Hippocampal neural stem cells facilitate access from circulation via apical cytoplasmic processes
Figures
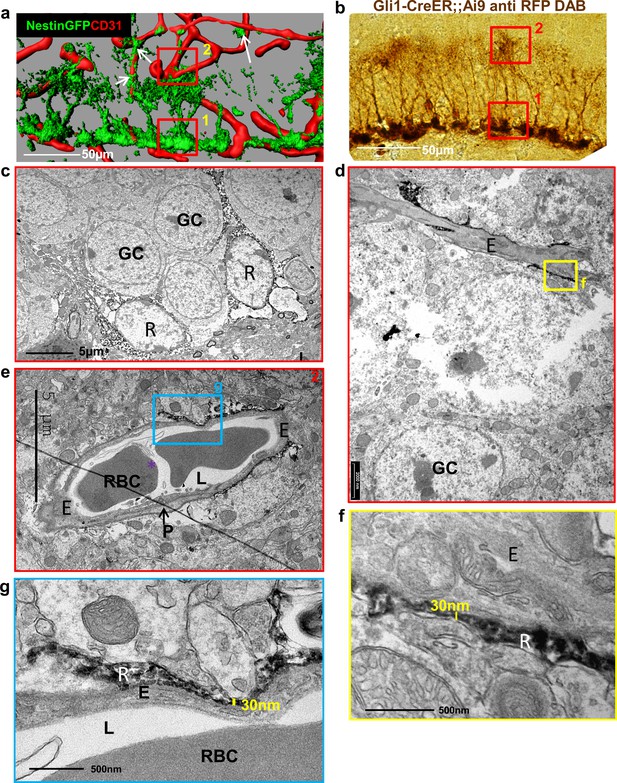
Fine apical processes of RGLs enfold capillary BVs in the inner molecular layer.
(a) 3D reconstruction of the DG of Nestin-GFP mouse co-stained with CD31 for blood vessels. In this line, both RGLs and pericytes (arrows) are GFP-labeled. Please note the contact points of RGLs and blood vessels at both the basal side at the SGZ (1) and the apical side at the inner molecular layer (2). (b) Gli1-CreERT2 crossed to Ai9 line (inducible TdTomato reporter) received tamoxifen three days before brain retrieval. Immunohistochemistry was done using anti-RFP antibody and DAB labeling. Note that only RGLs are labeled in the DG of this mouse line. (1) basal side, (2) apical side. Scale bar, 50 µm. (c) Transmission electron microscopy (TEM) images taken at the SGZ (basal side) showing the soma of two DAB-labeled RGLs (R). The cytoplasm of those cells is detected by DAB dark grains. GC-granule cells. (d and e) TEM images of the inner molecular layer (apical side) showing two representative examples of a capillary wrapped with DAB-labeled RGL processes. (f and g) Higher magnification of (d) and (e) showing that RGLs processes can reach up to 30 nm in thickness. R-RGL; GC-granule cell; E-endothelial cell, RBC-red blood cell; L-blood vessel lumen; P-pericyte.
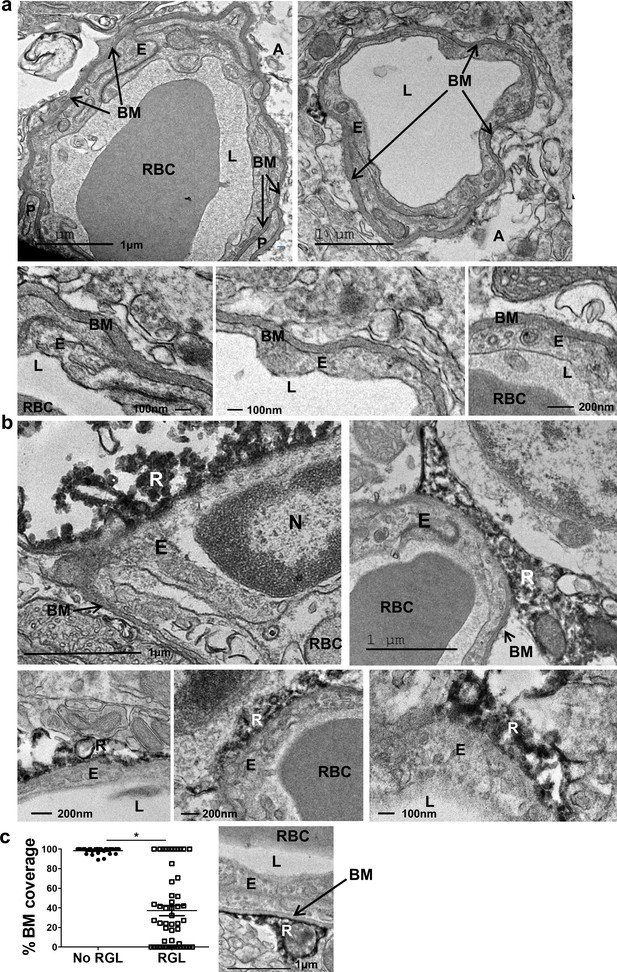
Absence of distinctive basement membrane at the contact points between RGLs and endothelial cells in the inner molecular layer.
TEM images of the same samples as in Figure 1. (a) Representative images of capillaries from the hilus area. Note BM covering the entire capillary abluminal membrane. Please note a double-layered BM at the pericyte (P) contact point and a clear BM at the contact point with an astrocyte (A). Bottom– higher magnifications. Scale bars: top - 1 µm, bottom- 100 nm. (b) Representative images of capillaries taken from the inner molecular layer that associate with DAB-labeled RGLs. Note no characteristic BM at the contact point of RGL (R) with the endothelial cell (E). (c) Quantification for the % of BM-covered endothelial abluminal surface with or without contact with RGL. N = 2 animals, 55 images. t(53)=11.51. p=2.37*10−5. Right: Image of BV-RGL contact with evident BM. R-RGL; E-endothelial cell; A-astrocyte; P-pericyte; RBC-red blood cell; L-blood vessel lumen; BM-basement membrane, N-endothelial nucleus.
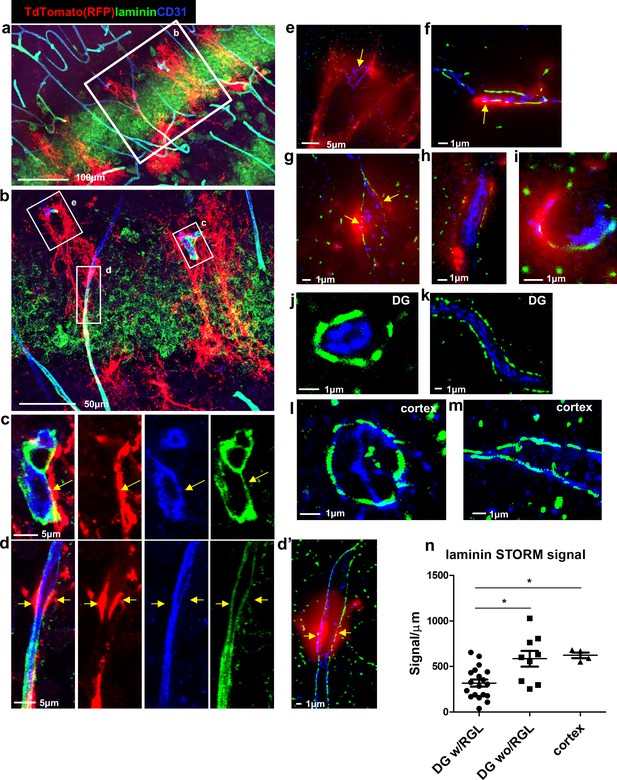
Reduced laminin expression at the BV-RGL contact point.
Brain slice of Gli1-CreERT2 mouse crossed to Ai9 reporter was immunostained with RFP, CD31 and pan-laminin antibodies. (a,b) Confocal image of the DG. (c,d) insets of b highlighting BV-RGL contact points. Arrows indicate areas of contact. (d’) Super-resolution image of d. The red (568 nm) channel was kept in epi-flaurescence mode to highlight areas of BV-RGL contact. (e–i) Representative super-resolution images of capillaries in contact with RGL (arrows). (J,k) Representative images of capillaries in the DG not accosicated with an RGL. (l,m) Cortical capillaries. (n,) Quantification of laminin signal (in dSTORM images) in blood vessels of the DG (with and without a contact with RGL) and in the cortex, normalized to the length of the blood vessel wall (in μm). F(2,29) = 8.221, p=0.001. Post hoc analysis: DG w/RGL vs. DG wo/RGL: p=0.005. DG w/RGL vs cortex: p=0.019.
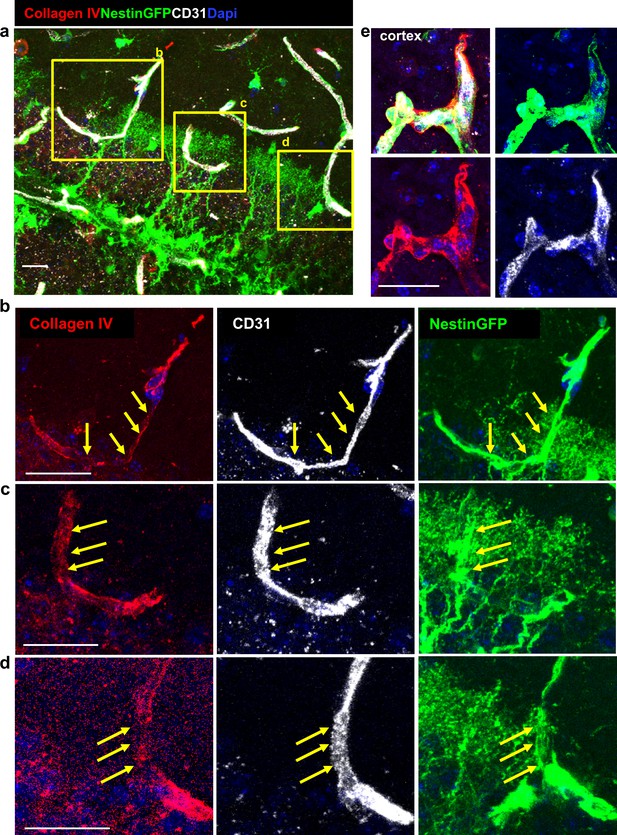
Reduced Collagen IV immunohistochemistry at the RGL-BV interface.
Brain sections taken from Nestin-GFP mouse crossed to Ai9 reporter, immunostained for CD31 and for the endothelial BM protein Collagen IV. (a) DG area (b–d) enlargement of the BV-RGL contact points. Note reduced Collagen IV staining at the contact points (arrows). (e) Blood vessels in the cortex. Scale bars, 20 µm.
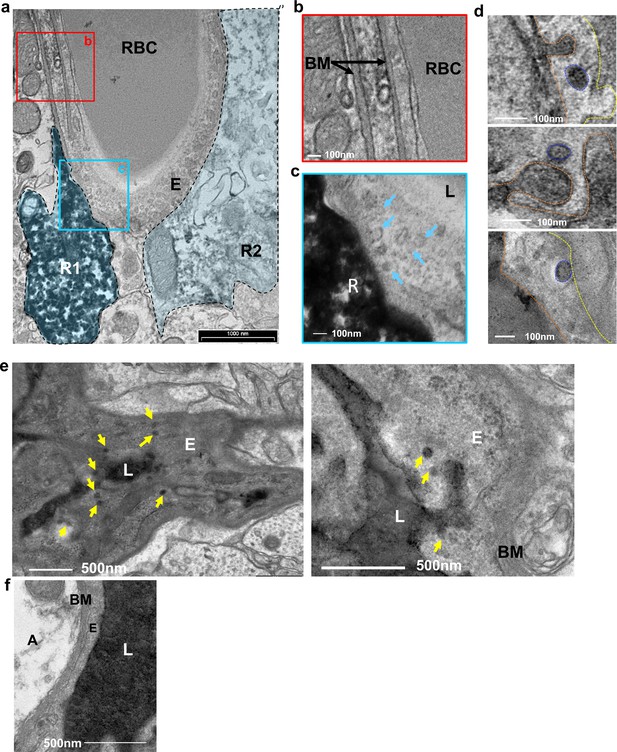
Vesicular activity at the BV-RGL interface.
(a–c) TEM image of a BV in the inner molecular layer which was stained with anti-RFP antibody as in Figure 1, to highlight RGLs (R1, R2). BV-RGL contacts were examined for the presence of vesicular-like structures (50–100 nm circular structures at the cytoplasm, luminal and abluminal surfaces). Arrows highlight abluminal membrane-connected and cytoplasmic endothelial vesicles. (d) HRP was injected i.v 30 min before brain disection. Representative examples for luminal, cytoplasmic and abluminal HRP-filled vesicles are shown, luminal aspect (dashed orange line), vesicle (blue line) and abluminal boundary (yellow line). Images were taken from endothelial cells at the inner molecular layer. (e) TEM of endothelial cells at the inner molecular layer, injected with systemic HRP. Arrows highlight multiple vesicles containing HRP. (f) Representative image of an endothelial cell aspect with a continuous BM and no vesicular activity. E-endothelial cell, R-RGL, L-BV lumen, RBC-red blood cell, BM-basement membrane, A- astrocyte.
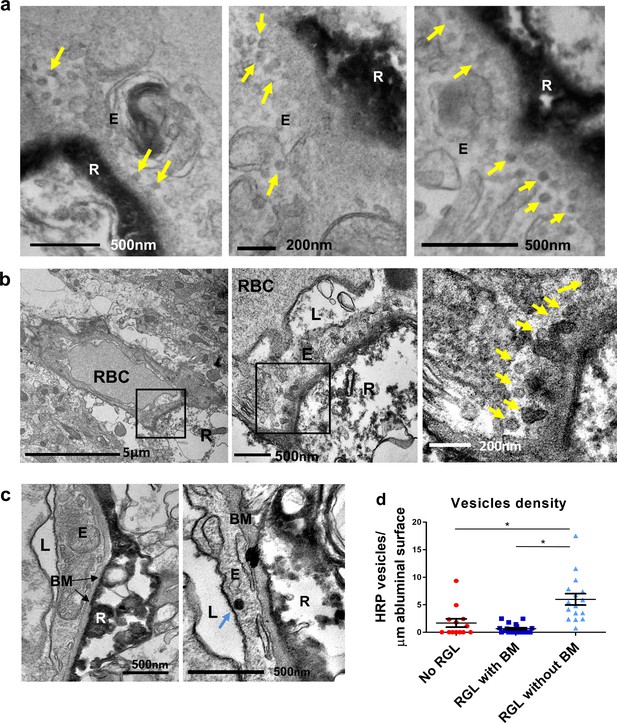
Vesicular activity in the BV-RGL contact point.
(a) TEM images from a Gli1-CreERT2 mouse crossed to Ai9 reporter and stained with anti RFP. Arrows highlight multiple endothelial vesicular structures at the contact points. (b–e) HRP was intravenously injected to a Gli1-CreERT2 mouse crossed to Ai9 reporter. Anti-RFP immunolabeling and DAB reaction for both the RGL and the luminal HRP content was done concomitantly. Due to the technical procedure, the majority of HRP was washed from the capillary lumen but was still found in vesicles and attached to endothelial luminal membrane. (b) Increased HRP-filled vesicles (arrows) at the contact point with an RGL (R) with modified\vague BM apparence. (c) Representative images of a vesicle-free endothelial segment (left) or a segment containing a single vesicle (right, arrow) both with full continuum of the BM. (d) Quantification of vesicle numbers per μm of abluminal aspect length in capillaries with no RGL, capillaries with RGL and BM and capillaries with direct contact with the RGL. F(2,46)=16.839 p=3.25*10−6. Post hoc analysis: No RGL vs. RGL without BM p=0.001. RGL with BM vs. RGL without BM p=3.19*10−6. E-endothelial cell, R-RGL, L-BV lumen, BM-basement membrane, RBC-red blood cell.
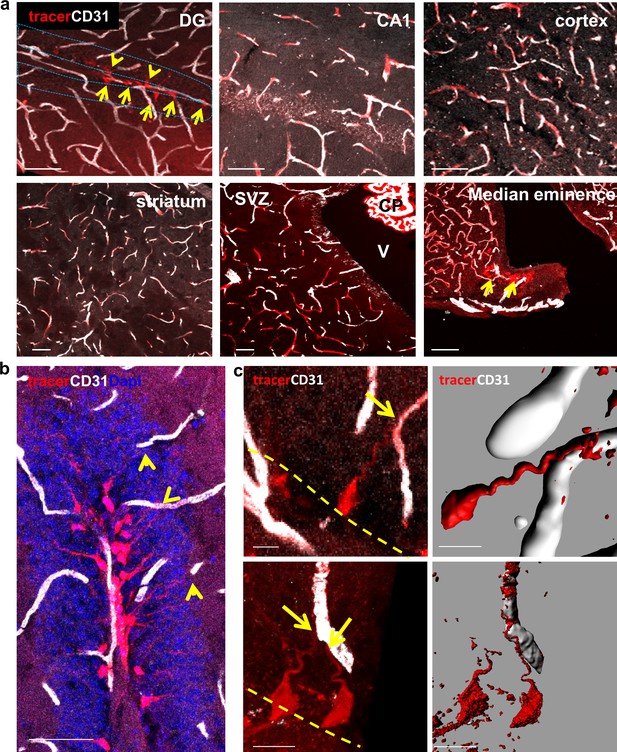
A unique population of cells in the SGZ is specifically labeled by systemically-injected 10Kd dextran tracer.
(a) 10Kd dextran TRITC-labeled tracer was injected to the left heart ventricle of an anesthetized mouse. One minute later the brain was retrieved and immediately fixed. Sections (co-stained with CD31) of the indicated brain areas are presented. Tracer did not infiltrate into cells outside of the vasculature except for a specific population of cells in the SGZ of the DG (arrows), the CP and the median eminence of the hypothalamus (arrow). Scale bars, 50 µm. V-cerebral ventricle. CP – choroid plexus. (b) Representative high magnification image of the DG area in TRITC- 10Kd dextran-injected animal co-immunostained with CD31. Labeled SGZ cells have a radial process that is associated with local blood vessels in the inner molecular layer (arrows). Scale bar, 50 µm. (c) Higher magnifications and 3D reconstruction of a dextran-labeled cell and a capillary illustrating mutual contact with no labeling of the extracellular space. The yellow dashed line represents the SGZ. Scale bar, 5 µm.
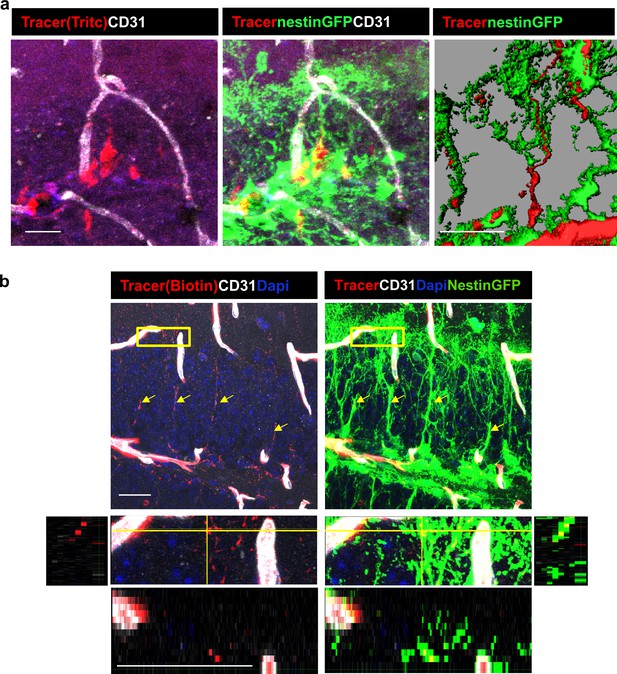
DG RGLs uptake a 10Kd systemically-injected dextran tracer.
(a) TRITC-labeled 10Kd tracer was injected into a Nestin-GFP mouse. 3D Imaris reconstruction (right) and confocal z-plane (left, middle) images showing accumulation of the tracer in the soma and major shaft of RGLs. Scale bar, 20 µM. (b) Biotin-labeled 10Kd tracer was injected as in (a). Sections were stained for CD31 and tracer was highlighted by cy3-labeled streptavidin. Note tracer distribution throughout all RGL processes and soma (arrows), although less strong than the fluorescent tracer. The inset demonstrates the localization of tracer within GFP+ RGL fine apical processes. Scale bar, 20 µM.
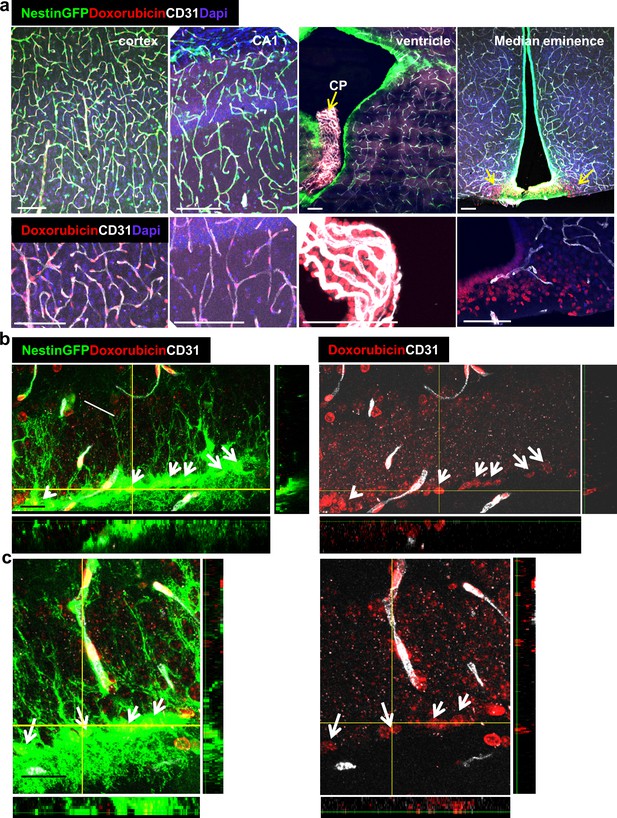
Doxorubicin has limited permeability through BBB capillaries but is being uptaken by RGLs.
Doxorubicin (200 µg) was injected into the left heart ventricle of Nestin-GFP mice. The brain was retrieved two minutes later. Doxorubicin is integrated into the DNA and detected by self red spectrum fluorescence. (a) Immunofluorescence images co-stained with CD31 showing integration of doxorubicin to the nuclei of various brain areas. In areas with intact BBB such as the cortex and the CA1, doxorubicin integrates solely to capillary nuclei. In areas with non-barriered endothelium such as the choroid plexus and the median eminence, doxorubicin is detected externally to blood vessels (arrows). Scale bar, 100 µM. (b and c) Two representative high-magnification images of the DG demonstrating the integration of doxorubicin to the nuclei of GFP+ RGLs. Z-plane projections are shown. Scale bar, 20 µM.
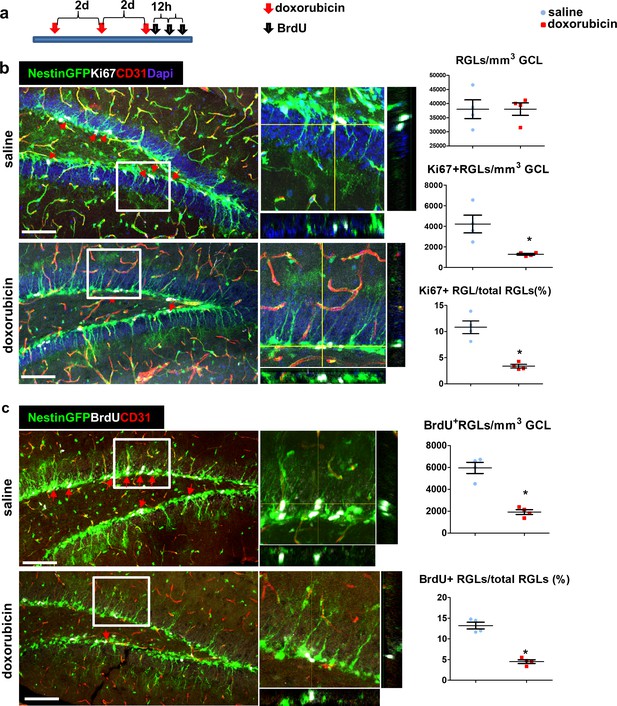
Doxorubicin treatment reduces the proliferation rates of Nestin-GFP+ RGLs.
(a) Experimental protocol. Doxorubicin (5 mg/kg) or saline were intraperitoneally (I.P) injected to Nestin-GFP mice every other day (3 injections). BrdU (50 mg/kg) was injected I.P. to all animals 3 times at 12 hr intervals before brain retrieval. N = 4 in each group. (b) Immunofluorescence images of nestin-GFP DG co-stained with Ki67 (a proliferation marker) and CD31. Arrows highlight Ki67+GFP+ cells that were verified to have a radial cytoplasmic process (see enlarged z-projection insets). Right: quantification of the total Nestin-GFP RGL population density (top, t(6)=-0.016 p=0.988), Ki67+ RGLs density (t(6)=3.398 p=0.015) and the percentage of Ki67+ RGLs among total RGLs (t(6)=5.923 p=0.001). (c) DG sections were co-stained with BrdU and CD31. Arrows highlight BrdU+GFP+ cells that were verified to have a radial cytoplasmic process (see enlarged insets). Right: quantification of BrdU+ RGLs density (t(6)=7.285 p=3.4*10−4) and the percentage of BrdU+ RGLs among total RGLs (t(6)=9.331 p=8.58*10−5). Scale bar for all images, 100 µM.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus C57Bl/6) | Nestin-GFP | Prof. Grigori Enikolopov, CSHL andStonybrook | ||
| Strain, strain background (Mus musculus C57Bl/6) | Gli1-CreERT2 | Jax mice | strain 007913 | |
| Strain, strain background (Mus musculus C57Bl/6) | Ai9 | Jax mice | strain 007909 | |
| Antibody | Mouse monoclonal anti BrdU | Serotec | PRID:AB_323427 | 1:400 |
| Antibody | Rat monoclonal anti-CD31 | Becton Dickinson | PRID:AB_393571 | 1:50 |
| Antibody | Rabbit polyclonal anti Laminin | Thermo Fischer Scientific | RRID:AB_60396 | 1:200 |
| Antibody | Rabbit polyclonal anti Collagen-IV | Abcam | PRID:AB_305584 | 1:200 |
| Antibody | Rabbit polyclonal Anti Ki67 | Thermo Fischer Scientific | PRID:AB_10979488 | 1:200 |
| Antibody | Rabbit polyclonal anti-GFP | Thermo Fischer Scientific | PRID:AB_221570 | 1:200 |
| Antibody | Rabbit polyclonal anti-RFP | Abcam | PRID:AB_945213 | 1:400 |
| Antibody | HRP Anti-Rabbit IgG | Vector | GZ-93951–40 | 1 drop |
| Antibody | Chick polyclonal anti RFP | EnCor | PRID:AB_2572308 | 1:300 |
| Antibody | Donkey anti-mouse Alexa647 | Jackson Immunoresearch | PRID:AB_234086 | 1:400 |
| Antibody | Donkey anti-rat Alexa647 | Jackson Immunoresearch | PRID:AB_2340694 | 1:400 |
| Antibody | Donkey anti- rabbit Alexa647 | Jackson Immunoresearch | PRID:AB_2492288 | 1:400 |
| Antibody | Donkey anti-rabbit Cy3 | Jackson Immunoresearch | PRID:AB_2307443 | 1:400 |
| Antibody | Donkey anti-rabbit Alexa488 | Jackson Immunoresearch | PRID:AB_2340619 | 1:400 |
| Antibody | Goat anti-hamster Alexa647 | Thermo Fischer Scientific | PRID:AB_2535868 | 1:400 |
| Antibody | Hamster polyclonal anti CD31 | Bio-Rad | PRID:AB_321653 | 1:50 |
| Commercial assay or kit | Streptavidin Alexa594 | Jackson Immunoresearch | PRID:AB_2337250 | 1:400 |
| Commercial assay or kit | Dab Plus | Abcam | ab103723 | |
| Chemical compound, drug | Florescent mounting medium | Thermo Fischer Scientific | TA-030-FM | |
| Chemical compound, drug | DAPI | Sigma | D9542 | |
| Chemical compound, drug | Tamoxifen | Sigma | T5648 | 8 mg/mouse PO |
| Chemical compound, drug | BrdU | Sigma | B5002 | 50 mg/Kg IP |
| Chemical compound, drug | Doxorubicin | Sigma | 44583 | 6 mg/kg IC 5 mg/kg IP |
| Chemical compound, drug | HRP | Sigma | P8250 | 10 mg/mouse IV |
| Chemical compound, drug | 10Kd Dextran, TRITC | Molecular Probes | D1817 | 6 mg/Kg IC |
| Chemical compound, drug | 10Kd Dextran, Biotin | Molecular Probes | D1956 | 6 mg/Kg IC |
| Chemical compound, drug | GLOX | Sigma | G2133 | 0.5 mg/ml |
| Chemical compound, drug | catalase | Sigma | C30 | 40 μg/ml |
| Chemical compound, drug | cysteamine MEA | Sigma | M9768 | 10 mM |
| Software, algorithm | ImageJ | NIH | ThunderSTORM plugin | |
| Software, algorithm | SPSS | IBM |





