Ser/Thr kinase Trc controls neurite outgrowth in Drosophila by modulating microtubule-microtubule sliding
Figures
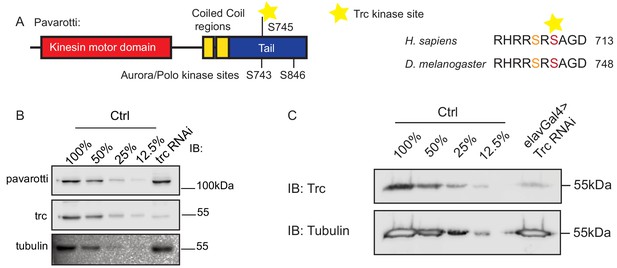
The kinase Trc regulates neurite outgrowth and microtubule sliding.
(A) Example images of timelapse imaging to measure microtubule sliding in Drosophila S2 cells. Microtubules are shown at time zero in green. Magenta indicates photoconverted region. Photoconverted region is highlighted by dotted yellow line at other timepoints. Red box indicates region shown in inset in A’ Scale bar 10 µm. (B) Quantification of microtubule sliding rate shows an increase upon Pavarotti or Trc depletion. n = 16–20 cells. Ctrl = 1.0 ± 0.1 upper 95% CI = 1.2, lower 95% CI = 0.8, Pav RNAi = 3.4 ± 1.0, 5.5, 1.4 Trc RNAi = 2.1 ± 0.4, 3.0, 1.2. Ctrl vs Trc RNAi p=0.03. (C) Representative images of 3rd instar larvae cultured neurons under control or elav > Trc RNAi conditions. (D) Quantification of total neurite length per cell over time in culture. The total neurite length is increased from control upon Trc depletion. 24 hr; ctrl = 137.8 ± 12.0 µm, Trc RNAi 254 ± 19.1 µm. p=0.0001. 48 hr; ctrl = 163.7 ± 15.3 µm, Trc RNAi = 251.2 ± 19.9 µm, p=0.001, 72 hr; ctrl = 156.3 ± 13.8 µm, TrcRNAi = 314.4 ± 36.5 µm, p=0.009. N = 11–23 cells from three independent experiments. (E) Example images from timelapse imaging of photoconverted microtubules in neurons under control conditions or upon Trc depletion. Tubulin was labeled with tdMaple3 alpha tubulin84b. After photoconversion, cells were imaged every minute for 10 min. Scale bar = 5 µm. (F) Quantification of microtubule sliding rates. Trc depletion leads to an increase in microtubule sliding rates in neurons. Ctrl = 1.0 ± 0.14, TrcRNAi = 1.69 ± 0.30. N = 20 cells from three independent experiments. p=0.04 Student’s T-test.
-
Figure 1—source data 1
Sliding rates for control, Pav RNAi and Trc RNAi-treated S2 cells.
Related to Figure 1A and B.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Neurite length for neurons cultured from control larvae or Trc RNAi larvae.
Related to Figure 1C and D.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Sliding rates for control, and Trc RNAi neurons.
Related to Figure 1E and F.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig1-data3-v2.xlsx
Domain structure of Pavarotti and demonstration of Trc knockdown.
(A) Schematic showing domain structure and location of proposed Trc phosphorylation site inDrosophila Pavarotti and human MKLP1. (B) Lysates from S2 cells demonstrate efficient knockdown of Trc with dsRNA treatment. Pavarotti protein levels are not affected. (C) Lysates from brains dissected from 3rd instarDrosophilalarvae confirm neuron-specific knockdown of Trc.
Trc regulates microtubule sliding in kinase-dependent manner.
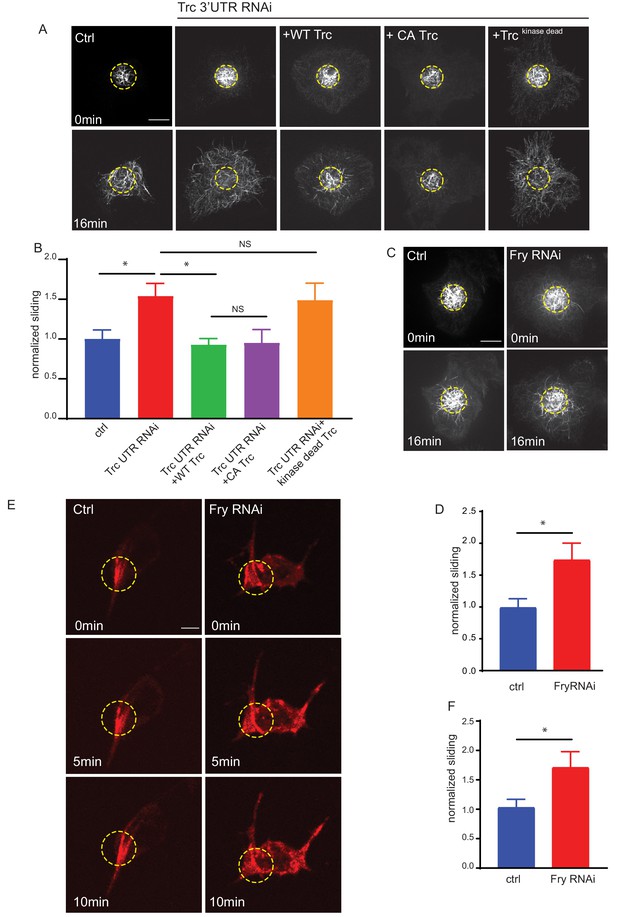
(A) Sliding experiments in S2 cells show increased sliding upon depletion of Trc with dsRNA targeting the non-coding region. The effect can be rescued with overexpression of WT Trc, but not kinase dead Trc. Scale bar 10µm. (B) Quantification of sliding experiments in A. n=18-50 cells from four independent experiments. Ctrl = 1.0 ± 0.11, Lower 95% CI 0.77, Upper 95% CI 1.23, Trc3’UTR RNAi = 1.54 ± 0.16, 1.218, 1.86, Trc3’UTR RNAi + WT Trc = 0.93 ± 0.08, 0.77, 1.09 Trc3’UTR RNAi + CA Trc = 0.95 ± 0.17, 0.59, 1.30, Trc3’UTR RNAi + kinase dead Trc = 1.49 ± 0.22, 1.05, 1.92. Ctrl vs Trc 3’UTR RNAi p = 0.03, Trc 3’UTR RNAi vs Trc 3’UTR RNAi + Trc WT p = 0.01. One-way ANOVA with Sidak’s post hoc correction. (C) Sliding experiments in S2 cells show increased sliding upon depletion of Fry. Scale bar 10 µmD. Quantification of sliding experiments in C. n=26-37 cells from four independent experiments. Ctrl = 1 ± 0.13, Fry RNAi = 1.75 ± 0.26. Lower 95% CI ctrl = 0.72, Fry RNAi 1.23. Upper 95% CI ctrl = 1.268, Fry RNAi = 2.265. p = 0.024 Student’s T-test. (E) Representative images from timelapse imaging of photoconverted microtubules in neurons under control conditions or upon Fry depletion. Tubulin was labeled with tdMaple3 alpha tubulin84b. After photoconversion, cells were imaged every 30s for 10 min. Scale bar = 5µm (F). Quantification of microtubule sliding rates. Fry depletion leads to an increase in microtubule sliding rates in neurons. n = 31-42 cells from four independent experiments. Ctrl = 1.0 ± 0.13, 0.78, 1.3, Fry RNAi = 1.7 ± 0.26, 2.3, 1.2. Ctrl vs Fry RNAi p = 0.02 Student’s T-test.
-
Figure 2—source data 1
Microtubule sliding rates in S2 cells for Trc knockdown and rescue experiments.
Related to Figure 2A and B.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Microtubule sliding rates in S2 cells for control and Fry knockdown S2 cells.
Related to Figure 2C and D.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Sliding rates for control and Fry RNAi neurons.
Related to Figure 2E and F.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig2-data3-v2.xlsx
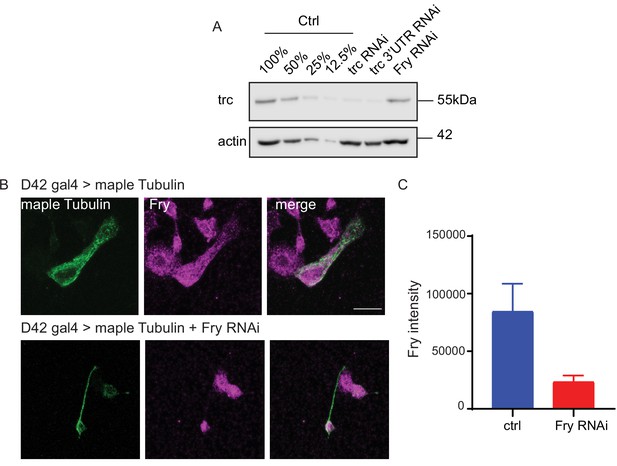
Confirming Fry knockdown.
(A) Western Blot from S2 lysate confirms Trc is knocked down upon treatment with dsRNA targeting a non-coding region. Trc levels remain unchanged upon treatment with Fry dsRNA. (B) Example images showing immunostaining against Fry in cultured Drosphila neurons expressing maple tubulin (control) or maple tubulin and Fry RNAi under control of D42 gal4. (C) Quantification of Fry signal confirms decrease in Fry protein level compared to control.
-
Figure 2—figure supplement 1—source data 1
Fry immunostaining intensities.
Related to Figure 2—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig2-figsupp1-data1-v2.xlsx
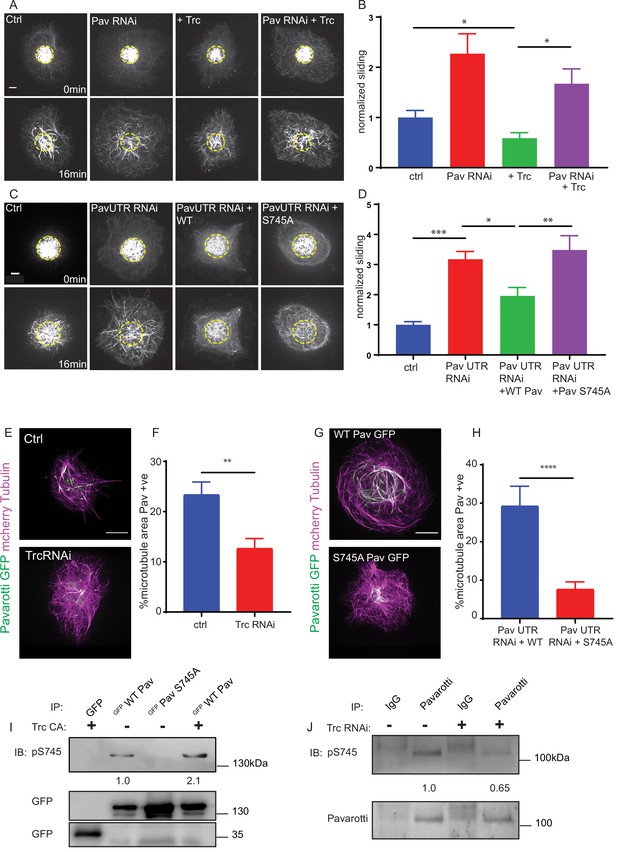
Trc regulates microtubule sliding via phosphorylation of Pavarotti.
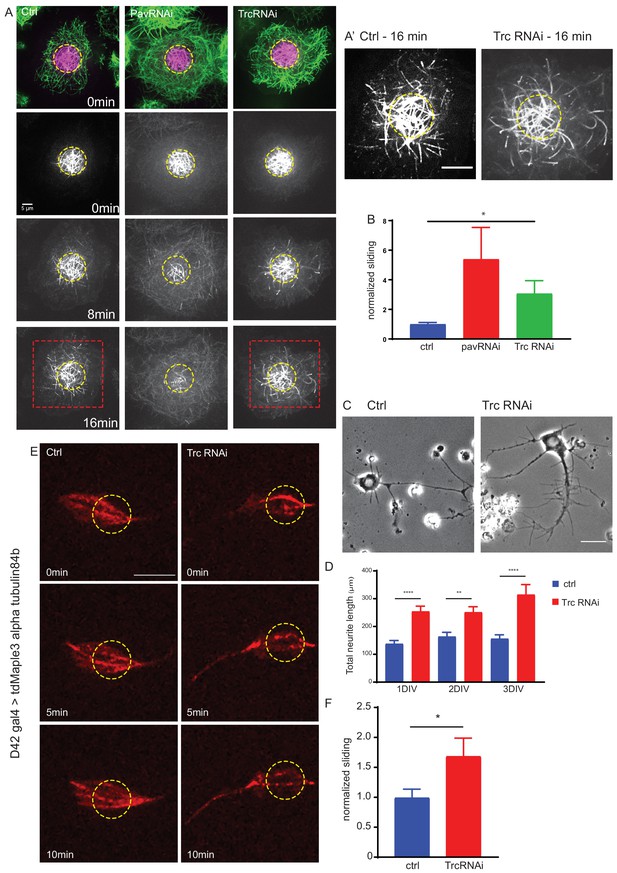
(A) Sliding experiments in S2 cells show a decrease in microtubule sliding with Trc overexpression. Trc overexpression in conjunction with depletion of Pavarotti increases sliding beyond control levels Scale bar 5 µm. (B) Quantification of sliding experiments in A. n = 27–41 cells per condition. Ctrl = 1.0 ± 0.11, Upper 95% CI = 1.2, Lower 95% CI = 0.8, Pav RNAi = 1.6 ± 0.25, 2.1, 1.0, Trc OE = 0.61 ± 0.0.8, 0.8, 0.44, Pav RNAi + Trc OE = 1.4 ± 0.22, 1.9, 0.9. Ctrl vs Trc OE p=0.04, Trc OE vs Pav RNAi + Trc OE p=0.003. One-way ANOVA with Sidak’s post-hoc test. (C) Sliding experiments in S2 cells show an increase in microtubule sliding with Pavarotti depletion. The effect can be rescued with WT Pavarotti but not with Phospho null mutant S745A. (D) Quantification of sliding experiments shown in C. n = 39–48 cells from 4 independent experiments. Ctrl = 1 ± 0.10 Upper 95% CI = 1.2, Lower 95% CI = 0.79, Pav RNAi = 3.16 ± 0.26, 2.65, 3.70, Pav RNAi + WT = 1.96 ± 0.28, 1.40, 2.52 Pav RNAi + Pav S745A = 3.49 ± 0.48, 2.53, 4.44. Ctrl vs Pav RNAi p=0.0001, Pav RNAi vs pav RNAi + WT p=0.04, Pav RNAi vs Pav RNAi + S745A p=0.94, Pav RNAi + WT vs Pav RNAi + S745A p=0.0025. One-way ANOVA with Sidak’s post hoc correction. (E) Example images of extracted S2 cells expressing mCherry Tubulin and WT Pavarotti GFP under control or Trc RNAi conditions. (F) Quantification of microtubule area colocalized with Pavarotti. n = 18–26 cells from three independent experiments. Ctrl = 23.5 ± 2.4%, Upper 95% CI = 28.6, Lower 95% CI = 18.3 Trc RNAi = 12.78 ± 1.9%, 16.62, 8.94. p=0.001 Student’s T-test Scale bar = 10 µm. (G) Example images of extracted S2 cells expressing mCherry Tubulin and WT Pavarotti GFP or Pavarotti S745A GFP. Endogenous Pavarotti was depleted with dsRNA targeting non-coding regions. (H) Quantification of microtubule area colocalized with Pavarotti. n = 12–17 cells from three independent experiments. Scale bar = 10 µm. WT = 29.4 ± 5.0%, Upper 95% CI = 40.4, Lower 95% CI = 18.4, S745A = 7.79 ± 1.8%, 11.5, 4.08. p=0.0001 Student’s T-test (I) Western blot of S745 phospho-Pavarotti from HEK cell lysates shows an increase in this species with Trc overexpression. (J) Western blot of immunoprecipitated Pavarotti prepared from dissected 3rd instar larvae brains shows a decrease in phospho Pavarotti at S745 upon depletion of Trc.
-
Figure 3—source data 1
Microtubule sliding rates for control, Pav knockdown and Trc overexpressing cells.
Related to Figure 3A and B.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Microtubule sliding rates for Pavarotti knockdown and rescue experiments.
Related to Figure 3C and D.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Pavarotti localization in control and Trc RNAi-treated cells.
Related to Figure 3E and F.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig3-data3-v2.xlsx
-
Figure 3—source data 4
Pavarotti WT and S745A localization.
Related to Figure 3G and H.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig3-data4-v2.xlsx
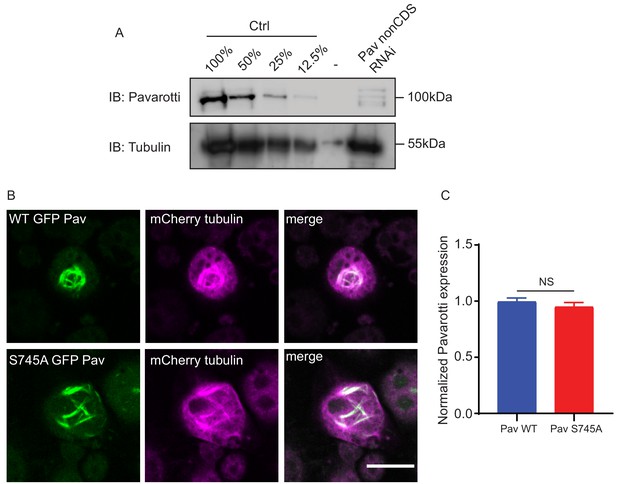
Pavarotti knockdown and rescue.
(A) Western blot of S2 cell lysate confirming Pavarotti knockdown with dsRNA targeting a non-coding region. (B) Immunofluorescence showing expression of WT or S745A GFP Pavarotti. (C) Quantification of Pavarotti GFP signal normalized to mCherry tubulin expression shows equal expression of WT and S745A Pavarotti.
-
Figure 3—figure supplement 1—source data 1
Pavarotti WT and S745A expression levels.
Related to B and C.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig3-figsupp1-data1-v2.xlsx
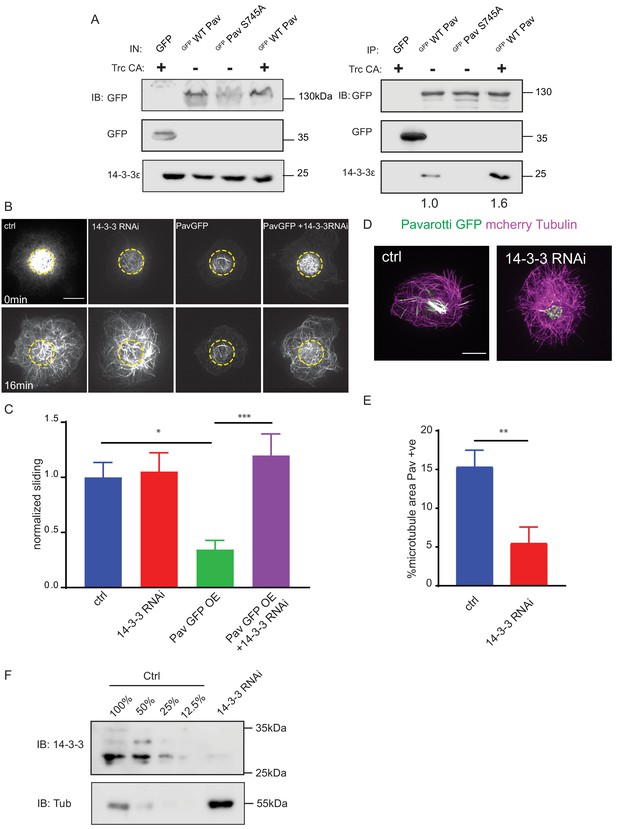
Phospho-Pavarotti brakes microtubule sliding via interaction with 14-3-3 proteins.
(A) Western blot from HEK cell lysate showing co immunoprecipitation of Pavarotti and 14-3-3. The interaction is lost upon mutation of S745 to Alanine and increased upon co-expression of the kinase Trc. (B) Sliding experiments in S2 cells show the ability of Pavarotti to brake microtubule sliding is dependent upon 14-3-3 proteins. Scale bar = 10µm. (C) Quantification of sliding experiments in B. n=21-26 cells from three independent experiments. Ctrl = 1 ± 0.14 Upper 95% CI = 1.28, Lower 95% CI = 0.72, PavOE = 0.35 ± 0.08, 0.52, 0.18 14-3-3 RNAi = 1.05 ± 0.17, 1.41, 0.70, 14-3-3RNAi + PavOE = 1.20 ± 0.20, 1.6, 0.8. ctrl vs pav OE p = 0.01, ctrl vs 1433RNAi p = 0.99, ctrl vs 1433 RNAi + PavOE p = 0.78, Pav OE vs 1433 RNAi p = 0.007, pav OE vs 1433 RNAi + Pav OE p = 0.0004, 1433 RNAi vs 1433 RNAi + Pav OE p = 0.91 One-way ANOVA with Tukey’s post-hoc correction. (D) Example images of extracted S2 cells expressing mCherry Tubulin and WT Pavarotti GFP. Depletion of 14-3-3s decreases microtubule area decorated with Pavarotti. Scale bar = 10µm (E). Quantification of microtubule area colocalized with Pavarotti. n = 22-26 cells from three independent experiments. Ctrl = 15.4 ± 2.1, Upper 95% CI = 19.72, Lower 95% CI = 11.07, 14-3-3 RNAi = 5.54 ± 2.0, 9.78, 1.30. p = 0.0017 Student’s T-test. (F) Western blot from S2 cell lysate demonstrating knockdown of 14-3-3.
-
Figure 4—source data 1
Sliding rates for 14-3-3 RNAi and Pavarotti overexpression experiments.
Related to Figure 4B and C.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Pavarotti localization with 14-3-3 RNAi.
Related to Figure 4D and E.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig4-data2-v2.xlsx
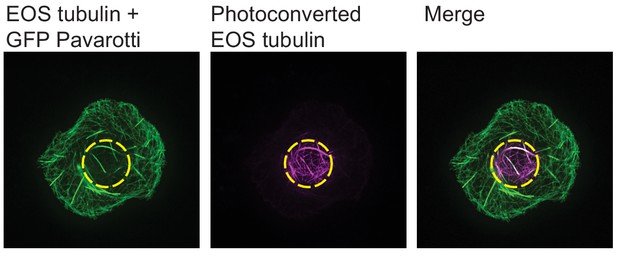
Confirming GFP Pavarotti expression.
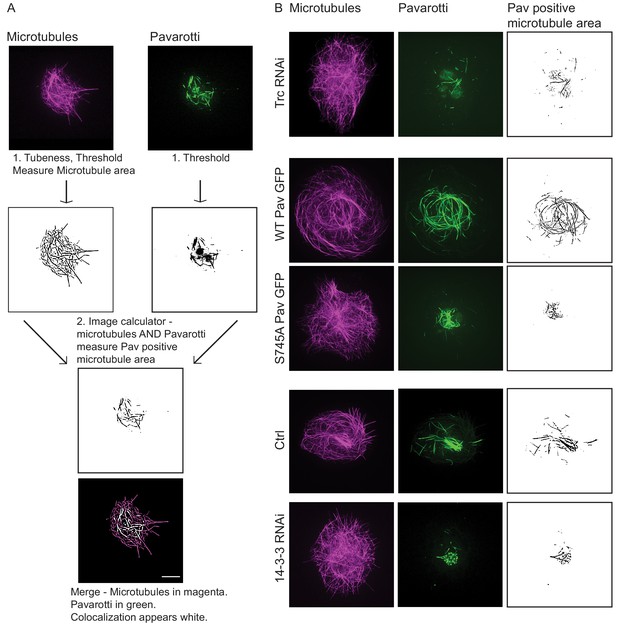
Expression of GFP Pavarotti can be detected after photoconversion of EOS tubulin. GFP Pavarotti does not convert upon exposure to UV light so is still visible in the 488 channel. See within photoconversion zone in dotted yellow circle. White in the merged image shows GFP Pavarotti colocalized with microtubules.
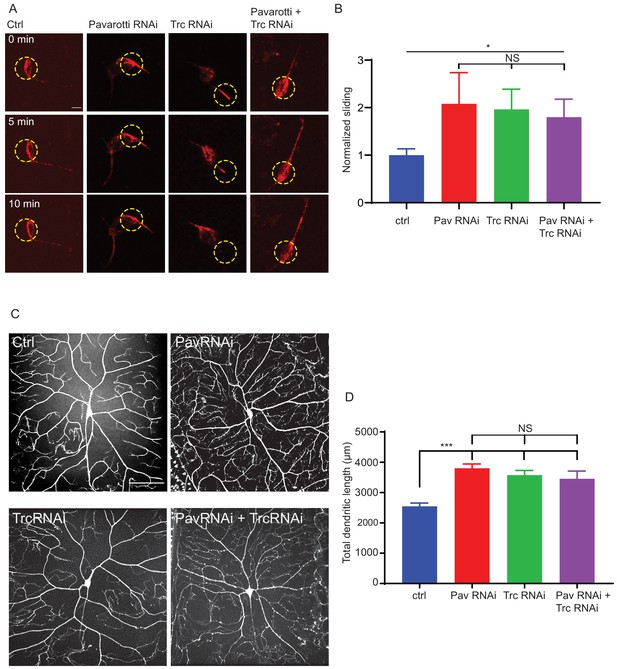
Pavarotti and Trc act in the same pathway to control dendrite outgrowth in vivo.
(A) Example images from timelapse imaging of photoconverted microtubules in neurons under control conditions or upon Pavarotti, Trc or Pavarotti and Trc depletion. Scale bar = 10 µm (B) Quantification of microtubule sliding rates. Ctrl = 1.0 ± 0.1, Upper 95% CI = 1.8, Lower 95% CI = 0.7, Pav RNAi = 2.1 ± 0.7, 3.5, 0.6, Trc RNAi = 2.0 ± 0.4, 2.8, 1.1, Pav and Trc RNAi = 1.8 ± 0.4, 2.6, 1.0. Ctrl vs Pav and Trc RNAi p = 0.014 Student’s T-test. (C) Example images showing DA neurons labeled with ppk::tdTomato in 3rd instar larvae under control conditions, with Pavarotti or Trc RNAi driven by elavGal4, or both RNAis together. Scale bar = 50 µm. n = 15–30 cells from at least 4 animals. Ctrl = 2549 ± 108 µm, Upper 95% CI = 2771, Lower 95% CI = 2328, Pav RNAi = 3803 ± 141 µm, 4099, 3507, Trc RNAi = 3577 ± 155 µm, 3904, 3250, Pav and Trc RNAi = 3455 ± 257 µm, 4005, 2905. (D) Pavarotti and Trc depletion cause an increase in dendritic length compared to control. Depletion of both proteins together has a non-additive effect compared with either RNAi. Ctrl vs Pav RNAi p<0.0001, Ctrl vs Trc RNAi, p<0.0001, Ctrl vs double RNAi p=0.0006, PavRNAi vs Trc RNAi p=0.76, Pav RNAi vs double RNAi p=0.48, Trc RNAi vs Double RNAi p=0.96. One-way ANOVA with Tukey’s post hoc correction.
-
Figure 5—source data 1
Sliding rates in neurons upon Trc, Pavarotti, or Trc and Pavarotti knockdown.
Related to Figure 5A and B.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Dendritic length of DA neurons.
Related to Figure 5C and D.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig5-data2-v2.xlsx
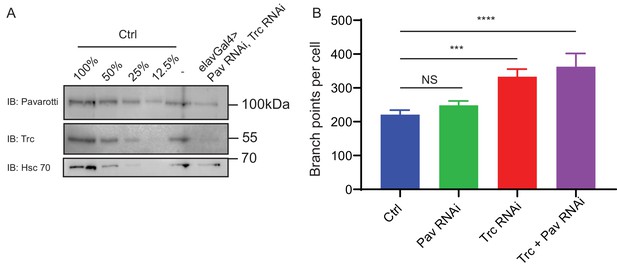
Analysis of branch points per cell in DA neurons.
(A) Western blot of Drososphila 3rd instar larvae brain lysate showing dual knockdown of Trc and Pavarotti driven by elav Gal4. (B) Quantification of number of branch points per DA neuron. Ctrl = 221 ± 13, Upper 95% CI = 248, Lower 95% CI = 194, Pav RNAi = 248 ± 13, 276, 221, Trc RNAi = 333 ± 22, 380, 287, Pav and Trc RNAi = 362 ± 39, 447, 278. Ctrl vs Pav RNAi, NS, Ctrl vs Trc RNAi p=0.0002, Ctrl vs Trc + Pav RNAi p<0.0001. One-way ANOVA with Dunnett’s post-hoc correction. Trc RNAi vs Trc + Pav RNAi NS p=0.5. Student’s T-test.
-
Figure 5—figure supplement 1—source data 1
Number of branch points of DA neurons.
Related to B.
- https://cdn.elifesciences.org/articles/52009/elife-52009-fig5-figsupp1-data1-v2.xlsx
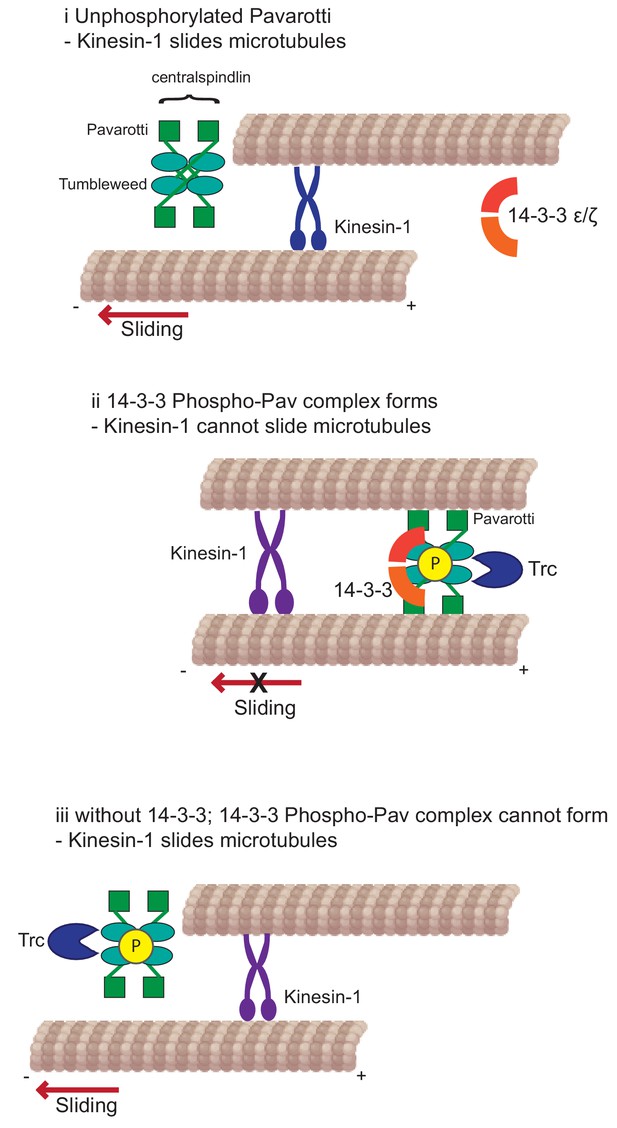
Kinesin-1 slides microtubules along one another to facilitate neurite outgrowth.
The kinase Trc inhibits this process by phosphorylation of Pavarotti – a part of the centralspindlin complex with tumbleweed. Phosphorylated Pavarotti forms a complex with 14-3-3 proteins and associates with microtubules. Under these conditions, microtubules are cross-linked and can no longer undergo sliding by kinesin-1. Therefore, neurite outgrowth is inhibited.
Videos
Pavarotti and Trc RNAi increase microtubule sliding in S2 cells.
Timelapse imaging of photoconverted microtubules in Drosophila S2 cells under control conditions or with depletion of Pavarotti or Trc. Microtubules were labeled with tdEOS alpha Tubulin 84b. 1 frame per minute.
Trc depletion increases microtubule sliding in neurons.
Timelapse imaging of photoconverted microtubules in Drosophila cultured neurons under control conditions or with depletion of Trc. Microtubules were labeled with UAS tdMaple alpha Tubulin 84b under control of a motor neuron specific D42 gal4 driver. Trc was depleted with UAS Trc RNAi under control of a motor neuron-specific D42 gal4 driver. 1 frame per minute. Scale bar 10 µm.
Microtubule sliding in S2 cells is dependent upon Trc kinase activity.
Timelapse imaging of photoconverted microtubules in Drosophila S2 cells under control conditions or with depletion of Trc using dsRNA against a non-coding region. Expression of WT Trc can rescue the effect of Trc RNAi. Expression of Kinase dead Trc is unable to rescue the effect of Trc RNAi. Microtubules were labeled with tdEOS alpha Tubulin 84b. 1 frame per minute.
Fry depletion increases microtubule sliding in S2 cells.
Timelapse imaging of photoconverted microtubules in Drosophila S2 cells under control conditions or with depletion of Furry (Fry). Microtubules were labeled with tdEOS alpha Tubulin 84b. 1 frame per minute.
Fry depletion increases microtubule sliding in neurons.
Timelapse imaging of photoconverted microtubules in Drosophila cultured neurons under control conditions or with depletion Fry. Microtubules were labeled with UAS tdMaple alpha Tubulin 84b under control of a motor neuron specific D42 gal4 driver. Fry was depleted with UAS Fry RNAi under control of a motor neuron specific D42 gal4 driver. 2 frames per minute. Scale bar 10 µm.
Trc requires Pavarotti to inhibit microtubule sliding in S2 cells.
Timelapse imaging of photoconverted microtubules in Drosophila S2 cells. Trc overexpression inhibits microtubule sliding only in the presence of Pavarotti. Microtubules were labeled with tdEOS alpha Tubulin 84b. 1 frame per minute.
Pavarotti phosphorylation at Ser745 is required to inhibit sliding.
Timelapse imaging of photoconverted microtubules in Drosophila S2 cells under control conditions or with depletion of Pavarotti using dsRNA against a non-coding region. Expression of WT Pav can rescue the effect of Pav RNAi. Expression of Phopsho-null Pavarotti is unable to rescue the effect of Pav RNAi. Microtubules were labeled with tdEOS alpha Tubulin 84b. 1 frame per minute.
14-3-3 proteins are required for Pavarotti to inhibit microtubule sliding.
Timelapse imaging of photoconverted microtubules in Drosophila S2 cells. Pavarotti can inhibit sliding only in the presence of 14-3-3 proteins. Microtubules were labeled with tdEOS alpha Tubulin 84b. 1 frame per minute.
Pavarotti and Trc control microtubule sliding together in neurons.
Timelapse imaging of photoconverted microtubules in Drosophila cultured neurons under control conditions or with depletion Fry. Microtubules were labeled with UAS tdMaple alpha Tubulin 84b under control of a motor-neuron-specific D42 gal4 driver. Trc was depleted with UAS Fry RNAi under control of a motor-neuron-specific D42 gal4 driver. 2 frames per minute. Scale bar 10 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | w; elav-Gal4 (III) | C. Doe University of Oregon | ||
| Genetic reagent (D. melanogaster) | yw; wg(Sp)/CyO;Dr(Mio)/TM3, Sb | E. Ferguson, University of Chicago | ||
| Genetic reagent (D. melanogaster) | yw; ppk-CD4-tdtomato (II) | Bloomington Drosophila Stock Center (BDSC) | Stock number 35844; FBst0035844; RRID:BDSC_35844 | Fly base genotype: w1118; P{ppk-CD4-tdTom}4a |
| Genetic reagent (D. melanogaster) | w; D42-Gal4 (III) | BDSC | Stock number 8816; FBst0008816; RRID:BDSC_8816 | Fly base genotype: w*; P{GawB}D42 |
| Genetic reagent (D. melanogaster) | y sc v; UAS-Trc-RNAi | BDSC | Stock number 41591; FBst0041591; RRID:BDSC_41591 | Fly base genotype y1 sc*v1 sev21; P{TRiP.GL01127}attP2 |
| Genetic reagent (D. melanogaster) | y sc v; UAS-Pav-RNAi | BDSC | Stock number 42573; FBst0042573; RRID:BDSC_42573 | Fly base genotype: y1 v1; P{TRiP.HMJ02232}attP40 |
| Genetic reagent (D. melanogaster) | y sc v; UAS-Fry-RNAi | BDSC | Stock number 60103; FBst0060103; RRID:BDSC_60103 | Fly Base Genotype: y1 sc*v1 sev21; P{TRiP.HMC05097}attP40 |
| Genetic reagent (D. melanogaster) | w; UASp-tdMaple3-alpha tubulin 84b | This lab | Described in Lu et al. (2016) Generated by injection of pUASp tdMaple3-alpha tubulin 84b. A second chromosome insertion was used in this study. | |
| Genetic reagent (D. melanogaster) | yw; UASp-tdMaple3-alpha tubulin 84b; UAS-Trc-RNAi | This lab | Generated using an insertion on the second chromosome | |
| Transfected construct (D. melanogaster) | pMT EOS tubulin | This lab | Described in Barlan et al. (2013). For S2 cell expression of EOS tubulin. | |
| Transfected construct (D. melanogaster) | pMT-BFP Trc WT | This lab | S2 cell expression of Trc. Generated from pUASt-Trc WT using EcoRI and NotI, a kind gift from P. Adler. | |
| Transfected construct (D. melanogaster) | pMT-BFP Trc CA (T253E) | This lab | S2 cell expression of Trc. Generated from pUASt-Trc T253E (a kind gift from P. Adler) and pMT vector using EcoRI and NotI. | |
| Transfected construct (D. melanogaster) | pMT-BFP Trc kinase dead (K122A) | This lab | S2 cell expression of Trc. Generated from pUASt-Trc K122A (a kind gift from P. Adler) and pMT vector using EcoRI and NotI. | |
| Transfected construct (D. melanogaster) | pMT-GFP Pavarotti | This lab | S2 cell expression. Generated from pMT-BFP Pavarotti (Del Castillo et al., 2015) | |
| Transfected construct (D. melanogaster) | pMT-GFP Pavarotti S745A | This lab | S2 cell expression. Generated by site directed mutagenesis of WT. | |
| transfected construct (D. melanogaster) | pEGFP Pavarotti WT | This lab | Mammalian expression. Pavarotti constructs were subcloned into pEGFP-C1 using EcoRI and SalI. | |
| Transfected construct (D. melanogaster) | pEGFP Pavarotti S745A | This lab | Mammalian expression. Pavarotti constructs were subcloned into pEGFP-C1 using EcoRI and SalI. | |
| transfected construct (D. melanogaster) | pcDNA Trc T253E | This lab | Mammalian expression. Trc T253E was subcloned into pcDNA 3.1+ using HindIII and NotI. | |
| Transfected construct (D. melanogaster) | pMT mCherry tubulin | This lab | For S2 cell expression. Described in del Castillo et al. (2015) | |
| Cell line (D. melanogaster) | S2 cells | DGRC | FlyBase Report: FBtc0000006 | Cell line maintained in this lab. |
| Cell line (Homo-sapiens) | HEK 293 FT | ATCC | RRID:CVCL_0045; ATCC: CRL-1573 | Cell line maintained in this lab. |
| Antibody | Single chain anti-GFP, GFP-Trap-M | Chromotek | ||
| Antibody | Anti Pavarotti (Rabbit polyclonal) | Scholey lab | Western blot 1:1000 | |
| Antibody | Anti Trc (Rabbit polyclonal) | Emoto lab | Western blot 1:1000 | |
| Antibody | Anti Tubulin (Rabbit polyclonal) | This lab | Affinity purified from immunized rabbit serum. Western blot 1:1000 | |
| Antibody | Anti phospho-Pavarotti S710 (Rabbit polyclonal) | Mishima lab | Western blot 1:1000 | |
| Antibody | Anti GFP (Rabbit polyclonal) | This lab | Affinity purified from immunized rabbit serum. Western blot 1:1000 | |
| Antibody | Anti Furry (Rabbit polyclonal) | Adler lab | Immunofluorescence 1:250 | |
| Antibody | Anti Hsc 70 (Goat polyclonal) | Santa cruz | K-19, RRID:AB_2120291 | Western blot 1:100 |
| Antibody | Anti 14-3-3ζ (Rabbit polyclonal) | Proteintech group | cat no. 14503–1-AP; RRID:AB_2218096 | Western blot 1:250 |
| Antibody | HRP conjugated Anti Rabbit | Jackson | Western blot 1:10,000 | |
| Antibody | HRP conjugated Anti Goat | Jackson | Western blot 1:10,000 |
Additional files
-
Source code 1
IJM macro used for sliding analysis.
- https://cdn.elifesciences.org/articles/52009/elife-52009-code1-v2.txt
-
Supplementary file 1
Primer sequences used for dsRNA generation.
- https://cdn.elifesciences.org/articles/52009/elife-52009-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52009/elife-52009-transrepform-v2.docx