Structural basis of transcription inhibition by the DNA mimic protein Ocr of bacteriophage T7
Figures
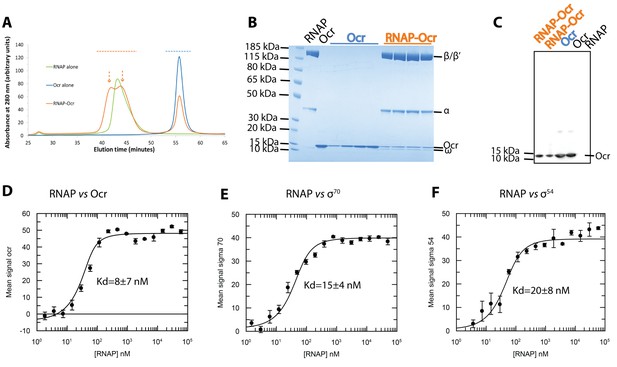
RNAP interacts with Ocr and can form a stable complex with Ocr.
(A) Gel filtration chromatography profiles of RNAP (green), Ocr (blue) and RNAP-Ocr complex (orange). Two peaks correspond to RNAP-Ocr complexes (B) SDS-PAGE of the corresponding fractions from (A) as indicated by coloured dashed lines in (A) and the purified RNAP and Ocr as comparison, the same below in (C). (C) Western blotting of samples from RNAP-Ocr gel filtration fractions to verify the presence of Ocr co-eluting with RNAP, here only Ocr is his-tagged. Two fractions under each peak (as coloured dash arrows shows) in gel-filtration is loaded. (D–F). MST experiments measure the dissociation constants (Kd) of Ocr, σ70 and σ54 with RNAP. These Kd values represent maximum values.
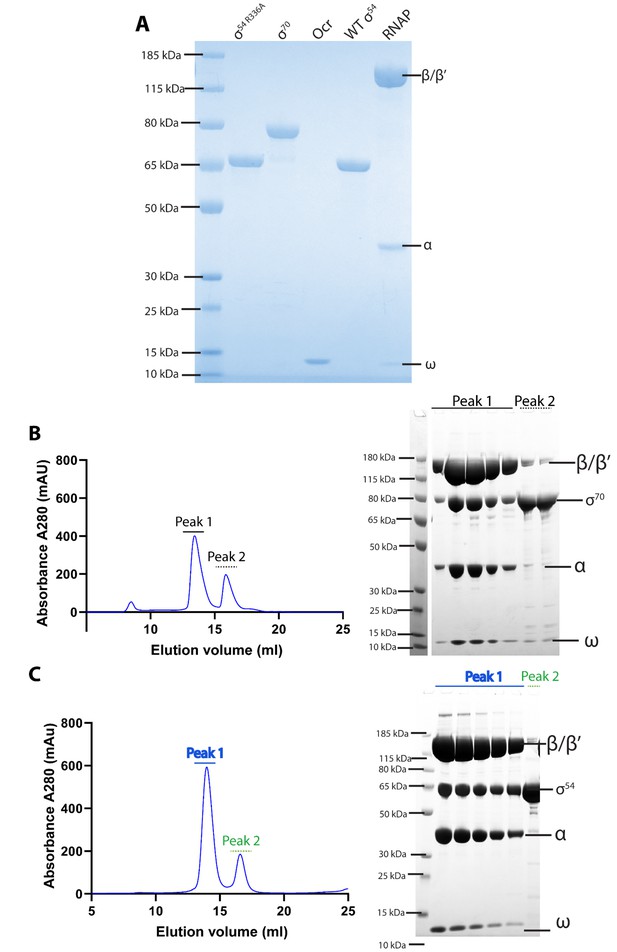
Protein purification and complex formation.
(A) Quality of proteins used in this study as judged by SDS-PAGE gels. (B–C) RNAP-σ70 and RNAP-σ54 holoenzymes are formed and purified by gel filtration chromatography and assessed by SDS-PAGE.
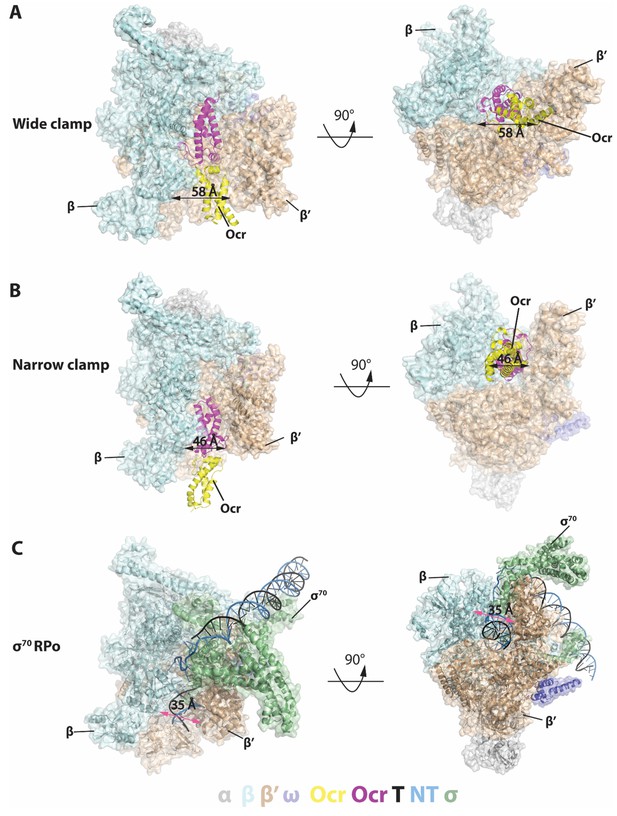
Structures of RNAP-Ocr in two different binding modes in two orthogonal views.
(A) ‘wide-clamp’ mode. (B) ‘narrow-clamp’ mode. RNAP and σ70 were shown as surface, Ocr shows as cartoon. The colour key is below the figure. α-grey, β-pale cyan, β’-wheat, ω-slate, σ70-palegreen, Ocr–magenta (proximal subunit) and yellow (distal subunit), non-template strand DNA(NT)-sky blue, template strand DNA (T)-black. (C) RNAP-σ70 open promoter complex (PDB code 6CA0).
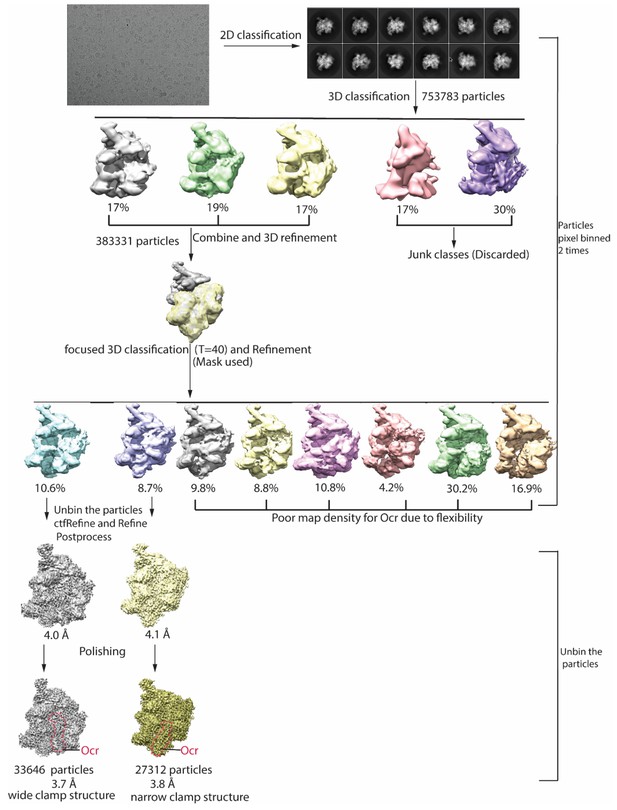
cryoEM data quality and Image processing flowchart.
Included are a typical micrograph, representative 2D classes and image processing flowchart, and the two distinct RNAP-Ocr complexes obtained.
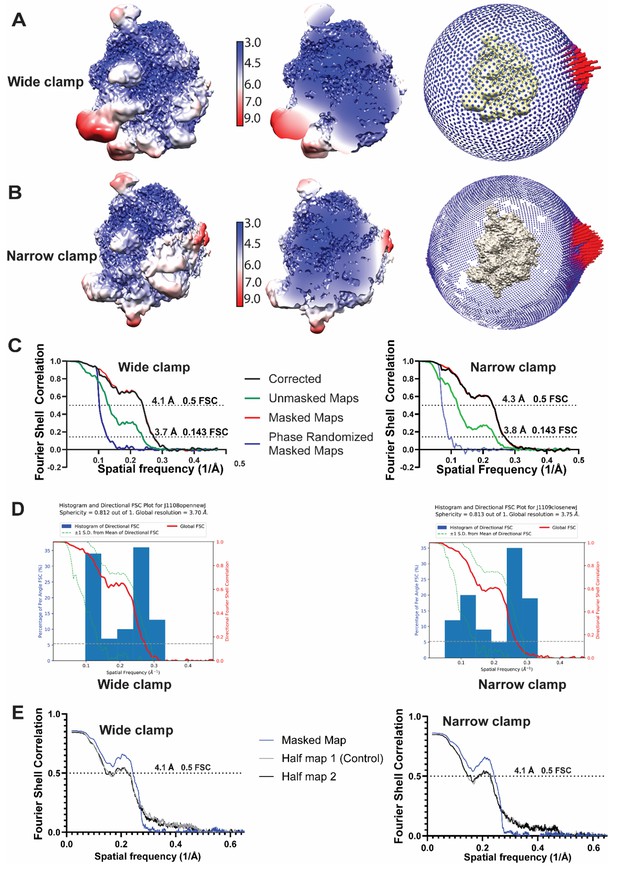
Quality of the two distinct RNAP-Ocr complex 3D reconstructions.
(A) ‘Wide-clamp’ (B) ‘narrow-clamp’. (A and B) Local resolution maps in surface view and cut-through views (separated by resolution indicator), and angular distribution of particles (the extreme right panel of (A) and (B)). (C) The corresponding FSC curves of wide-clamp and narrow-clamp structures including corrected, unmasked, masked and phase randomized masked maps). (D) Plots showing the spread of the directional resolutions (+/- 1 s.d., green dotted lines) from the global FSC (red line) and the histogram of directional resolutions (blue bars). Labels on the left Y-axis refer to directional FSC histograms. Labels on the right axis refer to global FSC. The grey dotted line indicates when FSC = 0.143. (E) FSC curves of model vs the two half maps. The model is only refined against half map 1.
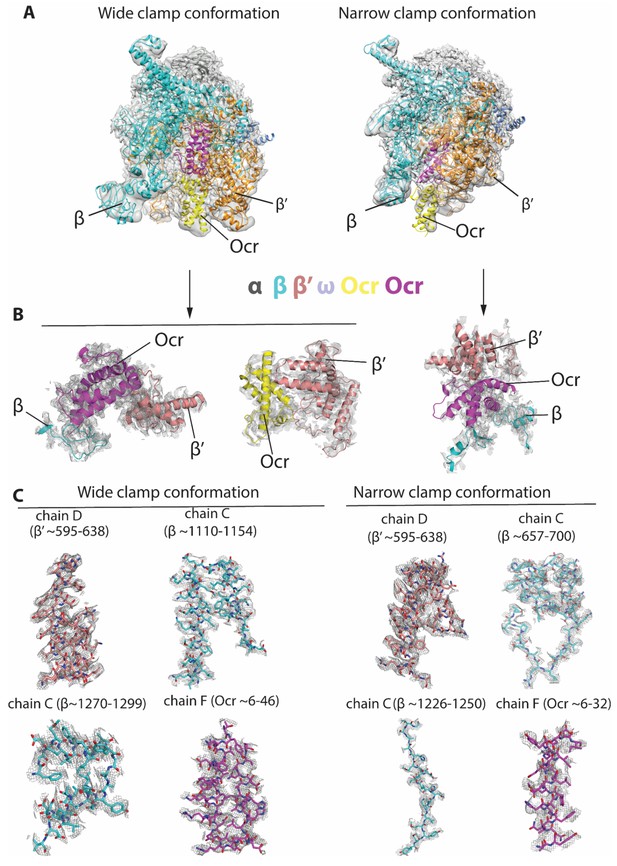
Representative electron density maps in the two reconstructions.
(A) Overall fit of the atomic structural models into the corresponding reconstructions. Maps are filtered to local resolutions. left panel - wide clamp conformation (global resolution at 3.7 Å), right panels - narrow clamp conformation (global resolution at 3.8 Å). (B) Views showing structural models of Ocr fitted into the electron density map and the surrounding RNAP. (C) Well defined regions showing clear density of RNAP and Ocr including side chains.
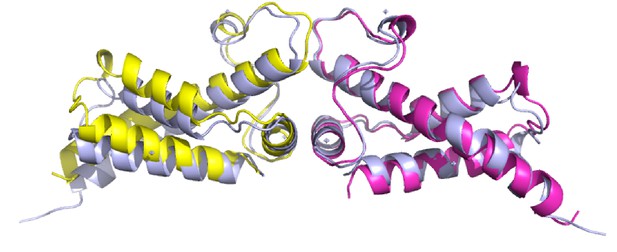
Structural comparison of Ocr from RNAP-Ocr complex (magenta and yellow) with that from crystal structure (blue, PDB code 1S7Z).
The structures are aligned on one Ocr monomer.
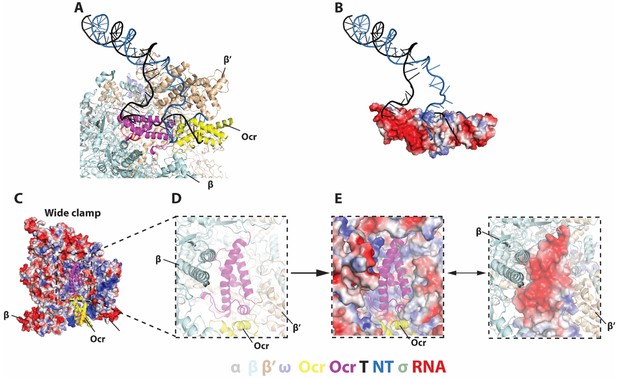
Detailed interactions between Ocr and RNAP and in the ‘wide-clamp’ conformation.
(A–B) Comparison with initial transcribing complex structure (PDB 4YLN). (C) Ocr resides in the positively charged RNAP channel. (D–E) Detailed charge distributions of RNAP and Ocr in the interacting regions. Blue – positive charge, red – negative charge.
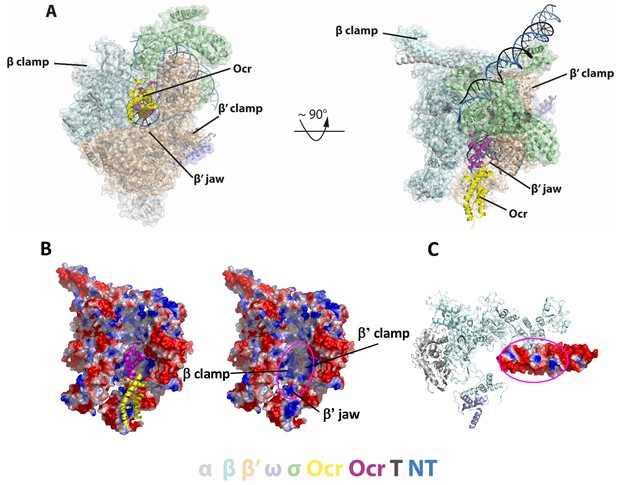
Detailed interactions between RNAP and Ocr in the ‘narrow-clamp’ conformation.
(A) Two orthogonal views of RNAP-Ocr narrow-clamp structure (RNAP shows as surface and Ocr shows as cartoon (magenta and yellow), overlaid with σ70 open complex structure (PDB code 6CA0). (B) Surface charge distribution of RNAP showing positively charged surfaces in the cleft where Ocr binds. Magenta ellipse – where Ocr binds. (C) Surface charge distribution of Ocr facing the bottom of the cleft including β’ jaw domain. Magenta ellipse – area of Ocr that is inside the cleft showing both negative and positively charged areas. Blue – positively charged, red – negatively charged.
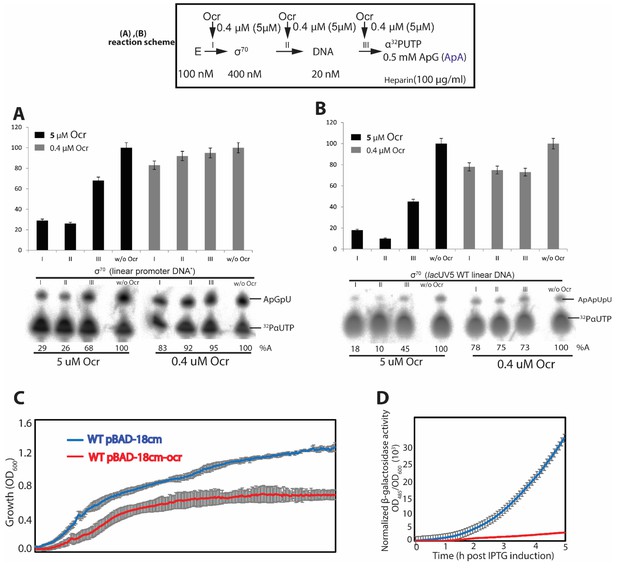
In vitro transcription and in vivo reporter assays.
(A-B) spRNA experiments on σ70 using two different promoter DNAs. Reaction schematics are shown with I, II, III representing experiments when Ocr is added during different stages of transcription initiation reaction. A control lane without Ocr is also shown. (C-D) using a reporter assay, we show that over-express of Ocr in E. coli cells inhibits cell growth (C) and β-galactosidase activities (D). At least three independent experiments were performed, each with 3–5 technical replicates.
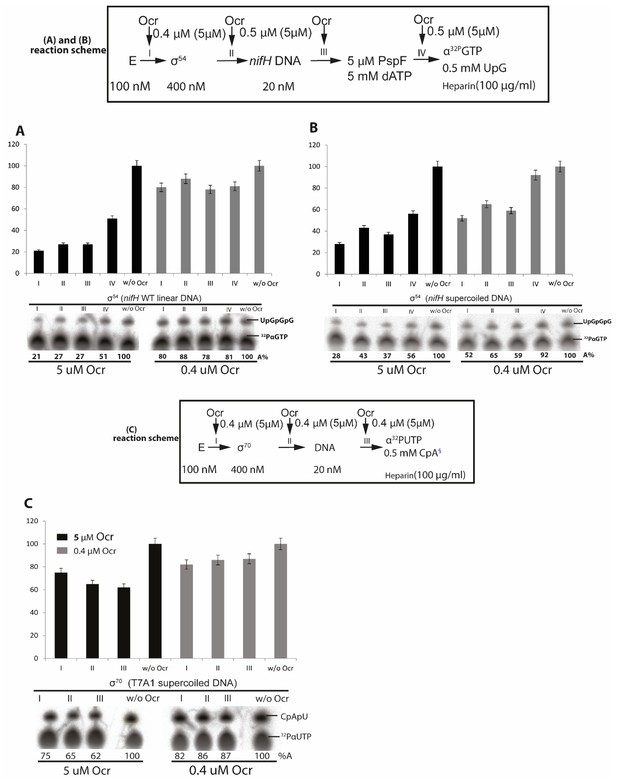
In vitro transcription assays of σ54-dependent systems using linear (A) and supercoiled DNA (B) and σ70 transcription using super-coiled DNA (C).
Reaction schematics are shown with I, II, III, IV representing experiments when Ocr is added during different stages of transcription initiation reaction. A control lane without Ocr is also shown. Data here show that Ocr can inhibit σ54 and σ70-dependent transcription during different stages of initiation and independent of promoters. At least three independent experiments were performed, each with 3–5 technical replicates.
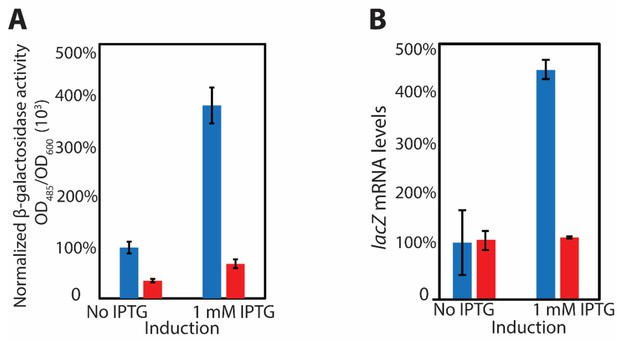
in vivo data showing Ocr can inhibit transcription.
(A) β-galactosidase activity and (B) lacZ mRNA levels at 15 hr post-induction in the presence and absence of Ocr. At least three independent experiments were performed, each with 3–5 technical replicates.
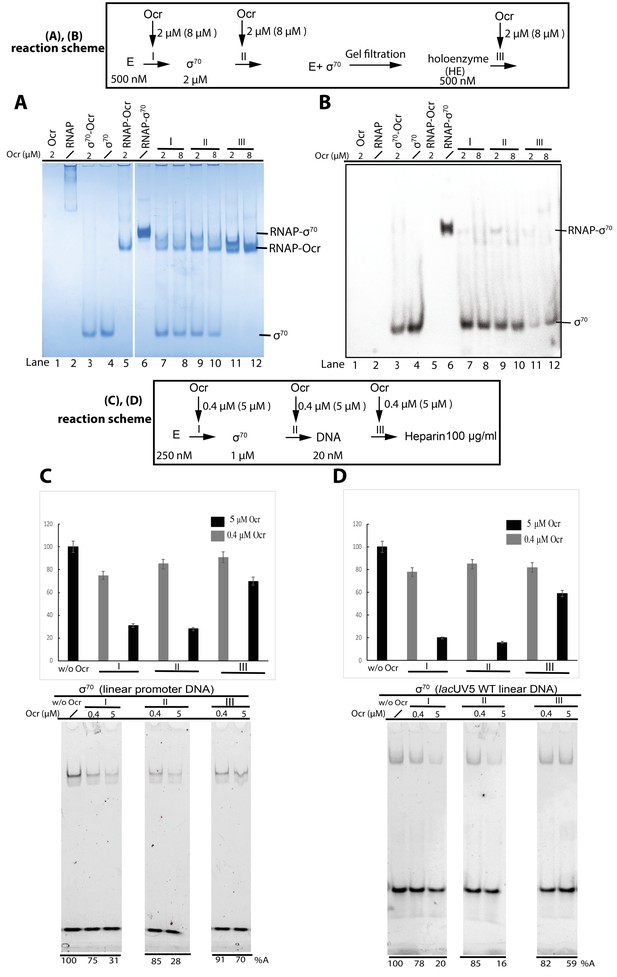
Ocr and its effect on σ70 holoenzyme and open complex formation.
(A and B) Native PAGE shows Ocr can inhibit and disrupt RNAP-σ70 holoenzyme formation. Coomassie blue staining (A) and Western blots against His-tagged σ70 (B). (A and B) reaction schemes are indicated above. E represents RNAP core enzyme; I, II, III indicate the point when Ocr was added during the reactions. In Reaction III, holoenzyme was preformed and purified by size exclusion chromatography first. Protein concentrations are shown in the reaction schemes. In lanes 7, 9, 11, a 1:1 molar ratio of Ocr to σ was used, whereas in lane 8, 10, 12, a 4:1 molar ratio of Ocr to σ was used. The exact Ocr concentrations are labelled above each lane. (C and D) σ70 open complex formation with different promoter DNA as assayed by native-PAGE gels and stained for DNA. Reaction schemes are indicated above. I, II, III indicate the point when Ocr was added during the reactions. The same experimental conditions were used as in vitro transcription assays in Figure 5, with either 0.4 µM or 5 µM final concentration Ocr added for each reaction, as indicated in each lane. Reaction in the absence of Ocr (w/o Ocr) was used as a control. All reactions include 100 µg/ml heparin to ensure only open complexes are captured.
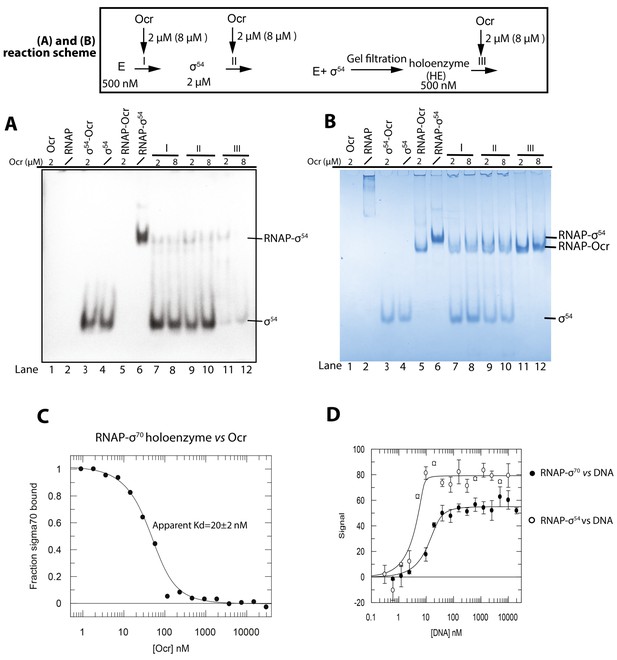
Ocr interferes with holoenzyme formation of σ54(A-B).
Native PAGE gels. (A) Western blots against His-tagged σ54 (B) Coomassie blue staining. Reaction schemes are indicated above. E represents core enzyme RNAP; I, II, III indicate the point when Ocr was added during the reactions. Proteins and DNA concentrations are shown. In lanes 7, 9, 11. a 1:1 molar ratio of Ocr to σ was used, whereas in lane 8, 10, 12, a 4:1 molar ratio of Ocr to σ was used. (C) Microscale thermophoresis (MST) experiment of Ocr binding to the RNAP-σ70 holoenzyme, supporting that Ocr competes with sigma in RNAP binding. (D) MST experiment of RNAP-σ70 and RNAP-σ54 binding to promoter DNA with high affinity. The bindings are too tight to give a reliable Kd.
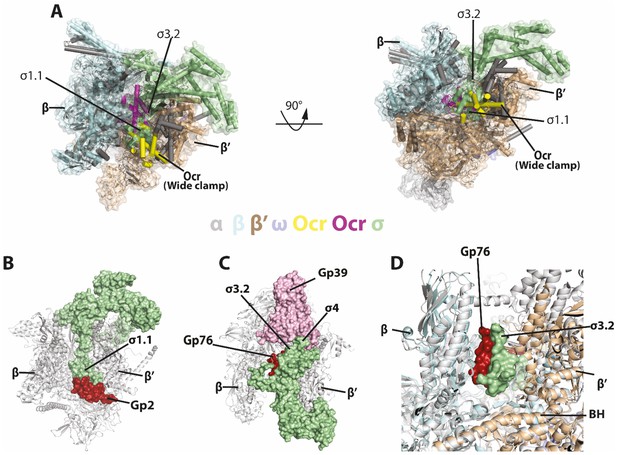
Comparisons with sigma and other phage proteins in binding to RNAP.
(A) Ocr overlay with RNAP-σ70 holoenzyme B) Complex structure of gp2 of T7 with σ70 holoenzyme (PDB 4LLG), (C) Complex structure of gp76-gp39 of P23-45 with σ70 holoenzyme (PDB 5XJ0), (D) Enlarged view of gp76 and σ70. RNAP shows as cartoon and coloured as grey, σ70 shows as surface and coloured pale green. Gp2, gp39, gp76 are show as surface and coloured as firebrick, light pink and firebrick, respectively.
Tables
Cryo-EM data collection and refinement statistics.
| RNAP-Ocr | ||
|---|---|---|
| Data collection and processing | ||
| Magnification | 47393 | |
| Total micrographs | 3543 | |
| Movie frames | 41 | |
| Pixel size (Å) | 1.055 | |
| Defocus range (µm) | −1.2 to −3 | |
| Voltage (kV) | 300 | |
| Electron dose (e-/Å−2) | 49.53 | |
| Total particles | 753783 | |
| FSC threshold | 0.143 | |
| Reconstruction (RELION) | Wide clamp | Narrow clamp |
| Particles | 33646 | 27312 |
| Resolution (Å) | 3.7 | 3.8 |
| Refinement | ||
| Resolution (Å) | 3.7 | 3.8 |
| R.m.s. deviations | ||
| Bond length (Å) | 0.003 | 0.003 |
| Bond angle (°) | 0.816 | 0.865 |
| Ramachandran plot | ||
| Favored regions (%) | 91.88 | 91.05 |
| Allowed regions (%) | 8.06 | 8.69 |
| Outlier | 0.06 | 0.27 |
| Validation | ||
| All-atom clashscore | 4.63 | 4.5 |
| Rotamer outliers (%) | 0.15 | 0.18 |
| C-beta deviations | 0 | 0 |
Additional files
-
Supplementary file 1
Pairwise comparisons of E. coli cells expressing Ocr (WT/pBAD18cm[ocr]) versus cells not expressing Ocr (WT/pBAD18cm).
- https://cdn.elifesciences.org/articles/52125/elife-52125-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52125/elife-52125-transrepform-v2.docx





