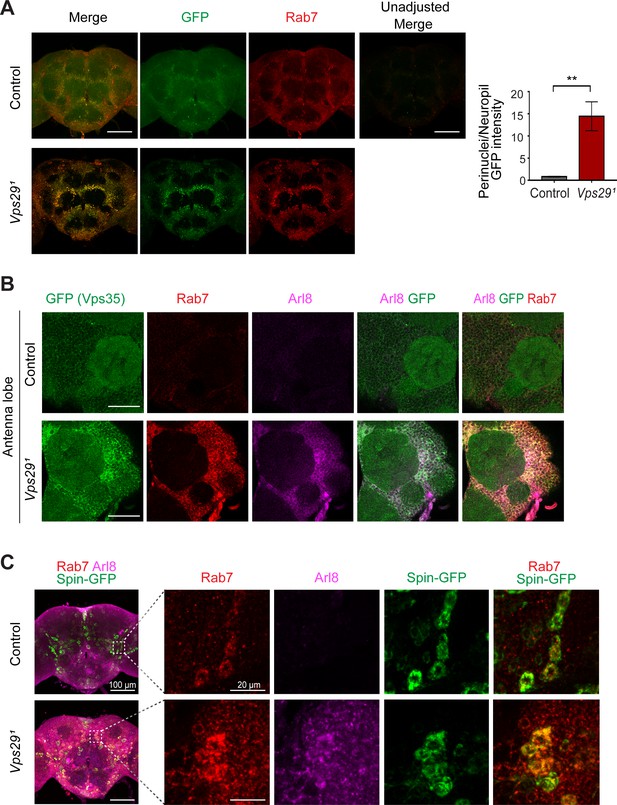
Loss of Vps29 causes mislocalization of Rab7.
(A) In the absence of Vps29, Rab7 protein is increased and mislocalized to neuronal soma. Vps291;Rab7EYFP homozygotes or control (Rab7EYFP) animals, harboring an endogenous, EYFP-tagged Rab7, were stained for Rab7 protein, using both anti-GFP (green) and anti-Rab7 (red). The fluorescence intensity of control animals was amplified for visualization of the weak signal (imaging using consistent settings for controls shown at right). The GFP intensity ratio (perinuclear to neuropil) was quantified (n = 4) within antennal lobe region in 2-day-old animals. Scale bars, 100 µm. (B) In Vps29 mutants, Vps35GFP (green) colocalizes with both Rab7 (anti-Rab7, red) and Arl8 (anti-Arl8, magenta). Scale bars, 50 µm. (C) In 1-day-old Vps29 mutants, the increased Rab7- (anti-Rab7, red) and Arl8-postive signal (anti-Arl8, magenta) exhibits a punctate, cytoplasmic distribution that further co-stains with Spinster-GFP, which was expressed in neurons. The following genotypes were examined: Vps291; nSyb-Gal4/UAS-Spinster-GFP and nSyb-Gal4/UAS-Spinster-GFP (control). Statistical analysis based on Student’s t test. All error bars denote SEM. **, p<0.01.
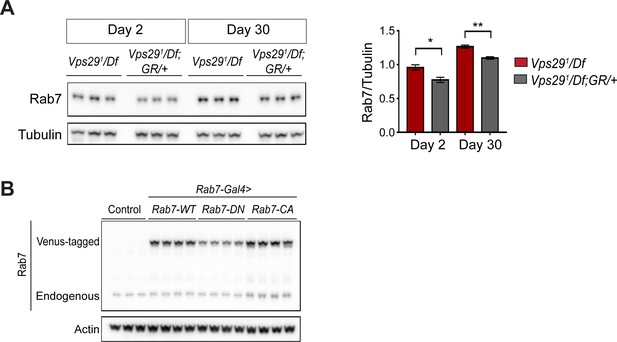
Increased Rab7 expression following loss of Vps29.
(A) Rab7 protein expression is increased in Vps29 null animals. Adult fly head homogenates were prepared from 2- and 30-day-old Vps291/Df or controls (Vps291/Df; GR/+). Western blots were probed with anti-Rab7 and anti-Tubulin (loading control), and quantified based on n = 3 replicate experiments. (B) Western blot showing increased expression of ‘GTP-locked’ Rab7Q67L along with corresponding wildtype (Rab7 WT) and ‘GDP-locked’ (Rab7 T22N) forms from 5-day-old fly head homogenates prepared from the following genotypes: (1, Rab7-Gal4 > Rab7 CA) UAS-Venus-Rab7 Q67L/+; Rab7-Gal4/+; (2, Rab7-Gal4 > Rab7 WT) UAS-Venus-Rab7 WT/+; Rab7-Gal4/+; (3, Rab7-Gal4 > Rab7 DN) UAS-Venus-Rab7 T22N/+; Rab7-Gal4/+; (4, control) w. Western blots were probed for anti-Rab7 and anti-Actin (loading control). Quantification shown in Figure 4G. Statistical analysis based on Student’s t test. All error bars denote SEM. *, p<0.05; **, p<0.01.
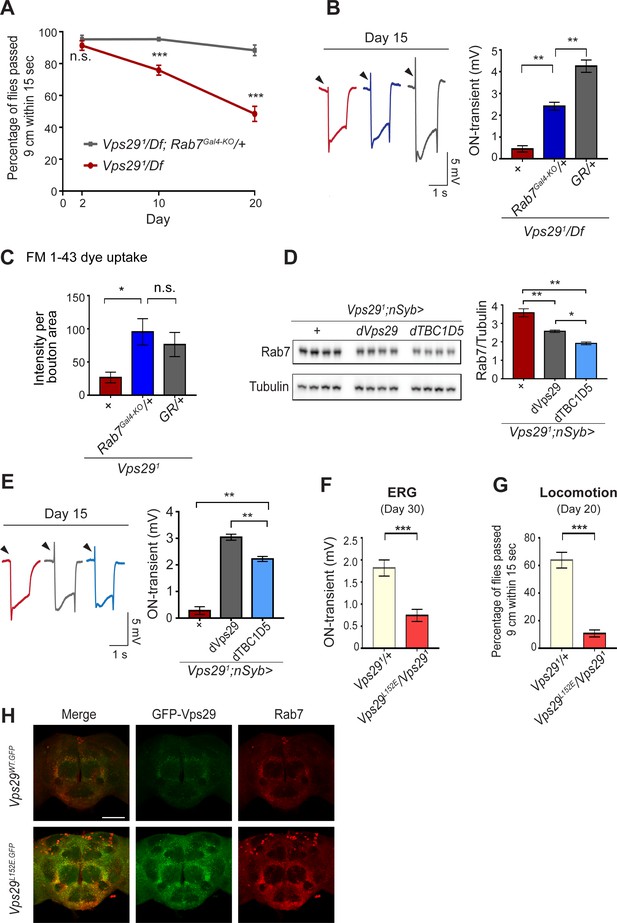
Reduction of Rab7 or overexpression of TBC1D5 suppresses Vps29 mutant phenotypes.
(A–C) Reduction of Rab7 rescues Vps29 mutant phenotypes, including progressive locomotor impairment (A), synaptic transmission (B), and synaptic vesicle endocytosis at the larval NMJ (C). (A) Quantification of locomotor behavior based on n = 4 groups, each consisting of 12–15 flies. (B) Quantification of electroretinogram (ERG) on-transients (n = 12–13) from adults raised using a 12 hr light/dark cycle. (C) FM 1-43 dye uptake signal intensity per NMJ bouton area was quantified (n = 7–9). (D) Pan-neuronal overexpression of dTBC1D5 (nsyb>dTBC1D5) restores Rab7 protein level in flies lacking Vps29. Western blots of adult fly head homogenates (30-day-old) were probed for Rab7 and Tubulin (loading control), and quantified based on n = 4 replicate experiments, including the following genotypes: (1, red) Vps291;nSyb-GAL4/+; (2, grey) Vps291;nSyb-GAL4/UAS-dVps29; (3, blue) Vps291;nSyb-GAL4/UAS-dTBC1D5. (E) Pan-neuronal overexpression of dTBC1D5 (nsyb>dTBC1D5) rescues synaptic transmission in Vps291 mutants. Quantification of ERG on-transients (n = 8) from adults raised using a 12 hr light/dark cycle. (F, G) The Vps29L152E mutation, predicted to disrupt the Vps29-TBC1D5 interaction, fails to complement Vps291, causing impaired synaptic transmission (F) and locomotor impairment (G). Quantification of ERG on-transients in 30-day-old flies raised in complete darkness (F) (n = 12–14) or locomotor behavior in 20-day-old flies (G) (n = 5–6 groups). See also Figure 5—figure supplement 1A. (H) The Vps29L152E mutation causes increased expression of Vps29 (anti-GFP, green) and Rab7 (anti-Rab7, red). Whole-mount brain immunofluorescence shown for Vps29L152E.GFP or Vps29WT.GFP homozygotes (2-day-old adults). Scale bars, 100 µm. See also Figure 5—figure supplement 1B. Statistical analysis based on Student’s t-test (A, F, G) or one-way ANOVA (B–E). All error bars denote SEM. n.s., not significant; *, p<0.05; **, p<0.01; ***, p<0.001.
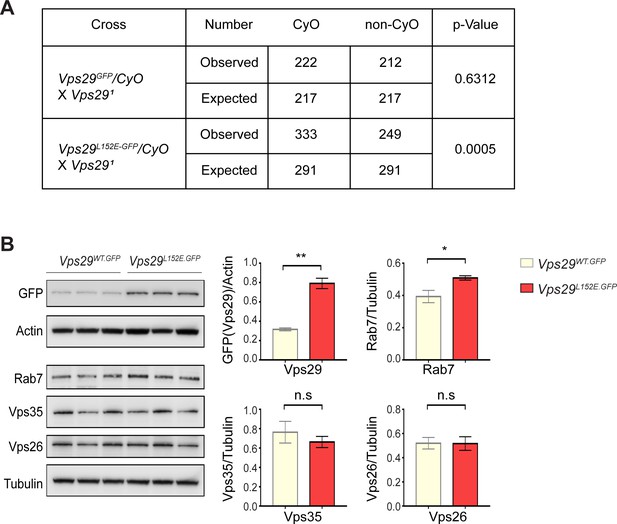
Additional characterization of the Vps29L152E.
(A) Vps29L152E/Vps291 transheterozygotes were recovered at ratios below Mendelian expectations (~43% vs. 50%). For statistical analysis, chi-square test was performed. (B) Vps29 (anti-GFP) and Rab7 protein expression are increased in 2-day-old Vps29L152E.GFP adult heads comparing to controls (Vps29WT.GFP). Vps35 and Vps26 protein levels were unchanged. Quantification based on n = 3 replicate experiments. For statistical analysis, Student’s t-test was performed. All error bars denote SEM. n.s., not significant; *, p<0.05; **, p<0.01.
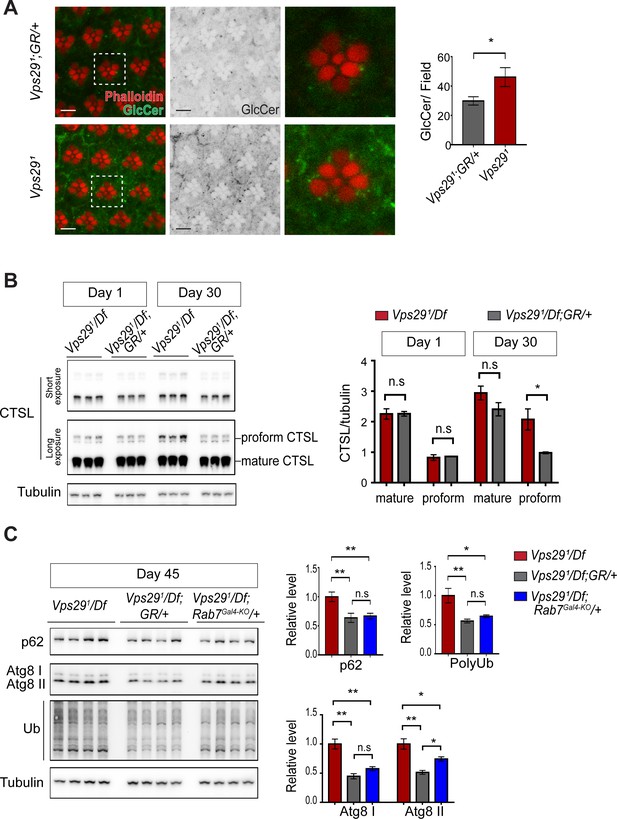
Loss of Vps29 causes age-dependent, progressive lysosomal dysfunction in the brain.
(A) In the absence of Vps29, glucosylceramide (anti-GlcCer, green) accumulates in the retina. Tissue is counterstained for actin (phalloidin, red) to highlight the photoreceptor rhabdomeres. Glucosylceramide signal was quantified from a 36.94 × 36.94 µm2 region encompassing ~10 ommatidia in 30-day-old animals (n = 10) raised in dark conditions, including GMR-w-RNAi/+; Vps291; GR/+ (controls) and GMR-w-RNAi/+; Vps291. Eye pigment was removed using RNAi against the w gene to reduce auto-fluorescence. A representative ommatidium (boxed) is shown at higher-power (right). Scale bars, 5 µm. (B) The uncleaved, CTSL proform is increased in aged animals lacking Vps29, consistent with diminished lysosomal proteolytic capacity. Western blots from adult fly head homogenates were probed for CTSL and Tubulin (loading control), and quantified based on n = 3 replicate experiments. See also Figure 6—figure supplement 1A. (C) Autophagic flux is reduced in aged animals lacking Vps29, and this phenotype is suppressed by reduction of Rab7. Western blots of adult head homogenates were probed for autophagic markers, including p62, Atg8, polyubiquitin (FK1), or Tubulin (loading control), and quantified based on n = 8 replicate experiments. See also Figure 6—figure supplement 1B,C. Statistical analysis based on Student’s t-test (A, B) and one-way ANOVA (C). All error bars denote SEM. n.s., not significant; *, p<0.05; **, p<0.01.
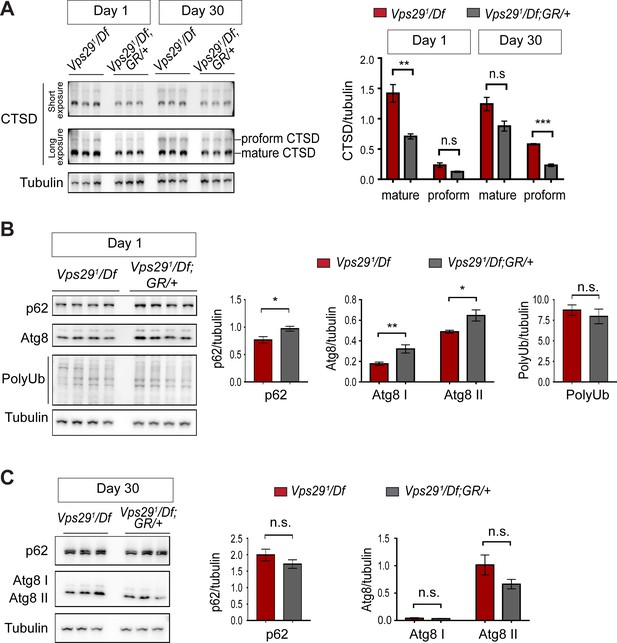
Additional studies of lysosomal proteolysis and autophagy.
(A) The uncleaved CTSD proform is increased in aged animals lacking Vps29. Western blots from adult fly head homogenates, including Vps291/Df and Vps291/Df; GR/+ (control), were probed with anti-CTSD, and quantified based on n = 3 replicate experiments. (B, C) Western blots from fly head homogenates were probed with for p62, Atg8, polyubiquitin (FK1) and Tubulin in 1- (B) or 30- (C) day-old animals. Quantification based on analysis of n = 4 (B) or n = 3 (C) replicate experiments. Statistical analysis (A–C) based on Student’s t-test. All error bars denote SEM. n.s., not significant. *, p<0.05; **, p<0.01; ***, p<0.001.
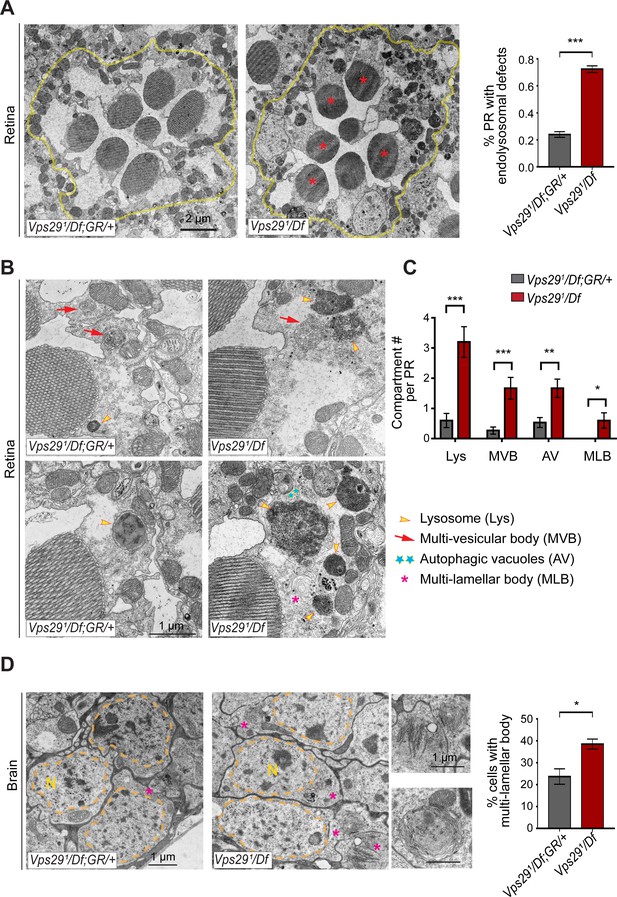
Loss of Vps29 disrupts lysosomal ultrastructure in the brain.
(A) Transmission electron microscopy (TEM) reveals overall preserved photoreceptor morphology, but aberrant endolysosomal structures in retinae from 30-day-old animals lacking Vps29. Within each ommatidium (yellow outline), the percentage of photoreceptors (PR) with endolysosomal defects was quantified. Asterisks denote photoreceptors with aberrant endolysosomal structures. Quantification based on n = 30 ommatidia from three animals per genotype. See also Figure 7—figure supplement 1A,B. (B) At higher magnification, TEM reveals an increase in lysosomes, multivesicular bodies, autophagic vacuoles, and multilamellar bodies in Vps29 mutant retinae. Lysosomes were frequently observed to be aberrantly enlarged and filled with granular, electron-dense material. (C) Distinct endolysosomal structures and/or compartments were quantified in n = 15 photoreceptors, including five photoreceptors from three animals for each genotype. (D) Increased numbers of multi-lamellar bodies (asterisks) are observed in cortical neurons from brains 30-day-old animals lacking Vps29. Neuronal nuclei (‘N’) are outlined. Quantification based on cell counts from n = 4 animals (50 cells per brain). See also Figure 7—figure supplement 1D. Statistical analysis (A, C, D) based on Student’s t-test. All error bars denote SEM. *, p<0.05; **, p<0.01; ***, p<0.001.
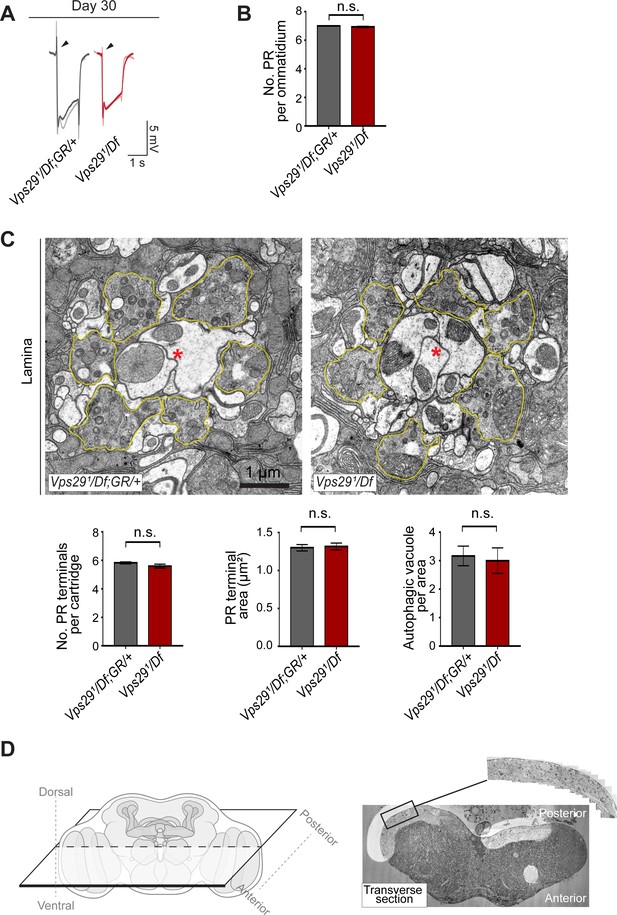
Additional ultrastructural analysis of Vps29 mutants.
(A) Electroretinogram (ERG) traces of Vps291/Df and Vps291/Df; GR/+ prior to transmission electron microscopy (TEM). Flies were raised under ambient light conditions (~500 lux) for 30 days. Representative traces for n = 2 animals are superimposed for each genotype. (B) Photoreceptor (PR) numbers are preserved in 30-day-old Vps291/Df animals, based on counts from retinal TEM. Quantification based on examination of n = 30 ommatidia (three independent animals) for each genotype. (C) In 30-day-old Vps29 mutants, no overt ultrastructural defects are detected in the lamina, which contains PR presynaptic terminals (yellow outline). Dendritic processes (postsynaptic) are indicated with red asterisks. Quantification based on examination of n = 30 cartridges (three independent animals) for each genotype. No changes were noted in either the number or area of PR terminals or the number of autophagic vacuoles. (D) Left: schematic showing orientation for thin sectioning prior to TEM analysis of fly brain. Right: Representative transverse section of fly brain at low magnification. Boxed region of interest (magnified inset) highlights the dorsal-posterior cortical region with densely packed neuronal cell bodies. This region was imaged at higher magnification in Figure 7D. Statistical analysis (B, C) based on Student’s t-test. All error bars denote SEM. n.s., not significant.
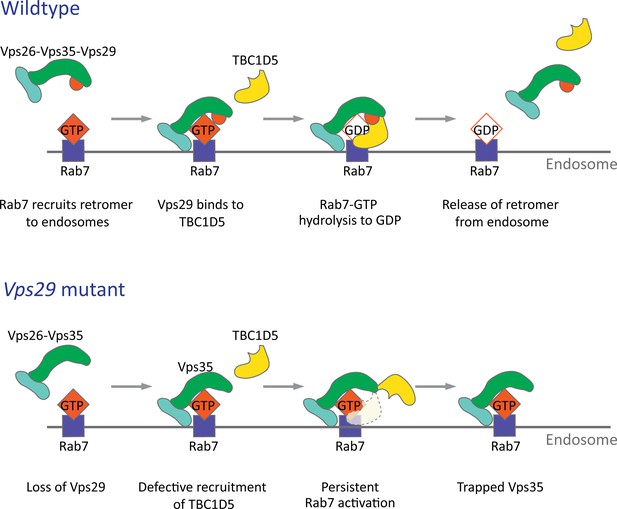
Model for Vps29-dependent retromer recruitment and release.
In neurons, the retromer core Vp26-Vps35-Vps29 is recruited by Rab7 to the endosomal membrane. Vps29 engages TBC1D5, which promotes inactivation of Rab7 and release of Vps35. In the absence of Vps29, the residual retromer complex is trapped at the endosomal membrane.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (D. melanogaster) | Vps35 | FLYB: FBgn0034708 | ||
| Gene (D. melanogaster) | Vps29 | FLYB: FBgn0031310 | ||
| Gene (D. melanogaster) | Vps26 | FLYB: FBgn0014411 | ||
| Gene (D. melanogaster) | TBC1D5 | FLYB: FBgn0038129 | ||
| Gene (D. melanogaster) | Rab7 | FLYB: FBgn0015795 | ||
| Genetic reagent (D. melanogaster) | FRT40A | Bloomington Drosophila Stock Center | FLYB: FBti000207; RRID:BDSC_1816 | y1 w1118; P{neoFRT}40A |
| Genetic reagent (D. melanogaster) | ey-FLP; FRT40A | Bloomington Drosophila Stock Center | FLYB: FBti0015982; RRID:BDSC_5622 | yd2, w1118, ey-FLP,GMR-lacZ;P{neoFRT}40A,w+,cl/CyO,y+ |
| Genetic reagent (D. melanogaster) | ey3.5-FLP; FRT40A | PMID:15848801 | y w ey3.5-FLP; FRT40A,cl, w+/CyO, Kr > GFP | |
| Genetic reagent (D. melanogaster) | nSyb-Gal4 | Bloomington Drosophila Stock Center | FLYB: FBti0150361; RRID:BDSC_51635 | y1 w*; P{nSyb-GAL4.S}3 |
| Genetic reagent (D. melanogaster) | nos-Cas9 | Bloomington Drosophila Stock Center | FLYB: FBti0159183; RRID:BDSC_54591 | y1, w*,M{nos-Cas9.P}ZH-2A |
| Genetic reagent (D. melanogaster) | Vps35MH20/CyO | Bloomington Drosophila Stock Center | FLYB: FBal0221635; RRID:BDSC_67202 | w; P{neoFRT}42D,Vps35MH20/CyO, Kr > GFP |
| Genetic reagent (D. melanogaster) | Df | Bloomington Drosophila Stock Center | FLYB: FBti0073468; RRID:BDSC_7491 | Df(2L)Exel6004 |
| Genetic reagent (D. melanogaster) | UAS-deGradFP | Bloomington Drosophila Stock Center | FLYB: FBti0147362; RRID:BDSC_38421 | w*; P{w[+mC]=UAS-Nslmb-vhhGFP4}3 |
| Genetic reagent (D. melanogaster) | GMR-w-RNAi | Bloomington Drosophila Stock Center | FLYB: FBti0074622; RRID:BDSC_32067 | GMR-w-RNAi13D |
| Genetic reagent (D. melanogaster) | UAS-Spin-GFP | Bloomington Drosophila Stock Center | FLYB:FBti0148827 RRID:BDSC_39668 | w*; P{UAS-spin.myc-EGFP} |
| Genetic reagent (D. melanogaster) | Vps35GFP | This study | Progenitor = w; Vps35TagRFP-T; TagRFP-T cassette was replaced by EGFP | |
| Genetic reagent (D. melanogaster) | Vps35RFP | PMID:26700726 | w; Vps35TagRFP-T | |
| Genetic reagent (D. melanogaster) | eyFLP;FRT42D | PMID:24781186 | y w,eyFLP,GMR-lacZ; P{neoFRT}42D,w+,cl/CyO,Kr > GFP | |
| Genetic reagent (D. melanogaster) | UAS-Vps35 | PMID:24781186 | y w; PBac{UAS-Vps35-HA} | |
| Genetic reagent (D. melanogaster) | Rab7EYFP | Bloomington Drosophila Stock Center | FLYB: FBst0062545 RRID:BDSC_62545 | w1118; TI{TI}Rab7EYFP |
| Genetic reagent (D. melanogaster) | Rab7Gal4-KO/TM3,Sb | PMID:24327558 | FLYB: FBal0294205 | FRT82B, Rab7Gal4-KO/TM3,Sb |
| Genetic reagent (D. melanogaster) | UAS-Venus-Rab7Q67L/CyO | PMID:24327558 | FLYB: FBal0294206 | PBac{UAS-Rab7.Q67L.Venus} |
| Genetic reagent (D. melanogaster) | UAS-Venus-Rab7T22N/CyO | PMID:24327558 | FLYB: FBal0294207 | PBac{UAS-Rab7.T22N.Venus} |
| Genetic reagent (D. melanogaster) | UAS-Venus-Rab7WT/CyO | PMID:24327558 | FLYB: FBal0294208 | PBac{UAS-Rab7.WT.Venus} |
| Genetic reagent (D. melanogaster) | Vps291/CyO,twi-GFP | This study | fly strain carrying the ywing2+ dominant marker replacing the gene Vps29 | |
| Genetic reagent (D. melanogaster) | Vps29WT.GFP/CyO,ubi-GFP | This study | fly strain carrying the Vps29 gene with EGFP…(GGS)4 sequence inserted at the N-terminus of Vps29 | |
| Genetic reagent (D. melanogaster) | Vps29L152E.GFP/CyO,ubi-GFP | This study | fly strain carrying the Vps29 gene with EGFP…(GGS)4 sequence inserted at the N-terminus of Vps29. L152 amino acid is mutated to E | |
| Genetic reagent (D. melanogaster) | Vps29-GR | This study | CH322-128G03 (FLYB: FBcl0761727) | |
| Genetic reagent (D. melanogaster) | UAS-dVps29 | This study | y w; PBac{UAS-dVps29-myc} | |
| Genetic reagent (D. melanogaster) | UAS-hVps29-1 | This study | y w; PBac{UAS-hVps29-1-FLAG} | |
| Genetic reagent (D. melanogaster) | UAS-hVps29-2 | This study | y w; PBac{UAS-hVps29-2-FLAG} | |
| Genetic reagent (D. melanogaster) | UAS-hVps29-3 | This study | y w; PBac{UAS-hVps29-3-FLAG} | |
| Genetic reagent (D. melanogaster) | UAS-dTBC1D5 | This study | y w; PBac{UAS-dTBC1D5-myc} | |
| Genetic reagent (D. melanogaster) | w; FRT40A, Vps291/CyO | This study | Recombinant betweenFRT40A and Vps291 | |
| Genetic reagent (D. melanogaster) | w;Vps291,Vps35GFP/CyO | This study | Recombinant betweenVps35GFP and Vps291 | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO; Vps29-GR/TM6B | This study | Derived by crosses | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO; Rab7EYFP/TM6B | This study | Derived by crosses | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO; nSyb-Gal4/TM6B | This study | Derived by crosses | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO; UAS-dVps29/TM6B | This study | Derived by crosses | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO; UAS-hVps29/TM6B | This study | Derived by crosses | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO;UAS-dTBC1D5/TM6B | This study | Derived by crosses | |
| Genetic reagent (D. melanogaster) | w;Vps291/CyO;UAS-dVps35/TM6B | This study | Derived by crosses | |
| Antibody | FITC-conjugated anti-GFP (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-9996 FITC; RRID:AB_627695 | 1:100 for IF |
| Antibody | Anti-Rab7 (mouse monoclonal) | Developmental Studies Hybridoma Bank | RRID:AB_2722471 | 1:100 for IF; 1:1000 for WB |
| Antibody | Anti-Elav (Rat monoclonal) | Developmental Studies Hybridoma Bank | DSHB: Elav-7E8A10; RRID:AB_528218 | 1:500 for IF |
| Antibody | Anti-tubulin Clone DM1A (mouse monoclonal) | Sigma Aldrich | Cat# T6199; RRID:AB_477583 | 1:1000 for WB |
| Antibody | Anti-Vps29 (goat polyclonal) | LifeSpan Biosciences | Cat# LS-C55674; RRID:AB_2214913 | 1:2000 for WB |
| Antibody | Anti-Vps35 (Guinea pig polyclonal) | This study | against C-terminal 338 amino acids of fly Vps35; 1:2000 for WB | |
| Antibody | Anti-Vps26 (Guinea pig polyclonal) | PMID:24781186 | 1:2000 for WB 1:500 for IF | |
| Antibody | Anti-CTSL (mouse monoclonal) | R and D Systems | Cat# MAB22591; RRID:AB_2087830 | 1:1000 for WB |
| Antibody | Anti-CTSD (goat polyclonal) | Santa Cruz Biotechnology | Cat# sc-6487; RRID:AB_637895 | 1: 500 for WB |
| Antibody | Rabbit anti-Atg8 | PMID:27068460 | 1:1000 for WB | |
| Antibody | Anti-Polyubiquitinylated conjugates Clone FK1 (mouse monoclonal) | Enzo Life Sciences | Cat# BML-PW8805-0500; RRID:AB_2052280 | 1:1000 for WB |
| Antibody | Rabbit anti-p62/Ref(2)p | PMID:25686248 | 1:2000 for WB | |
| Antibody | Rabbit anti-Arl8 | Developmental Studies Hybridoma Bank | DSHB Cat# Arl8, RRID:AB_2618258 | 1:1000 for IF |
| Antibody | Anti-actin clone C4 (mouse monoclonal) | Millipore | Cat# MAB1501; RRID:AB_2223041 | 1:1000 for WB |
| Antibody | Cy3-conjugated anti-HRP | Jackson ImmunoResearch | Cat# 123-165-021; RRID:AB_2338959 | 1:150 for IF |
| Antibody | Cy3-conjugated Goat anti-mouse IgG | Jackson ImmunoResearch | Cat# 115-165-146; RRID:AB_2338690 | 1:500 for IF |
| Antibody | Cy3-conjugated Goat anti-Rat IgG | Jackson ImmunoResearch | Cat# 112-165-003; RRID:AB_2338240 | 1:500 for IF |
| Antibody | Mouse anti-goat IgG-HRP | Santa Cruz Biotechnology | Cat# sc-2354, RRID:AB_628490 | 1:5000 for WB |
| Antibody | Goat anti-mouse IgG-HRP | Santa Cruz Biotechnology | Cat# sc-2005; RRID:AB_631736 | 1:5000 for WB |
| Antibody | Goat anti-rabbit IgG-HRP | Santa Cruz Biotechnology | Cat# sc-2004; RRID:AB_631746 | 1:5000 for WB |
| Antibody | Rabbit anti-Glc-Cer | Glycobiotech | Cat# RAS_0010 | 1:250 for IF |
| Recombinant DNA reagent | Plasmid: pUASTattb_dVps29-myc | This study | Progenitors: GH25884 (cDNA) | |
| Recombinant DNA reagent | Plasmid: pUASTattb_dTBC1D5-myc | This study | Progenitors: BS16827 (cDNA) | |
| Recombinant DNA reagent | Plasmid: pUASTattb_hVps29-1-FLAG | This study | Progenitors: OHu00442D (cDNA) | |
| Recombinant DNA reagent | Plasmid: pUASTattb_hVps29-2-FLAG | This study | Progenitors: OHu02289D (cDNA) | |
| Recombinant DNA reagent | Plasmid: pUASTattb_hVps29-3-FLAG | This study | Progenitors: OHu05688D (cDNA) | |
| Commercial assay or kit | Subcloning Efficiency DH5α competent cells | Thermo Fisher Scientific | Cat# 18265017 | |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | NEB | Cat# E0554S | |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Cloning Kit | NEB | Cat# E5520S | |
| Commercial assay or kit | PureLink Genomic DNA Kits | Thermo Fisher Scientific | Cat# K182001 | |
| Commercial assay or kit | GFP-Trap agarose beads | Allele Biotechnology | Cat# ABP-NAB-GFPA100 | |
| Chemical compound, drug | 2X Laemmli Sample Buffer | Bio-Rad | Cat# 161–0737 | |
| Chemical compound, drug | 4% paraformaldehyde in 1XPBS | ChemCruz | Cat# sc-281692 | |
| Chemical compound, drug | RapiClear | SunJin Lab Co. | ||
| Chemical compound, drug | Vectashield | Vector Laboratories | Cat# H-1000 | |
| Chemical compound, drug | Protein A/G agarose | Thermo Fisher | Cat# 20421 | |
| Chemical compound, drug | 8% glutaraldehyde | EMS | Cat# 16020 | |
| Chemical compound, drug | Cacodylic Acid, Trihydrate Sodium 100 g | EMS | Cat# 12300 | |
| Chemical compound, drug | EM-grade glutaraldehyde, 25% Aq solution | EMS | Cat# 16221 | |
| Chemical compound, drug | Osmium tetroxide4% Aq solution | EMS | Cat# 19191 | |
| Chemical compound, drug | Paraformaldehyde 16% Aq Solution | EMS | Cat# 15711 | |
| Chemical compound, drug | Propylene Oxide | EMS | Cat# 20411 | |
| Chemical compound, drug | Koptec 200 Proof100% ethanol Anhydrous | VWR | Cat# 89125–186 | |
| Chemical compound, drug | Embed-812 | EMS | Cat# 14901 | |
| Chemical compound, drug | NMA | EMS | Cat# 19001 | |
| Chemical compound, drug | DDSA | EMS | Cat# 13711 | |
| Chemical compound, drug | DMP-30 | EMS | Cat# 13600 | |
| Chemical compound, drug | Uranyl Acetate | EMS | Cat# RT22400 | |
| Chemical compound, drug | Lead Nitrate | EMS | Cat# RT17900-25 | |
| Chemical compound, drug | Phalloidin 488 nm | ThermoFisher | Cat# AB_2315147 | |
| Chemical compound, drug | FM 1-43FX | Invitrogen | Cat# F35355 | |
| Chemical compound, drug | Western Lightning Plus-ECL | PerkinElmer | Cat# 121001EA | |
| Software, algorithm | Leica Application Suite X | Leica | RRID:SCR_013673 | |
| Software, algorithm | LabChart Reader | ADInstruments | https://www.adinstruments.com/products/labchart-reader | |
| Software, algorithm | ImageJ | National Institute of Health | RRID:SCR_003073 | |
| Sequence-based reagent | dVps29-myc-F | This study | 5’-GAAGATCTTCATGCTCGTTCTGGTACTCGGCGA-3’ | |
| Sequence-based reagent | dVps29-myc-R | This study | 5’-GCTCTAGACTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGATCTTCTTGTACTCGATGCGCTCCA-3’ | |
| Sequence-based reagent | dTBC1D5-myc-F | This study | 5’-GAAGATCTATCAACATGACTGTTTGGGGAATAGAAGCCATCA-3’ | |
| Sequence-based reagent | dTBC1D5-myc- R | This study | 5’-GCTCTAGATCACAGATCCTCTTCTGAGATGAGTTTTTGTTCACTCGATTCGTTTCGATGCCGT-3’ | |
| Sequence-based reagent | hVps29-1-F | This study | 5’-CCGCCTCGAGGCCACCATGTTGGTGTTGGT-3’ | |
| Sequence-based reagent | hVps29-2-F | This study | 5’-CCGCCTCGAGGCCACCATGGCTGGGCACA-3’ | |
| Sequence-based reagent | hVps29-3-F | This study | 5’-CCGCCTCGAGGCCACCATGAGCAGGTGTGCT-3’ | |
| Sequence-based reagent | hVps29-R | This study | 5’-CTAGTCTAGATTATCACTTATCGTCGTCATCCTTGTAATCAGGT-3’ | |
| Sequence-based reagent | Vps29-P1-F | This study | 5’-GAACCTGACGTATCCGGAGC-3’ | |
| Sequence-based reagent | Vps29-P1-R | This study | 5’-TCGCCGATCAGTTGGTACAC-3’ | |
| Sequence-based reagent | Vps29-P2-F | This study | 5’-CTCGTTCTGGTACTCGGCG-3’ | |
| Sequence-based reagent | Vps29-P2-R | This study | 5’-ACGAACGAAGGCACCACATT-3’ | |
| Sequence-based reagent | Vps29-P3-F | This study | 5’-GGCCGCATACATCACATCCT-3’ | |
| Sequence-based reagent | Vps29-P3-R | This study | 5’-GAATTTGTTGCCGTGCTCGT-3’ | |
| Sequence-based reagent | Vps35-F (Internal control) | This study | 5’-TTGTACCTCCTCATAACAGTGGG-3’ | |
| Sequence-based reagent | Vps35-R (Internal control) | This study | 5’-TCGTTCTCCTCAACCATCACAT-3’ | |
| Other | Leica SP8 confocal microscope | Leica | ||
| Other | Leica DM 6000 B system | Leica | ||
| Other | Zeiss LSM 880 with Airyscan | Zeiss |





