The speed of GTP hydrolysis determines GTP cap size and controls microtubule stability
Figures
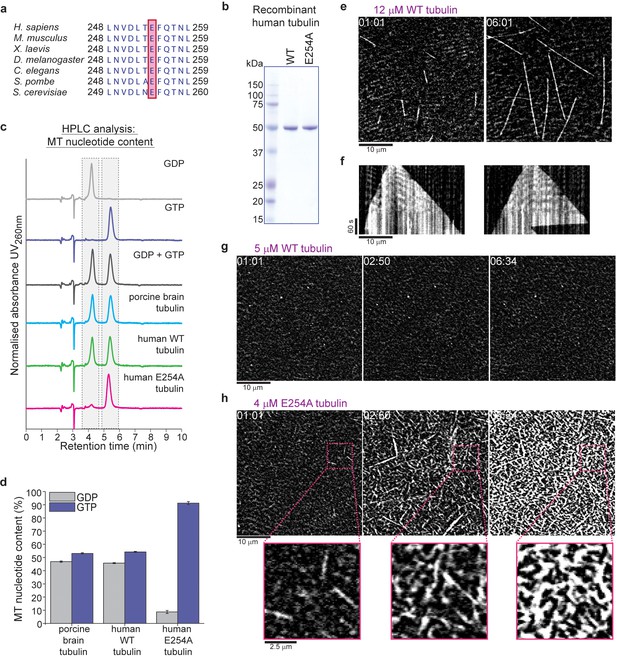
Recombinant human tubulin lacking GTPase activity strongly nucleates microtubules.
(a) The evolutionarily conserved catalytic glutamic acid of α-tubulin (E, pink). (b) Coomassie Blue-stained SDS gel of purified wildtype (WT) and E254A mutant recombinant human tubulin. (c) HPLC chromatograms of pure GDP and GTP compared to nucleotides extracted from microtubules polymerized from porcine brain tubulin or wildtype and E254A mutant human tubulin. (d) Quantification of the nucleotide content of different microtubules. Bar graphs depict the means of 3 independent experiments, error bars represent standard deviation (SD). (e) iSCAT microscopy images of unlabeled wildtype human tubulin (12 µM) growing from immobilized GMPCPP-stabilized biotinylated seeds (GMPCPP-seeds). (f) Kymographs showing wildtype microtubule growth, conditions as in (e). (g, h) iSCAT microscopy images showing (g) lack of microtubule nucleation at 5 µM wildtype tubulin and (h) strong nucleation at 4 µM E254A tubulin at a surface with an immobilized rigor kinesin (Kin1rigor). Bottom: magnified E254A microtubule views. Scale bars as indicated, time is always min:sec.
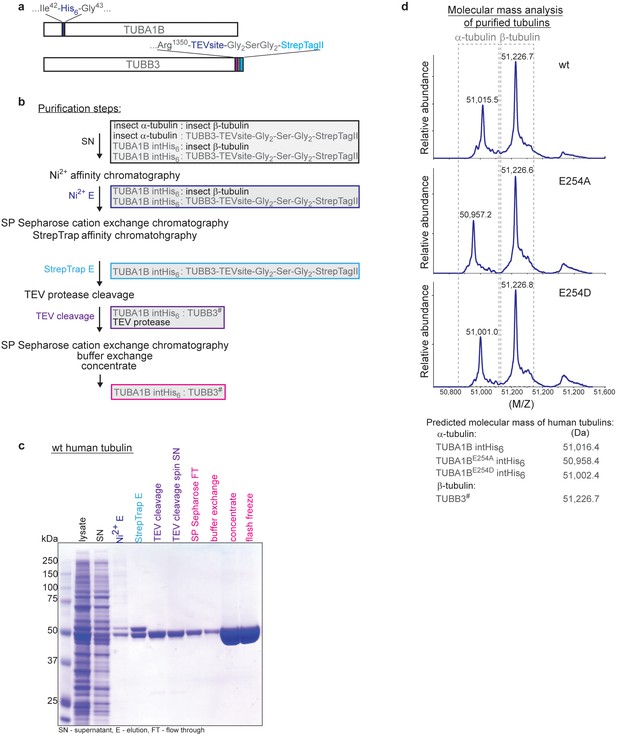
Expression and purification of recombinant human α/β tubulin.
(a) Schematic of the human TUBA1B α-tubulin and TUBB3 β-tubulin expression constructs. (b) Flow chart of the purification steps to obtain recombinant human α/β-tubulin from insect cells. The hash (#) sign on TUBB3 marks the TEV-protease cleavage site. (c) Coomassie Blue-stained SDS gel showing the step-wise purification of recombinant wildtype (wt) human tubulin. Coloring of the labels as for the corresponding purification steps in the flow chart (b). (d) Molecular mass determination of purified recombinant tubulin subunits confirms the lack of insect cell tubulin contamination and uncleaved TUBB3. Note that the predicted masses for TUBA1B and its mutants correspond to N-terminally acetylated versions of the protein. This is a common modification of proteins expressed in insect cells.
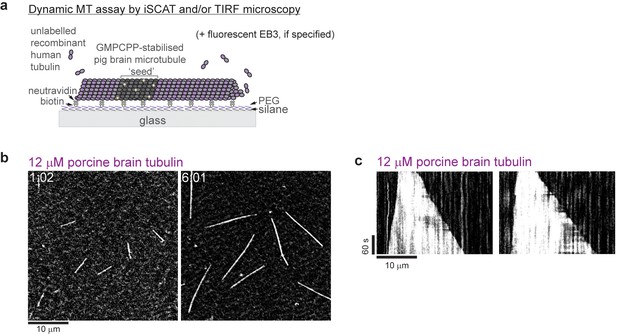
iSCAT microscopy of unlabeled porcine brain microtubules.
(a) Schematic of a dynamic microtubule assay. (b) iSCAT microscopy images of unlabeled porcine brain microtubules growing from GMPCPP-seeds at 12 µM porcine brain tubulin. (c) Kymographs showing the characteristic fast plus and slow minus end growth of dynamic porcine brain microtubules. Conditions as in (b). Scale bars as indicated, time is in min:sec.
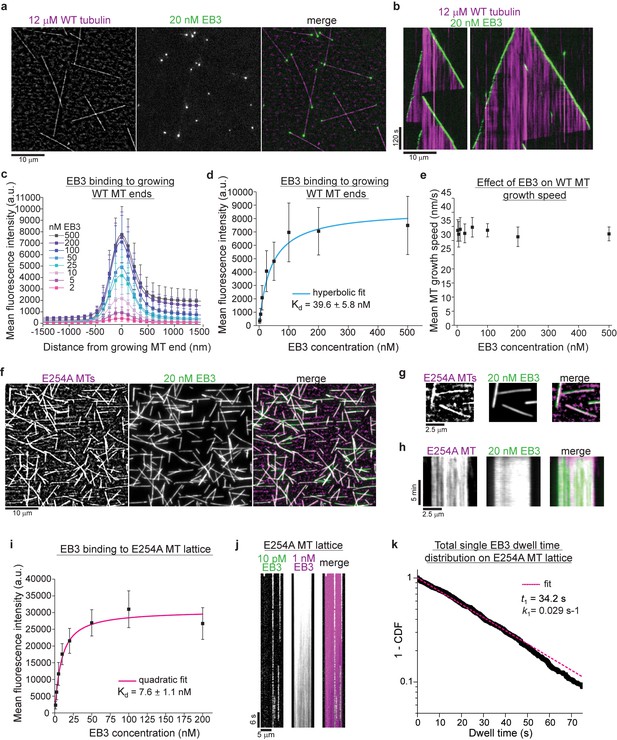
GTPase deficient microtubules are stable and bind EB3 with high affinity.
(a) iSCAT/TIRF microscopy image of unlabeled wildtype microtubules (WT, magenta) growing from GMPCPP-seeds at 12 µM wildtype human tubulin in the presence of 20 nM human mGFP-EB3 (green). (b) Kymographs showing mGFP-EB3 (green) tracking the ends of growing wildtype microtubules, conditions as in (a). (c) Mean mGFP-EB3 intensity profiles at the ends of human wildtype microtubules (WT MT) growing at 12 µM wildtype tubulin in the presence of varying mGFP-EB3 concentrations (as indicated). Number of microtubules (and frames) averaged for each mGFP-EB3 concentration: 2 nM – 64 (7360), 5 nM – 87 (9222), 10 nM – 93 (8742), 25 nM – 89 (8900), 50 nM – 56 (5600), 100 nM – 58 (4756), 200 nM – 56 (4088), 500 nM – 46 (4140). Error bars are SD. (d) Quantification of mGFP-EB3 binding affinity to microtubule ends growing at 12 µM wildtype tubulin (WT MT). Black symbols depict the averaged maximal mGFP-EB3 intensities at growing plus ends at each condition, error bars represent SD. Experimental data as in (c). A hyperbolic fit (cyan) was used to calculate the Kd. (e) mGFP-EB3 does not affect significantly the growth speed of human wildtype microtubules (WT MT). Experimental data as in (c). (f) iSCAT/TIRF microscopy images of 20 nM mGFP-EB3 (green) binding to unlabeled E254A mutant microtubules (magenta) attached to a Kin1rigor surface. (g) Magnified images and (h) kymographs of stable unlabeled E254A microtubules (magenta) decorated with 20 nM mGFP-EB3 (green), conditions as in (d). (i) Quantification of mGFP-EB3 binding affinity to E254A mutant microtubules (E254A MTs). Black symbols depict the averaged mGFP intensities measured all along the microtubules at each condition, error bars represent SD. A quadratic fit (magenta) was used to calculate the Kd. Number of microtubules measured for each mGFP-EB3 concentration: 1 nM – 96, 2.5 nM – 126, 5 nM – 86, 10 nM – 69, 20 nM – 78, 50 nM – 61, 100 nM – 64, 200 nM - 75. (j) Kymograph of mGFP-EB3 (green) single molecule imaging at 10 pM on an individual E254A microtubule (MT) in the presence of 1 nM Alexa647-EB3 (magenta). (k) Dwell time analysis of mGFP-EB3 molecules on E254A microtubule (MT) lattice plotted as a survival function (1 - CDF, cumulative density function). Dashed line (magenta) is a mono-exponential fit to the data (black dots). Number of microtubules analyzed - 126, number of mGFP-EB3 binding events - 807. Scale bars as indicated.
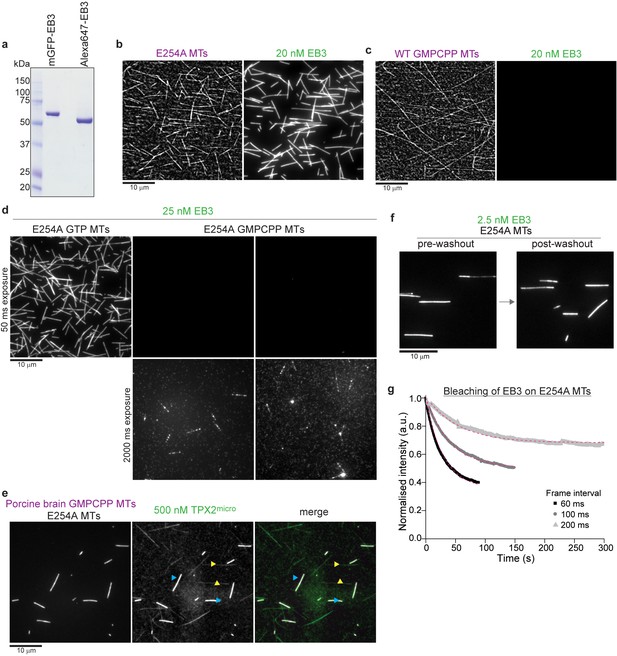
Characterization of EB3 binding to wildtype and GTPase deficient human microtubules.
(a) Coomassie Blue-stained SDS gel of purified recombinant mGFP-EB3 and Alexa647-labeled SNAP-EB3 (Alexa647-EB3). (b, c) Comparative iSCAT/TIRF microscopy images of 20 nM mGFP-EB3 binding to (b) unlabeled E254A microtubules (E254A MTs) and (c) wildtype microtubules polymerized in the presence of GMPCPP (WT GMPCPP MTs), attached to a Kin1rigor surface. EB3 binds strongly to E254A microtubules, but not to GMPCPP microtubules. (d) Comparative TIRF microscopy images of 25 nM mGFP-EB3 binding to unlabeled E254A microtubules polymerized in the presence of GTP (E254A GTP MTs, ) or in the presence of GMPCPP (E254A GMPCPP MTs). Microtubules were attached to a Kin1rigor surface. EB3 binds strongly to E254A microtubules polymerized in the presence of GTP, but not to E254A microtubules polymerized in the presence of GMPCPP. Two different exposure conditions (50 ms and 2000 ms) are shown for GMPCPP-bound E254A microtubules to visualize the faintly bound mGFP-EB3. (e) TIRF microscopy images of 500 nM mGFP-TPX2micro binding to a mixture of fluorescently labeled GMPCPP-bound porcine brain microtubules (porcine brain GMPCPP MTs, magenta) and unlabeled GTP-bound E254A microtubules (E254A MTs). mGFP-TPX2micro binds strongly to GMPCPP microtubules (cyan arrowheads) and faintly to GTP-bound E254A microtubules (yellow arrowheads). (f) EB3 washout experiment: TIRF microscopy images of mGFP-EB3 at 2.5 nM bound to unlabeled E254A microtubules (E254A MTs) immobilized on a Kin1rigor surface before (left) and after (right) washing out mGFP-EB3 from the flow chamber. (g) Steady-state bleaching curves for 2.5 nM mGFP-EB3 bound to Kin1rigor surface-immobilized E254A microtubules (E254A MTs), imaged with a 60 ms exposure time and different frame intervals, as indicated. Dashed magenta lines are a fit to the data (Materials and methods) revealing a dwell time of mGFP-EB3 molecules of 83 s (and bleaching times of 45 s, 83 s, and 171 s for imaging with a time lapse of 60 ms (single molecule imaging condition), 100 ms, and 200 ms, respectively). The obtained dwell time and bleaching time for the 60 ms time lapse are consistent with the measured apparent mean dwell time of 1/ (0.029 s−1) = 34 s for single mGFP-EB3 molecules on E254A microtubules (Figure 2k). This single molecule dwell time is affected by bleaching. Its inverse value is expected to be the sum of the inverse real dwell time and the inverse bleaching time obtained from the steady state bleaching experiment here in (g), which is the case. Scale bars as indicated, time is in min:sec.
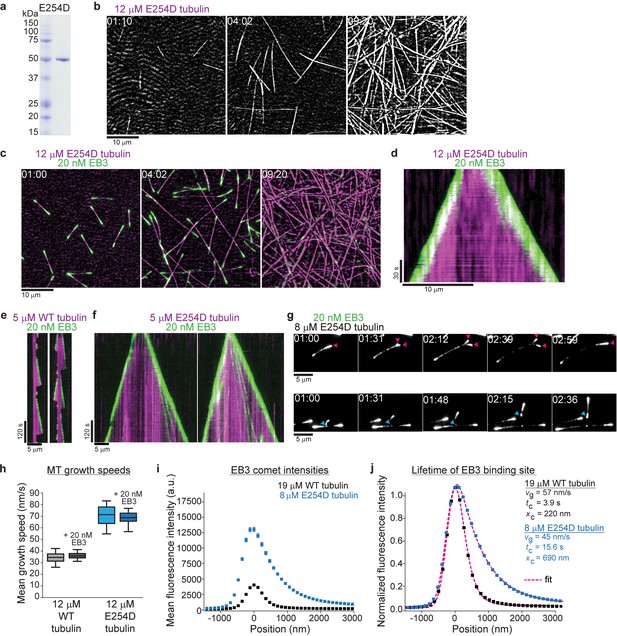
Slowing down GTP hydrolysis extends the GTP cap and stabilizes growing microtubules.
(a) Coomassie Blue-stained SDS gel with purified E254D mutant recombinant human tubulin. (b, c) iSCAT/TIRF microscopy images of unlabeled E254D mutant microtubules growing from GMPCPP-seeds and also spontaneously nucleating at 12 µM E254D mutant human tubulin (b) in the absence and (c) presence of 20 nM human mGFP-EB3 (green). (d) Kymograph showing mGFP-EB3 (green) tracking the ends of persistently growing E254D microtubules, conditions as in (c). (e, f) iSCAT/TIRF microscopy kymographs depicting (e) wildtype (WT) and (f) E254D microtubules (magenta) growing at 5 µM tubulin in the presence of 20 nM mGFP-EB3 (green). E254D microtubules display very few catastrophes under conditions where wildtype human microtubules undergo frequent catastrophes. (g) TIRF microscopy images of protofilament bending, and re-association (arrowheads, magenta) or splitting (arrowheads, cyan) at unlabeled E254D microtubule ends growing at 8 µM tubulin visualized by mGFP-EB3. (h) Quantification of wildtype (WT) and E254D microtubule growth speeds at 12 µM respective tubulin concentrations in the presence and absence of mGFP-EB3. The boxes extend from 25th to 75th percentiles, the whiskers extend from 5th to 95th percentiles, and the mean value is plotted as a line in the middle of the box. Number of microtubules measured for each condition: 12 µM wildtype tubulin – 95, 12 µM wildtype tubulin and 20 nM mGFP-EB3 – 86, 12 µM E254D tubulin – 35, 12 µM E254D tubulin and 20 nM mGFP-EB3 - 83. (i) Mean mGFP-EB3 intensity profiles at growing wildtype (black) and E254D (blue) microtubule plus ends. Number of microtubules (and frames averaged) for each condition: wildtype – 68 (36960), E254D – 74 (41720). Error bars are SE. (j) Normalized comet profiles from (h) with dashed lines (magenta) representing exponentially modified Gaussian fits (see Materials and methods) yielding the comet length xc and, knowing the measured growth speed vg, the life time tc = xc/vg of the EB binding sites. Scale bars as indicated, time is min:sec.
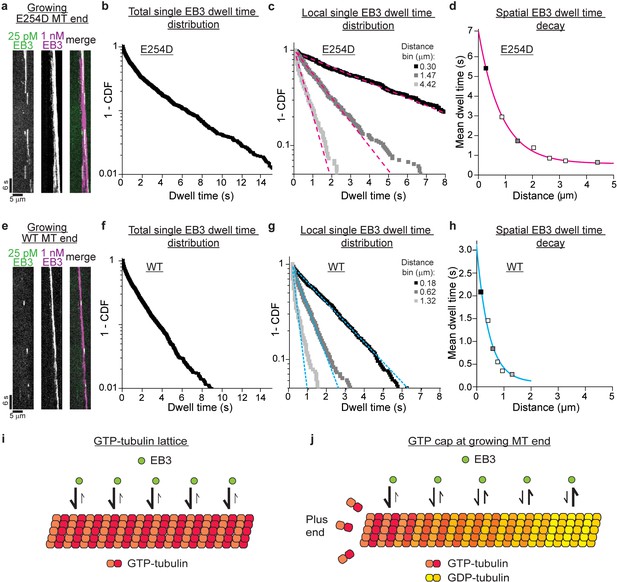
Conformational gradient in the GTP cap revealed by spatially resolved single EB3 dwell time distributions.
(a) TIRF microscopy kymograph of single mGFP-EB3 molecules (green) at 25 pM in the plus end region of an E254D microtubule (E254D MT) growing at 10 µM E254D tubulin (vg = 58.5 nm/s) in the additional presence of 1 nM Alexa647-EB3 (magenta) (for end region visualization). (b) Dwell time distribution of single mGFP-EB3 molecules plotted as a survival function (1- CDF, cumulative density function). Number of microtubules analyzed - 150, number of mGFP-EB3 binding events - 1834. (c) Local dwell time distributions at distinct distances from the growing microtubule plus end. Dashed magenta lines are mono-exponential fits. The distance bins are 0–0.59, 1.18–1.77 and 3.84–5.02 µm; bin centers shown in the legend. Experimental data as in (b). (d) Local mean EB3 dwell times as a function of distance from the growing E254D microtubule end. Filled symbols correspond to the local mean dwell times calculated based on data in (c). The solid magenta line is a mono-exponential fit with a decay length of 850 nm. (e) TIRF microscopy kymograph of single mGFP-EB3 molecules (green) at 25 pM in the plus end region of a wildtype (WT) microtubule growing at 19 µM wildtype tubulin (vg = 59.8 nm/s) in the presence of additional 1 nM Alexa647-EB3 (magenta). (f) mGFP-EB3 dwell time distribution. Number of microtubules analyzed - 548, number of mGFP-EB3 binding events - 2425. (g) Local mGFP-EB3 dwell time distributions. Dashed cyan lines are mono-exponential fits. Experimental data as in (f). (h) Local mean EB3 dwell times in the wildtype microtubule plus end region. Filled symbols correspond to the local mean dwell times calculated based on data in (g). The solid cyan line is a mono-exponential fit to the data with a decay length of 450 nm. (i) Schematic of high affinity EB3 binding to the GTP lattice of a GTPase-deficient microtubule. (j) Schematic of a growing end of a GTPase-competent microtubule displaying a gradually decreasing EB3 binding affinity (illustrated by a color gradient) as the lattice conformation changes as a consequence of GTP hydrolysis.
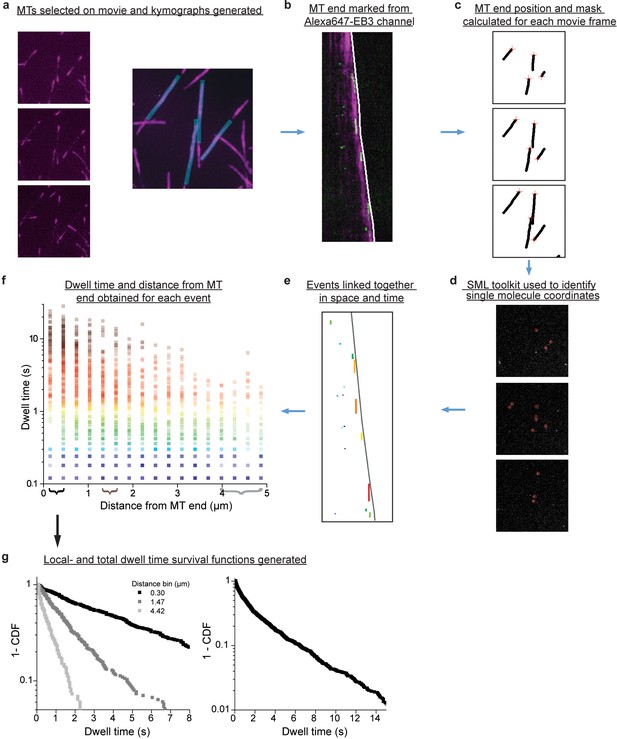
Flow chart of single molecule localization and spatially-resolved dwell time analysis.
For single molecule analysis of mGFP-EB3 binding to microtubules, TIRF microscopy movies were recorded in the presence of 25 pM mGFP-EB3 and additional 1 nM Alexa647-EB3 to visualize the growing microtubule end region. (a) Left: Single TIRF microscopy frames of a movie showing the Alexa647-EB3 channel. Right: Microtubule (MT) positions were marked by hand on a maximum-intensity projection of all frames. (b) Kymographs were generated for each marked microtubule in (a), and the growing plus-end position traced by hand in the kymographs (white line). (c) The x-y position of each microtubule end was calculated for each frame in the original movie (red crosses). The moving end position and the positions of the static microtubule projection from (a) were used to create a dynamic binary mask movie. (d) For each frame of the original movie, the mGFP-EB3 channel was analyzed using a Single Molecule Localization (SML) plugin, to determine the coordinates of each potential single molecule event (red circles). The mask from (c) was then used to exclude all events from outside the microtubules of interest. (e) Events were linked together in time and space using specific linking parameters. The resulting kymograph corresponding to (b) shows automatically detected and linked single molecule events, color-coded by duration. (f) For each linked event, the dwell time and initial distance from the nearest microtubule end were calculated. (g) Data from all movies were binned at specific distances from the growing microtubule end (braces in f), to give spatially-resolved dwell time survival functions (1- CDF, cumulative density function), or binned over all distances to give a total survival function. The example plots in (g) are the same as displayed on Figure 4b–c.
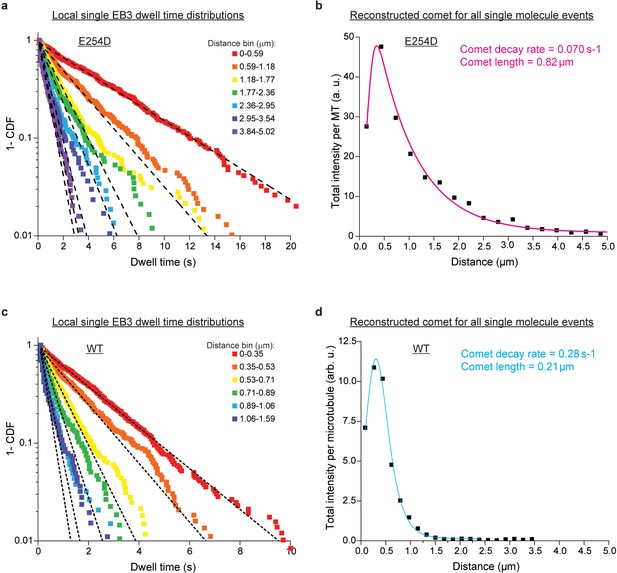
Spatially-resolved EB3 dwell time distributions at the ends of E254D and wildtype microtubules.
(a) Local dwell time survival functions at specific distance bins from the microtubule end for E254D microtubules. Dashed black lines are mono-exponential fits. The global reduced χ2 value for all the fits is half that of a mono-exponential fit to the pooled data in Figure 4b. (b) Reconstructed comet for all single molecule events, from summing the complete emission of each binding event in each distance bin from the microtubule end. The total intensity is normalized to the number of microtubules analyzed. (c) and (d) depict the same information as (a) and (b) but for wildtype (WT) microtubule ends. Intensities in (b) and (d) can be compared. Good agreement of the reconstructed comets based on single molecule data with the measured comets at higher EB3 concentrations (Figure 3i and j) demonstrate equivalence of single molecule and ensemble analysis.
Wildtype and E254D microtubules grow with similar speeds under single molecule experiment conditions.
Quantification of microtubule growth speeds at 19 µM wildtype (WT) and 10 µM E254D tubulin concentrations in the presence of 1 nM Alexa647-EB3 and 25 pM mGFP-EB3. The boxes extend from 25th to 75th percentiles, the whiskers extend from 5th to 95th percentiles, and the mean value is plotted as a line in the middle of the box. The experimental data is the same as analysed and presented in Figure 4.
Videos
Pure recombinant human wildtype tubulin polymerizes into dynamic microtubules.
iSCAT microscopy time-lapse movie of unlabeled microtubules growing in the presence of 12 µM human wildtype tubulin from immobilized stable microtubule seeds at 30°C. Scale bar is 5 µm. Time stamp is in min:s.
Human wildtype tubulin does not nucleate microtubules at low tubulin concentration.
iSCAT microscopy time-lapse movie showing lack of microtubule nucleation at 5 µM human wildtype tubulin on a surface with an immobilized rigor kinesin (Kin1rigor) at 30°C. Scale bar is 5 µm. Time stamp is in min:s.
Human E254A tubulin mutant lacking GTPase activity strongly nucleates microtubules.
iSCAT microscopy time-lapse movie showing strong microtubule nucleation at 4 µM human E254A tubulin on a surface with an immobilized rigor kinesin (Kin1rigor) at 30°C. Scale bar is 5 µm. Time stamp is in min:s.
EB3 binds to the growing ends of human wildtype microtubules.
iSCAT/TIRF microscopy time-lapse movie of unlabeled wildtype microtubules (magenta) growing from immobilized stable microtubule seeds in the presence of 12 µM human wildtype tubulin and 20 nM human mGFP-EB3 (green) at 30°C. Scale bar is 5 µm. Time stamp is in min:s.
Slowing down GTP hydrolysis extends the GTP cap and stabilizes growing microtubules.
iSCAT/TIRF microscopy time-lapse movie of unlabeled E254D mutant microtubules (magenta) growing from immobilized microtubule seeds and also spontaneously nucleating in the presence of 12 µM human E254D mutant tubulin and 20 nM human mGFP-EB3 (green) at 30°C. Scale bar is 5 µm. Time stamp is in min:s.
Wildtype microtubules are very dynamic at low tubulin concentrations.
iSCAT/TIRF microscopy time-lapse movie of human wildtype microtubules (magenta) undergoing frequent catastrophes when growing from immobilized microtubule seeds in the presence of 5 µM human wildtype tubulin and 20 nM human mGFP-EB3 (green) at 30°C. Scale bar is 5 µm. Time stamp is in min:s.
Microtubules with reduced GTPase activity are more stable than wildtype microtubules.
iSCAT/TIRF microscopy time-lapse movie of human E254D microtubules (magenta) show persistent growth when growing from immobilized microtubule seeds in the presence of 5 µM E254D tubulin and 20 nM human mGFP-EB3 (green) at 30°C. Scale bar is 5 µm. Time stamp is in min:s.





