Deciphering anomalous heterogeneous intracellular transport with neural networks
Figures
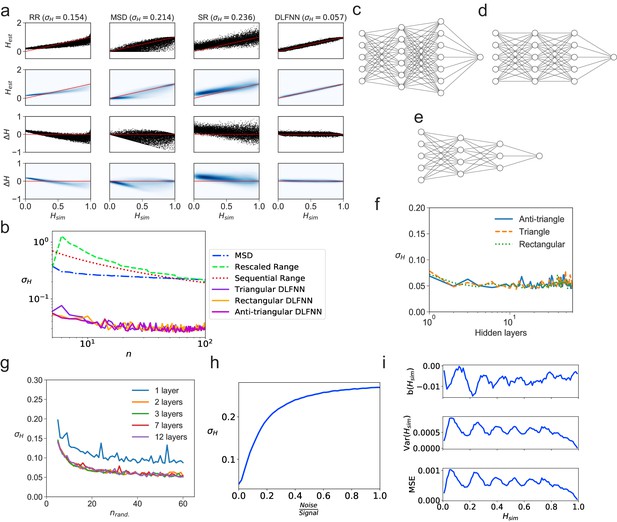
Tests of exponent estimation for the DLFNN using N = 104 simulated fBm trajectories.
(a) Plots showing the Hurst exponent estimates of fBm trajectories with data points by a triangular DLFNN with three hidden layers compared with conventional methods. Plots are vertically grouped by Hurst exponent estimation method: (left to right) rescaled range, MSD, sequential range and DLFNN. values are shown in the title. Top row: Scatter plots of estimated Hurst exponents and the true value of Hurst exponents from simulation . The red line shows perfect estimation. Second row: Due to the density of points, a Gaussian kernel density estimation was made of the plots in the top row (see Materials and methods). Third row: Scatter plots of the difference between the true value of Hurst exponents from simulation and estimated Hurst exponent . Last row: Gaussian kernel density estimation of the plots in the third row. (b) as a function of the number of consecutive fBm trajectory data points for different methods of exponent estimation. Example structures for two hidden layers and time series input points of the anti-triangular, rectangular and triangular DLFNN are shown in (c, d and e), respectively. (f) as a function of the number of hidden layers in the DLFNN for triangular, rectangular and anti-triangular structures. (g) as a function of the number of randomly sampled fBm trajectory data points with different number of hidden layers in the DLFNN shown in the legend. (h) as a function of the noise-to-signal ratio () (NSR) from Gaussian random numbers added to all data points in simulated fBm trajectories. (i) Plots of bias , variance and mean square error (MSE) as functions of . For each value of , fBm trajectories with points were simulated and estimated by a triangular DLFNN.
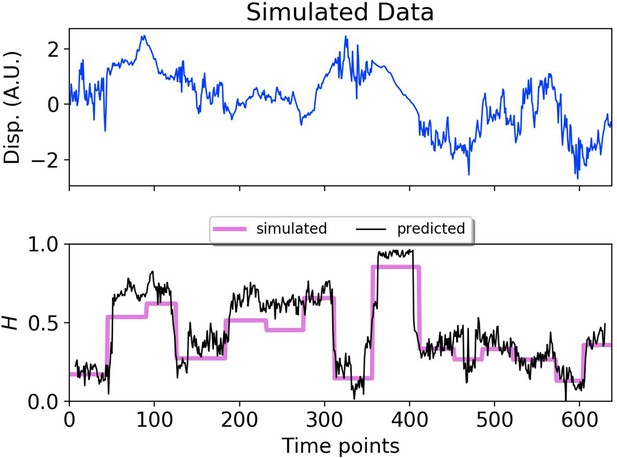
DLFNN analysis of a simulated trajectory.
Top: Plot of displacement as a function of time from a simulated fBm trajectory (blue) with multiple exponent values. Bottom: Hurst exponent values used for simulation (magenta), and the DLFNN exponent predictions of the neural network using a 15 point moving window (black).
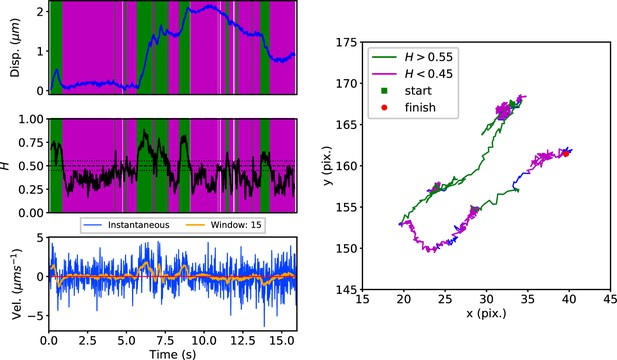
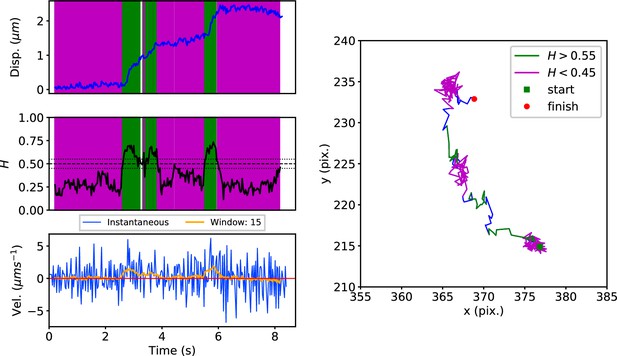
DLFNN analysis of a GFP-Rab5 endosome trajectory.
Top: Plot of displacement from a single trajectory in an MRC-5 cell (blue). Shaded areas show persistent (0.55 < H < 1 in green) and anti-persistent (0 < H < 0.45 in magenta) behaviour. Middle: A 15 point moving window DLFNN exponent estimate for the trajectory (black) with a line (dashed) marking diffusion H = 0.5 and two lines (dotted) marking confidence bounds for estimation marking H = 0.45 and 0.55. Bottom: Plot of instantaneous and moving (15 point) window velocity. Right: Plot of the trajectory with start and finish positions. Persistent (green) and anti-persistent (magenta) segments are shown. For sections that were 0.45 < H < 0.55 were not classified as persistent or anti-persistent and are depicted in blue.
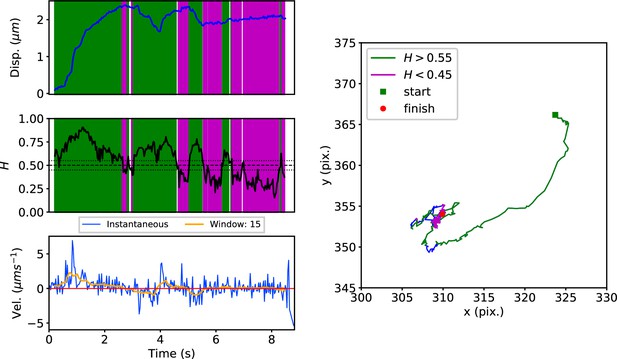
DLFNN analysis of a GFP-SNX1-labeled endosome trajectory, depicted as in Figure 3.
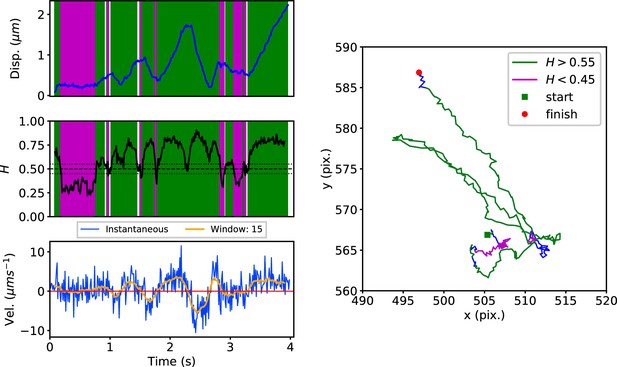
DLFNN analysis of a lysosome trajectory, depicted as in Figure 3.
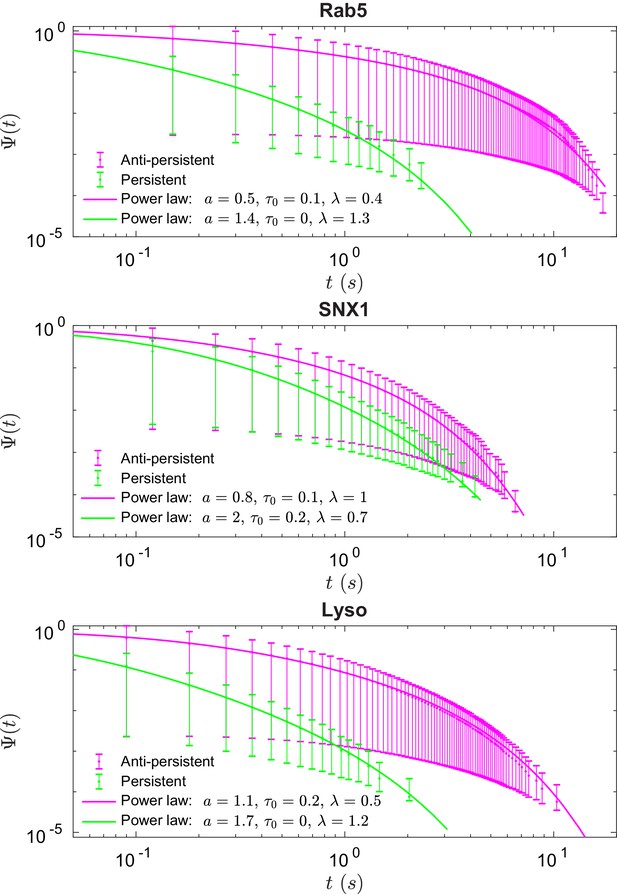
Survival functions plotted with error bars for persistent and anti-persistent segments for Rab5-positive endosomes, SNX1-positive endosomes and lysosomes with the power-law fits.
Fit parameters can be found in Appendix 3—table 1.
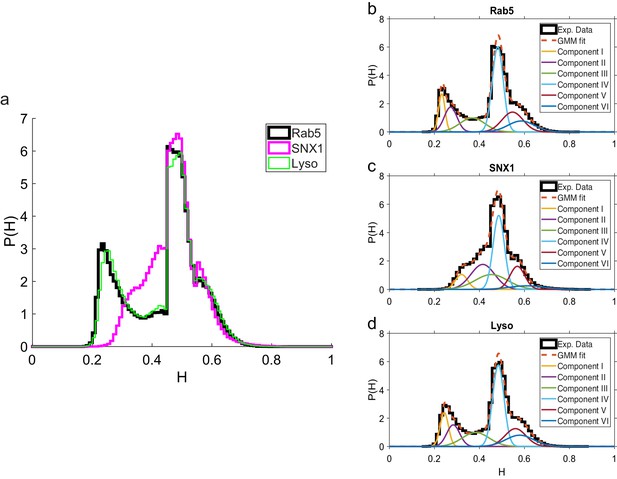
Comparison of Hurst exponent distributions for GFP-Rab5, GFP-SNX1 and lysosomes.
(a) Histograms of Hurst exponents for GFP-Rab5 (black), GFP-SNX1 (magenta) endosomes and lysosomes (green) plot on the same axes for comparison. The individual histograms of Hurst exponents (black solid) for GFP-Rab5-tagged endosomes, GFP-SNX1-tagged endosomes and lysosomes are shown in (b, c and d) respectively. For each histogram, the Gaussian mixture model fit for six components (red dashed) and individual Gaussian distribution components are shown on the same plot. The number of components were chosen through the Bayes information criterion shown in Appendix 4—figure 1.

MRC-5 cells stably expressing GFP-Rab5, GFP-SNX1 or stained with Lysobrite with tracking data overlaid.
The colours show the value of estimated by the neural network using a 15 point window. The scalebar is 10 µm.
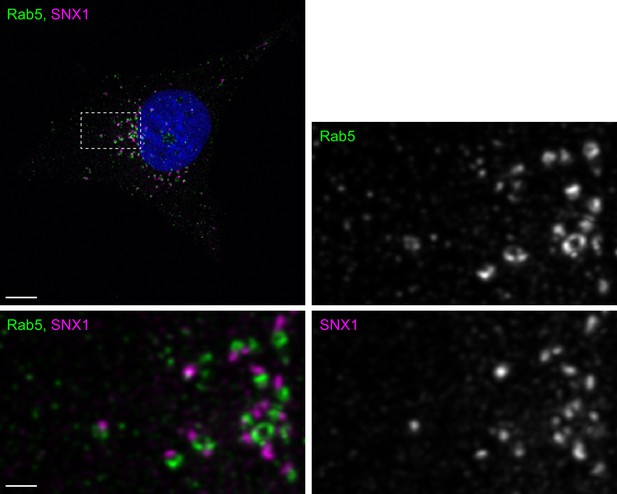
Distribution of endogenous Rab5 and SNX1.
MRC-5 cells were fixed and labeled with antibodies to Rab5 and SNX1, then imaged by confocal microscopy. A maximum-intensity z-projection of deconvolved images is shown. The boxed region is enlarged and presented as grey-scale single channels and a two color merged image. The scale bar is 10 µm (main image) and 2 µm (enlargements).
Video of MRC-5 cell stably expressing GFP-Rab5.
The field of view is 62.88 µm by 62.88 µm (600 × 600 pixels) and playback is in real time.
Video of MRC-5 cell stably expressing GFP-SNX1.
The field of view is 62.88 µm by 62.88 µm (600 × 600 pixels) and playback is in real time.
Video of MRC-5 cell stained with Lysobrite.
The field of view is 62.88 µm by 62.88 µm (600 × 600 pixels) and playback is in real time.
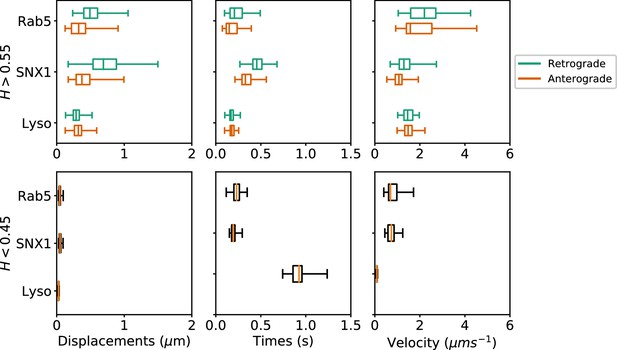
Box and whisker plots of displacements, times and velocities of persistent retrograde, persistent anterograde and anti-persistent segments in experimental trajectories.
Any segment with was classed as persistent and as anti-persistent. These H values were chosen as a precaution against the mean error of the neural network estimation. Each data point within the box and whisker plots are averages of all trajectory segments in a single cell. A total of 65 MRC-5 cells for GFP-Rab5-tagged endosomes, 63 MRC-5 cells for SNX1-GFP-tagged endosomes and 71 MRC-5 cells for lysosomes were analysed with at least 5 to 500 (average 54) anterograde or retrograde segments for each cell.
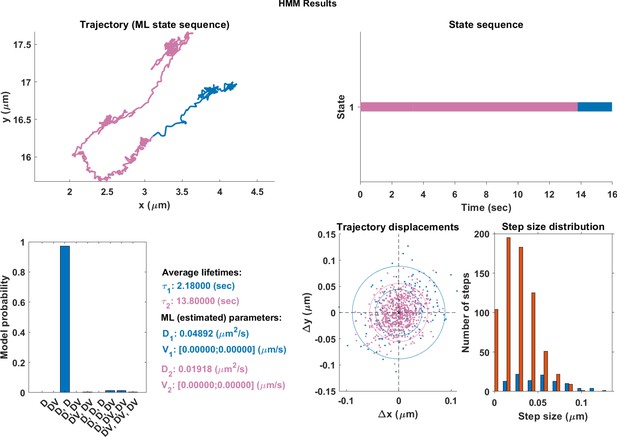
The same single trajectory as in Figure 3 but processed using the HMM-Bayes package described in Monnier et al. (2015).
The plot shows: the original trajectory (top left); the inferred state sequence of the most likely model (top right); the model probabilities given a maximum of three possible states (bottom left); the average lifetime of each state and estimated parameters of the most likely model (bottom center left), which in this case is two different diffusive states; individual increment displacements (bottom center right); and the step size distribution of those increments classed into the two different states.
Top: Plot of displacement from a single GFP-SNX1 endosome trajectory in an MRC-5 cell (blue).
Shaded areas show persistent ( in green) and anti-persistent ( in magenta) behaviour. Middle: A 15 point moving window DLFNN exponent estimate for the trajectory (black) with a line (dashed) marking diffusion and lines (dotted) marking the confidence bounds and 0.45. Bottom: Plot of instantaneous and moving (15 point) window velocity. Right: Plot of the trajectory of a GFP-SNX1 endosome in an MRC-5 cell with start and finish positions, and persistent (green) and anti-persistent (magenta) segments indicated.
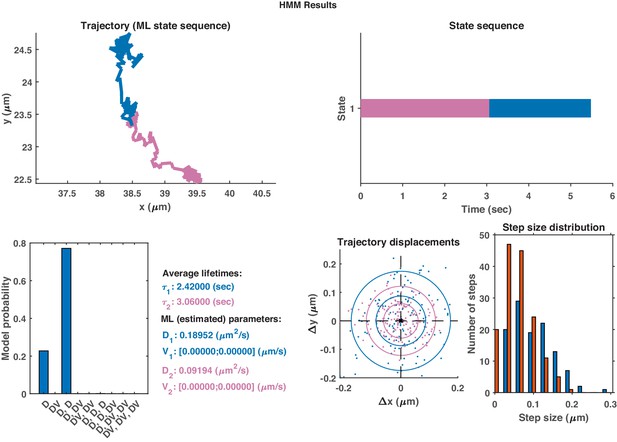
The same HMM-Bayes analysis as shown in Appendix 1—figure 1 applied to the trajectory in Appendix 1—figure 2.
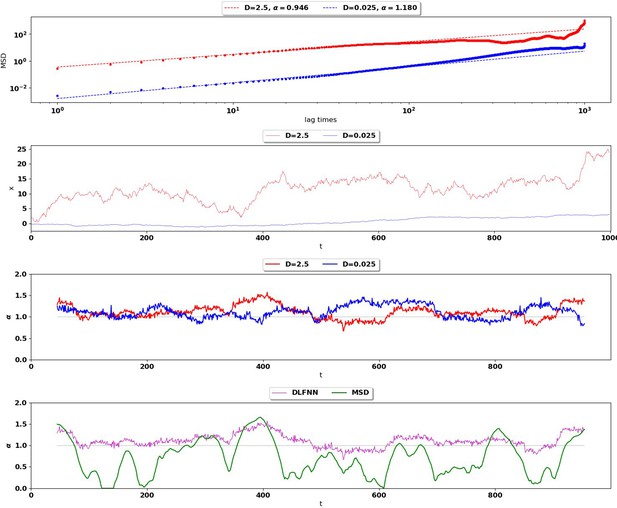
Top: MSD (points) and power-law fits (dashed) for two different Brownian trajectories containing 1000 data points with diffusion coefficient 2.5 (red) and 0.025 (blue).
The Hurst exponent should be for both trajectories. Second Row: Simulation of the two Brownian trajectories with diffusion co-efficient 2.5 (red) and 0.025 (blue). Third Row: Local Hurst exponent estimates given by the DLFNN for the two different trajectories using a 90 point window. The averages of DLFNN Hurst exponent estimates are (red) and (blue). Bottom: Local Hurst exponent estimates of the track given by DLFNN and MSDs using a 90 point window. The average of DLFNN Hurst exponent estimates is and the average of MSD estimates is .
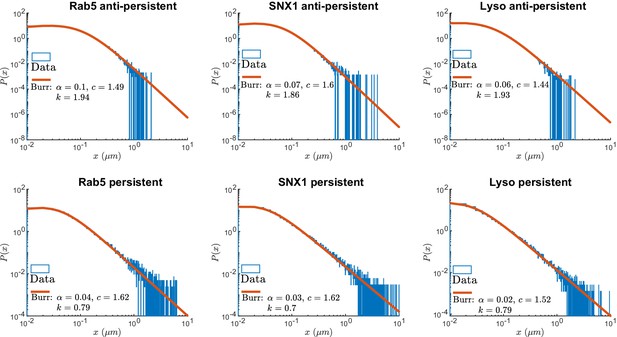
Normalized histograms (blue) and corresponding maximum likelihood estimation for Burr distributions (line) of segment displacements from lysosome and endosome experimental trajectories segmented using DLFNN.
Parameter estimates are shown the legend.
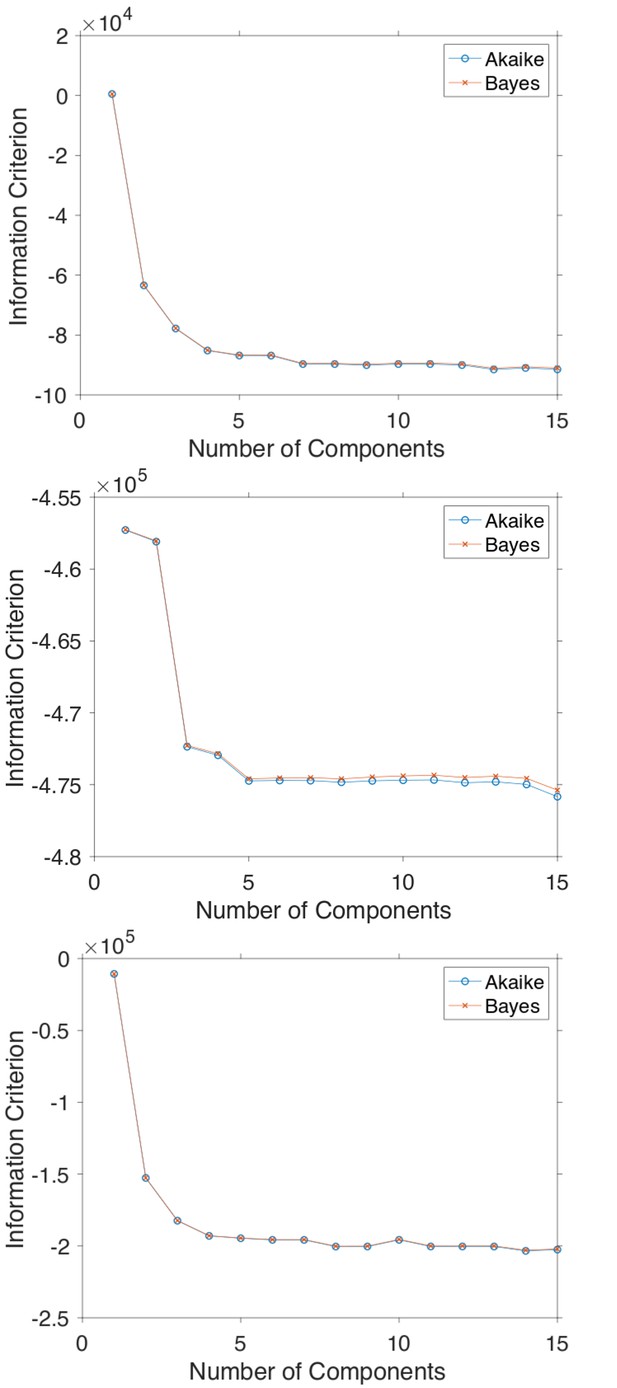
The Akaike and Bayes information criterion against number of components in the Gaussian mixture model shown in Figure 5 for GFP-Rab5 tagged endosomes (top), SNX1-GFP tagged endosomes (middle) and lysosomes (bottom).
Tables
Statistics of experimental trajectory segments.
The persistent and anti-persistent segments in this table are: from trajectories that travelled over 0.5 µm at any point from their initial starting positions; contained more points than the window size; and switched behavior more than twice in the trajectory. Note that these conditions are much stricter than those to generate Figures 4 and 5. Each persistent segment was then further subdivided into retrograde and anterograde segments (see Materials and methods).
| Rab5 | SNX1 | Lyso | ||
|---|---|---|---|---|
| Number of persistent segments | 2369 | 2099 | 7645 | |
| Number of anti-persistent segments | 6983 | 3947 | 19,320 | |
| Number of retrograde segments | 2925 | 2343 | 5882 | |
| Number of anterograde segments | 2303 | 1609 | 6827 | |
| Anti-persistent displacement (µm) | Mean | 0.05 | 0.05 | 0.03 |
| Median | 0.04 | 0.05 | 0.03 | |
| St. Dev | 0.02 | 0.01 | 0.004 | |
| Anti-persistent speed (µms-1) | Mean | 0.82 | 0.75 | 0.10 |
| Median | 0.70 | 0.73 | 0.09 | |
| St. Dev | 0.31 | 0.19 | 0.02 | |
| Anti-persistent time (s) | Mean | 0.23 | 0.20 | 0.93 |
| Median | 0.23 | 0.19 | 0.92 | |
| St. Dev | 0.05 | 0.03 | 0.11 | |
| Retrograde displacement (µm) | Mean | 0.53 | 0.74 | 0.29 |
| Median | 0.49 | 0.69 | 0.29 | |
| St. Dev | 0.19 | 0.28 | 0.08 | |
| Retrograde speed (µms-1) | Mean | 2.29 | 1.35 | 1.49 |
| Median | 2.21 | 1.29 | 1.46 | |
| St. Dev | 0.87 | 0.39 | 0.25 | |
| Retrograde time (s) | Mean | 0.22 | 0.46 | 0.17 |
| Median | 0.21 | 0.45 | 0.17 | |
| St. Dev | 0.09 | 0.09 | 0.03 | |
| Anterograde displacement (µm) | Mean | 0.35 | 0.43 | 0.31 |
| Median | 0.33 | 0.37 | 0.32 | |
| St. Dev | 0.17 | 0.20 | 0.08 | |
| Anterograde speed (µms-1) | Mean | 2.06 | 1.10 | 1.51 |
| Median | 1.71 | 1.08 | 1.48 | |
| St. Dev | 0.95 | 0.30 | 0.27 | |
| Anterograde time (s) | Mean | 0.18 | 0.34 | 0.18 |
| Median | 0.15 | 0.33 | 0.18 | |
| St. Dev | 0.08 | 0.08 | 0.03 | |
| Reagent type or resource | Designation | Source | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Lung fibroblast line | Allison et al., 2017 https://doi.org/10.1083/jcb.201609033 | GFP-SNX1-MRC5 | MRC5 cell line stably expressing GFP-SNX1. Mycoplasma free. |
| Cell line (H. sapiens) | Lung fibroblast line | Other | GFP-Rab5-MRC5 | MRC5 cell line stably expressing GFP-Rab5 generated by retroviral transduction by G. Pearson and E. Reid, University of Cambridge. Mycoplasma free. |
| Cell line (H. sapiens) | MRC-5 SV1 TG1 Lung fibroblast line | ECACC | MRC-5 SV1 TG1 cells, cat no. 85042501 | Mycoplasma free. |
| Antibody | Anti-human Rab5A Rabbit monoclonal | Cell Signalling Technology | 3547S | IF(1/200) |
| Antibody | Anti-human sorting nexin 1 (mouse monoclonal) | BD Biosciences | 611482 | IF(1/200) |
| Antibody | Alexa594-conjugated anti-mouse IgG (donkey polyclonal) | Jackson ImmunoResearch | 715-585-150 | IF(1/400) |
| Antibody | A488-conjugated donkey anti-rabbit IgG | Jackson Immunoresearch | 711-545-152 | IF(1/400) |
| Recombinant DNA reagent | pLXIN-GFP-Rab5C-I-NeoR | Other | Used by G. Pearson and E. Reid, University of Cambridge to generate retrovirus containing GFP-Rab5C | |
| Sequence-based reagent | Hpa1 GFP Forward | Other | PCR primer | Used by G. Pearson and E. Reid, University of Cambridge to generate retrovirus containing GFP-Rab5C. TAGGGAGTTAACATGGTGAGCAAGGGCGAGGA |
| Sequence-based reagent | Not1 Rab5C Reverse | Other | PCR primer | Used by G. Pearson and E. Reid, University of Cambridge to generate retrovirus containing GFP-Rab5C . ATCCCTGCGGCCGCTCAGTTGCTGCAGCACTGGC |
| Chemical compound, drug | DAPI | Biolegend | 422801 | IF (1 µg/mL) |
| Chemical compound, drug | Prolong Gold | ThermoFisher | P36930 | |
| Chemical compound, drug | Lysobrite Red | AAT Bioquest | 22645 | (1/2500) |
| Chemical compound, drug | Geneticin (G418) | Sigma-Aldrich | G1397 | 200 µg/mL to maintain GFP-Rab5-MRC5 and GFP-SNX1-MRC5 cells in culture. |
| Chemical compound, drug | Formaldehyde solution, 37% (wt/v) | Sigma-Aldrich | 252549 | |
| Chemical compound, drug | Triton X-100 | Anatrace | T1001 | |
| Software, algorithm | NNT (aitracker.net) | Newby et al., 2018 | AITracker | Web-based automated tracking service |
| Software, algorithm | Metamorph | Molecular Devices LLC | Metamorph | Metamorph Microscopy Automation and Image Analysis Software |
| Software, algorithm | FIJI | Schindelin, J.; Arganda-Carreras, I. and Frise, E. et al. (2012) ,‘Fiji: an open-source platform for biological-image analysis’, Nature methods 9 (7): 676–682, PMID22743772, doi:10.1038/nmeth.2019 | FIJI/ImageJ | |
| Software, algorithm | DLFNN Exponent Estimator | Han, Daniel. (2020, January 20). DLFNN Exponent Estimator (Version 0). http://doi.org/10.1101/777615 | DLFNN/DLFNN Exponent Estimator | Hurst exponent estimator with Deep Learning Feed-forward Neural Network application for Windows 10. Documentation included. |
| Software, algorithm | Python3 | Python Software Foundation.Python Language Reference 3.7. Available at www.python.org | Python/Python3 | |
| Software, algorithm | SciPy | Virtanen et al. (2020) SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods, in press. | SciPy/scipy | |
| Software, algorithm | Tensorflow | Abadi et al., 2016 | Tensorflow | |
| Software, algorithm | Keras | Chollet, François and others. ‘Keras.' (2015). Available from https://keras.io | Keras | |
| Software, algorithm | fbm | Flynn, Christopher, fbm 0.3.0 available for download at https://pypi.org/project/fbm/ or https://github.com/crflynn/fbm | FBM package in Python | Exact methods for simulating fractional Brownian motion (fBm) or fractional Gaussian noise (fGn) in python. Approximate simulation of multifractional Brownian motion (mBm) or multifractional Gaussian noise (mGn). |
| Other | 35 mm glass-bottomed dishes (µ-Dish) | Ibidi | Cat. No. 81150 |
Results for the fits of survival time probabilities shown in Figure 4.
The parameters and the analytical survival functions used to fit the Kaplan-Meier estimator survival curves.





