An advanced cell cycle tag toolbox reveals principles underlying temporal control of structure-selective nucleases
Figures
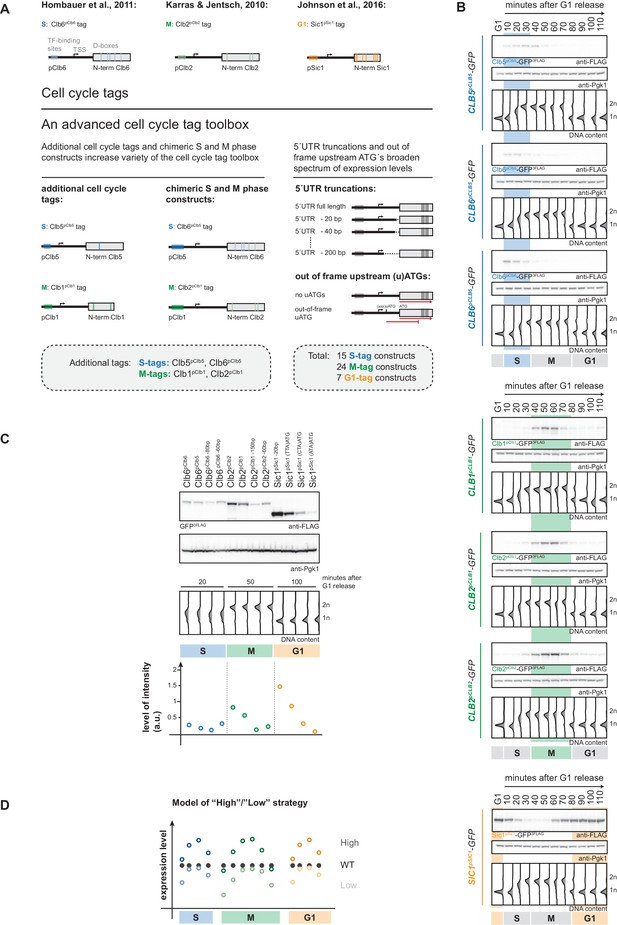
An advanced toolbox of cell cycle tag constructs.
(A) Schematic representation of the applied strategies for improved cell cycle tag methodology. Upper panel: conventional cell cycle tag methodology was limited by only one construct for each cell cycle phase. Lower panel: the advanced cell cycle tag toolbox was expanded to 46 constructs. Therefore, we used new promoters and degrons from Clb5 and Clb1, chimeric promoter-degron combinations and protein expression was crippled using 5´UTR truncations and upstream out of frame ATGs. Vertical bars indicate location of cell cycle regulatory elements in the promoter and N-terminal degron sequence (see Figure 1—figure supplement 1 for detailed description of the tagging procedure). (B) New cell cycle tag constructs allow cell cycle-restricted expression of GFP at varied peak expression levels. Anti-FLAG westerns of cells expressing Clb5pClb5-, Clb6pClb5-, Clb6pClb6-, Clb1pClb1-, Clb2pClb1-, Clb2pClb2-3FLAG-tagged versions of GFP after G1 arrest with α-factor and synchronous release through the cell cycle up to the next G1 phase. Pgk1 western was used as control and DNA content measurements indicate cell cycle progression at the individual time points below (see Figure 1—figure supplement 2 for G1 release experiments of the corresponding 5´UTR truncations and for constructs containing upstream out of frame ATGs). (C) New promoters and degrons, chimeric promoter-degron combinations, 5’UTR truncations and upstream out of frame ATGs allow a broad spectrum of peak expression levels of cell cycle-restricted GFP. Western blot analysis of peak expression levels of cell cycle-tagged 3FLAG-GFP variants at indicated time points after G1 release (20 min = S phase, 50 min = M phase, 100 min = G1 phase). DNA content measurements below indicate cell cycle progression. Graph: peak expression levels were quantified using Image-J and signals of the individual constructs were divided by the corresponding Pgk1 sample to normalize to overall protein levels (see Figure 1—figure supplement 3 for an overview of all cell cycle-tagged GFP versions in cells arrested in the corresponding cell cycle phase). (D) Schematic representation of the suggested cell cycle tag strategy using two sets of constructs with matching ‘low’ and ‘high’ peak expression levels. ‘Low’ expressing constructs (light colours) are chosen by matching peak expression levels similar to the endogenous protein but will show underexpression at cell cycle phase transitions. ‘High’ expressing tags generally show higher expression compared to the wildtype protein with the advantage of broader timeframes of action (timeframes in which protein levels are similar or higher than endogenous protein levels).
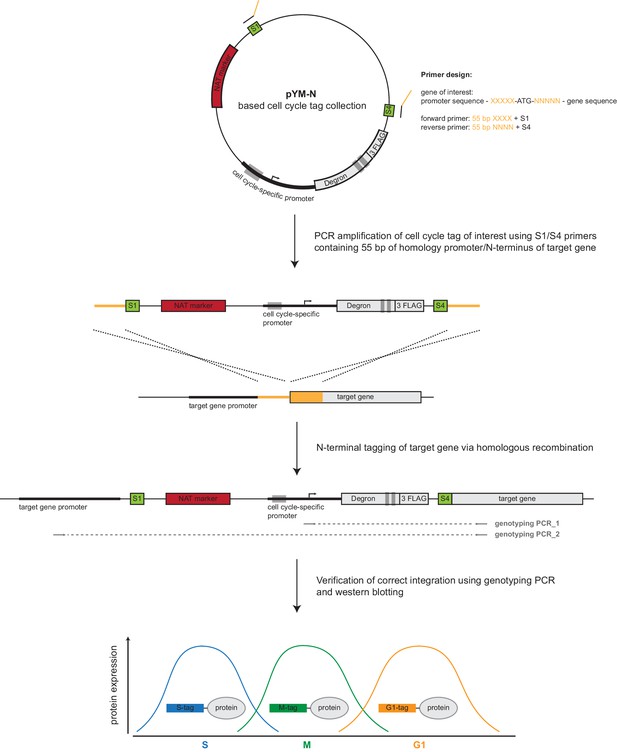
Schematic representation of the cell cycle tagging procedure.
A construct from the pYM-N-based (Janke et al., 2004) collection of cell cycle tags is used as a template for PCR amplification and homologous recombination-based tagging of a gene of interest. A detailed description of the workflow can be found in Materials and methods (construction of cell cycle-tagged strains).

5'UTR truncations and upstream out of frame ATGs do not interfere with cell cycle restriction.
Western blot analysis of Clb5pClb5-, Clb6pClb6-, Clb1pClb1-, Clb2pClb2- and Sic1pSic1-tagged GFP expressed from 33 constructs with truncated 5’UTR or upstream out of frame ATGs during a single cell cycle. Cells were arrested in G1 and synchronously released to the cell cycle up to the next G1 as in Figure 1B. Anti-FLAG western shows cell cycle-tagged constructs, anti-Pgk1 western is used as control. Bottom: DNA content measurement by flow cytometry.
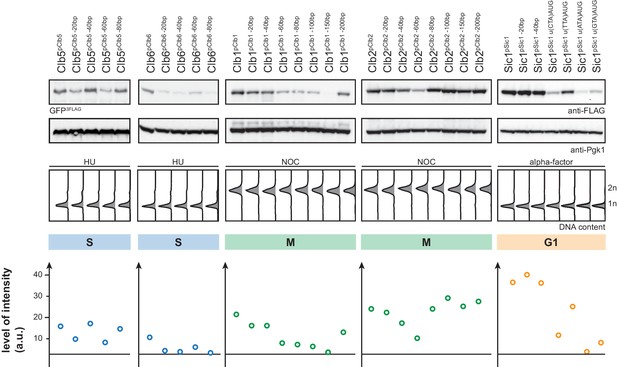
5'UTR truncations and upstream out of frame ATGs cripple peak expression levels of cell cycle tag constructs.
Western blot analysis of Clb5pClb5-, Clb6pClb6, Clb1pClb1-, Clb2pClb2- and Sic1pSic1-tagged GFP expressed from constructs with truncated 5’UTR and upstream out of frame ATGs in HU (S), NOC (M) and α-factor (G1) arrested cells. Middle: DNA content measurements of corresponding samples by flow cytometry. Bottom: Expression levels were quantified using Image-J.
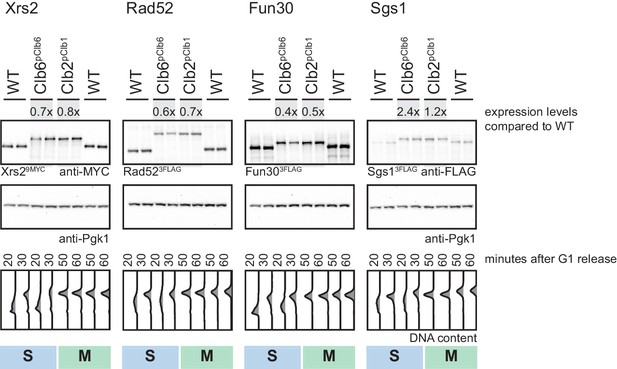
Clb6pClb6- and Clb2pClb1-tag induce similar peak expression levels for several cell cycle-tagged proteins.
Clb6pClb6 (S)- and Clb2pClb1 (M)-tagged Xrs2, Rad52, Fun30 and Sgs1 show similar peak expression levels. Western blot analysis of peak expression levels at indicated time points after G1 release. Expression levels were quantified using Image-J and signals of the individual constructs were divided by the corresponding Pgk1 sample to normalize to overall protein levels. Normalized protein levels of Clb6pClb6 and Clb2pClb1-tagged Xrs2, Rad52, Fun30 and Sgs1 were compared to WT expression at the corresponding time point (fold difference) and the two analysed time points (20 and 30 min time points for S phase and the 50 and 60 min time points for M phase) for each construct were averaged to result in a mean fold difference that is indicated above the western blots. Bottom: DNA content analysis by flow cytometry.
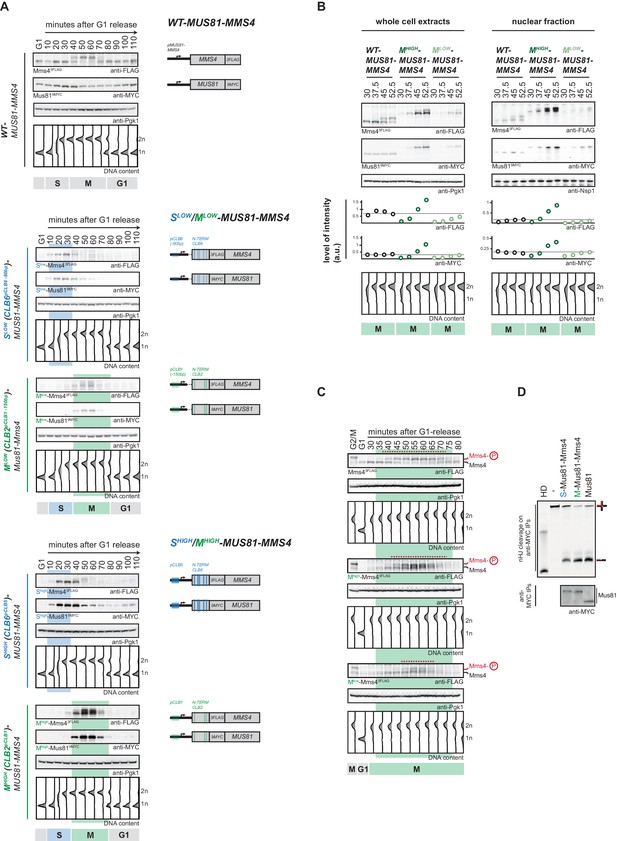
Cell cycle-restricted expression of Mus81-Mms4.
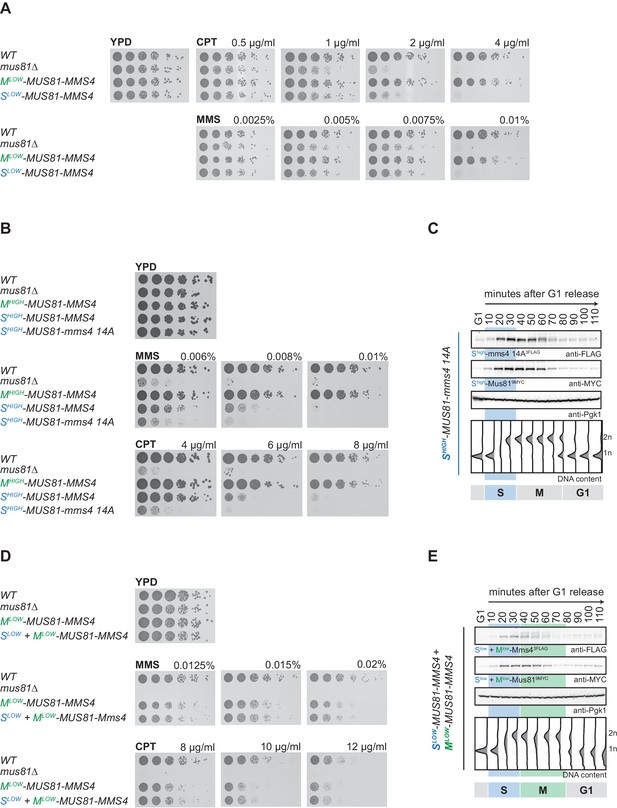
(A) Restriction of Mus81-Mms4 expression to S or M phase of matched pairs of ‘low’ and ‘high’ expressing cell cycle tag constructs. (Left) Western blot and DNA content analysis of strains expressing WT, S phase-restricted (Slow (Clb6pClb6 -80bp)-/Shigh (Clb6pClb5)-Mus81-Mms4) and M phase-restricted (Mlow (Clb2pClb1 -150bp)-/Mhigh (Clb2pClb1)-Mus81-Mms4) alleles of Mus81 (9MYC-tagged)-Mms4 (3FLAG-tagged) during a single cell cycle as in Figure 1D (see Figure 2—figure supplement 1 for quantification of peak expression levels of the Slow-/Mlow -Mus81-Mms4 and Shigh-/Mhigh-Mus81-Mms4 constructs). (Right) Schematic representation of WT, S (Slow-/Shigh-Mus81-Mms4) and M phase (Mlow-/Mhigh-Mus81-Mms4) restricted Mus81-Mms4 constructs. Blue and green bars indicate location of cell cycle regulatory elements in the promoter and N-terminal degron sequence. (B) Different constructs (‘high’ and ‘low’ peak expression levels) of Mus81-Mms4 lead to underexpression of the Mlow-Mus81-Mms4 construct or to overexpression of the Mhigh-Mus81-Mms4 construct in early M phase and similar trends are seen in the nuclear fraction. Western blot analysis of protein levels in whole cell extracts (left panel) and after nuclei separation (right panel) at indicated time points after a G1-release (early M phase; see DNA content profile depicted at the bottom). While immediately with entry into M phase the Mhigh-Mus81-Mms4 construct reaches similar or higher protein levels than endogenous Mus81-Mms4, the Mlow-Mus81-Mms4 construct shows a 10–15 min delay in reaching comparable expression levels and this holds true for both, whole cell extracts and the nuclear fraction. Expression levels were quantified using Image-J and signals of the individual time points were divided by the corresponding Pgk1 (whole cell extracts) or Nsp1 (nuclear fraction) signal to normalize to overall protein levels (graphs below contain normalized values for every construct). (see Figure 2—figure supplement 2 for control western blots of the nuclear fractionation) (C) Different constructs (‘high’ and ‘low’ peak expression levels) of Mus81-Mms4 lead to different windows of Mus81-Mms4 phosphorylation in Mlow-Mus81-Mms4 and Mhigh-Mus81-Mms4. Western blot analysis of the phosphorylation states of Mms4 at indicated time points after a G1-release (M phase; see DNA content profile depicted below the western blots). While Mhigh-Mus81-Mms4 shows a similar timeframe of Mms4 phosphorylation to endogenous Mus81-Mms4, Mlow-Mus81-Mms4 is phosphorylated and stimulated during a shortened window of time only (compare red lines above the Mms4-3FLAG western blots: 15–20 min of phosphorylation in Mlow-Mus81-Mms4 compared to 30 min in Mhigh-Mus81-Mms4 and 30–35 min in Mus81-Mms4). (D) N-terminal tagging does not alter Mus81-Mms4 activity. Resolution assay using a nicked HJ (nHJ) substrate and immunopurified Mus81-Mms4, S-Mus81-Mms4 and M-Mus81-Mms4 (note that WT and cell cycle-tagged proteins were expressed from pGal1-10 promoter). Myc-tagged Mus81-Mms4 was purified from cycling cells, dephosphorylated using λ-Phosphatase and incubated with the nHJ substrate for 2 hr. Upper panel: nHJ cleavage assay with heat DNA substrate (HD) as control. Lower panel: western blot analysis of Mus81-9MYC IP after nHJ cleavage assay (see Figure 2—figure supplement 3 for a western blot analysis of Mms4 dephosphorylation by λ-Phosphatase).
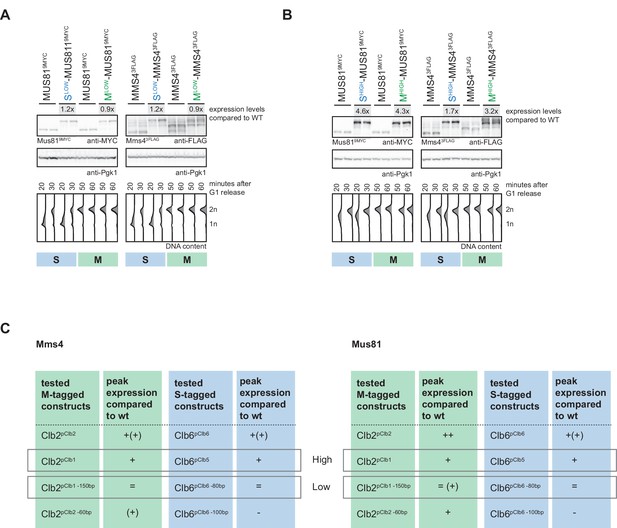
Analysis of ‘low’ and ‘high’ S- and M-tagged Mus81-Mms4 peak expression levels.
(A/B) Sets of Slow-Mus81-Mms4 (Clb6pClb6 -80bp)/Mlow-Mus81-Mms4 (Clb2pClb1 -150bp) (A) and Shigh-Mus81-Mms4 (Clb6pClb5)/Mhigh-Mus81-Mms4 (Clb2pClb1) (B) display similar peak expression levels. Western blot analysis of peak expression levels at individual time points after G1 release. Expression levels were quantified using Image-J and signals at the individual time points were divided by the corresponding Pgk1 sample to normalize to overall protein levels. Normalized protein levels of Slow-/Mlow-Mus81-Mms4 and Shigh-/Mhigh-Mus81-Mms4 were compared to wt expression at the corresponding time point (fold difference) and the two analysed time points (20 and 30 min time points for S phase and 50 and 60 min time points for M phase) for each construct were averaged to result in a mean fold difference that is indicated above the western blots. Bottom: DNA content measurement by flow cytometry. (C) Summary of peak expression levels of various cell cycle-tagged Mus81/Mms4 constructs. Depicted are relations of peak expression values in comparison to WT, which lead to selection of ‘high’ and ‘low’ expressing constructs.
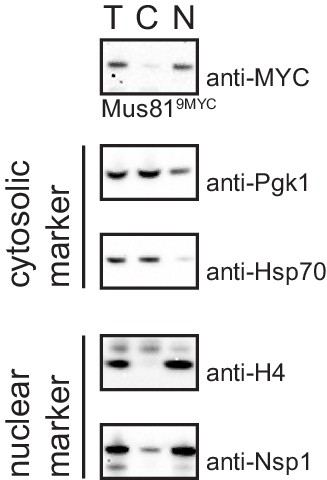
Nuclear/cytoplasmic fractionation.
Western blot analysis of different fractions obtained after nuclei separation of a representable sample from the experiment in Figure 2B shows efficient separation into cytoplasmic and nuclear fractions. Pgk1 and chaperone Hsp70 serve as cytosolic markers, histone H4 and the nuclear pore complex protein NSP1 serve as nuclear markers. T = total lysate, C = cytoplasmic fraction, N = nuclear fraction.

λ-Phosphatase treatment leads to efficient Mms4 dephosphorylation of WT, S- and M-Mus81-Mms4 used for activity assays.
Western blot analysis of Mms4 shows efficient dephosphorylation of Mms4 after λ-Phosphatase treatment of WT, S- and M-Mus81-Mms4 preparations used in activity assays (see Figure 2D for activity assay done with S-, M- and WT Mus81-Mms4).
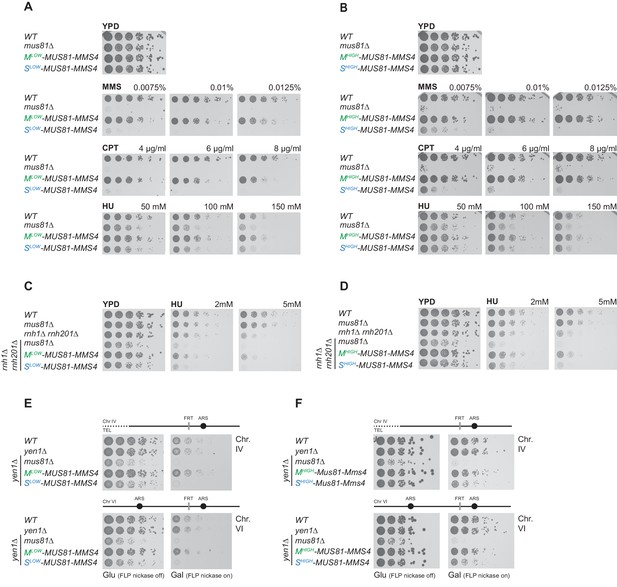
Mus81-Mms4 restricted to M phase, but not S phase is sufficient for the response to genotoxic insults.
(A/B) M phase-restricted Mus81-Mms4 is sufficient to confer viability to replication fork stalling drugs. Viability of cells with Mlow (Clb2pClb1 -150bp)-Mus81-Mms4/Slow (Clb6pClb6 -80bp)-Mus81-Mms4 constructs at low peak expression levels (A) or Mhigh (Clb2pClb1)-Mus81-Mms4/Shigh (Clb6pClb5)-Mus81-Mms4 constructs at high peak expression levels (B) is compared to that of WT and mus81Δ cells. Strains were plated in 5-fold serial dilutions on YPD plates containing the indicated amounts of MMS, CPT or HU and incubated at 30°C for 2 days. (C/D) Mitotic function of Mus81-Mms4 is sufficient to confer viability upon induction of RNA-DNA-hybrids in the absence of RNAse H enzymes and mild replication stress (HU). Cell cycle-tagged versions of Mus81-Mms4 were integrated in the rnh1∆ rnh201∆ background. Strains were spotted in 5-fold serial dilutions on YPD containing indicated concentrations of HU and incubated at 30°C for 2 days. (C) Spotting containing Mlow-Mus81-Mms4/Slow-Mus81-Mms4 cells. (D) Spotting of Mhigh-Mus81-Mms4/Shigh-Mus81-Mms4 cells. (E/F) Repair of Flp-nickase induced DNA lesions requires the M phase function of Mus81-Mms4. Galactose-induced DNA nicking is presumed to be followed by replication run-off in S phase to form single-ended DSBs and repair by BIR (Nielsen et al., 2009; Mayle et al., 2015). Location of the corresponding FRT sites on chromosome IV and VI are indicated relative to replication origins. Cells were spotted in 5-fold serial dilutions in presence of glucose or galactose (FLP-induction) and incubated at 30°C for 2 days. (E) Spottings of Mlow-Mus81-Mms4/Slow-Mus81-Mms4 cells. (F) Spottings of Mhigh-Mus81-Mms4/Shigh-Mus81-Mms4 cells.
Residual Shigh-Mus81-Mms4 function in response to genotoxic agents is explained by insufficient restriction to S phase; slight Mlow-Mus81-Mms4 defect in response to genotoxic agents is due to underexpression during M phase, respectively.
(A) Slow-Mus81-Mms4 cells show comparable DNA damage sensitivity to mus81∆ even at low CPT and MMS concentrations. Indicated strains were chronically exposed to CPT and MMS as in Figure 3A/B. (B/C) Residual Mms4 phosphorylation is responsible for the observed difference between mus81∆ and Shigh-Mus81-Mms4. While Shigh-Mus81-Mms4 cells show better survival after MMS and CPT treatment compared to mus81∆ cells, additional removal of phosphorylation sites targeted during M phase (mms4 14A) from the Shigh-Mus81-Mms4 construct (Shigh-Mus81-mms4 14A) lead to a phenotype comparable to mus81∆ indicating this residual functionality comes from leakage of the Shigh tagged protein in M phase (Shigh-Mus81-mms4 14A). (B) Spotting of indicated strains as in Figure 3A/B. (C) Western blot analysis of the Shigh-Mus81-mms4 14A construct at indicated time points after G1 release as in Figure 1D. (D/E) The observed survival defect of Mlow-Mus81-Mms4 cells upon exposure with high MMS and CPT concentrations is not derived from a missing S phase function, but rather from underexpression of the Mlow-Mus81-Mms4 construct. Additional expression of Slow-Mus81-Mms4 as a second copy together with Mlow-Mus81-Mms4 (Slow+Mlow-Mus81-Mms4) does not rescue the defect of Mlow-Mus81-Mms4 cells at high MMS and CPT concentrations and thereby rules out a contribution of S phase Mus81-Mms4. (D) Spotting of indicated strains as in Figure 3A/B. (E) Western blot analysis of cells expressing both Slow-Mus81-Mms4 and Mlow-Mus81-Mms4 (Slow+Mlow-Mus81-Mms4) constructs at indicated time points after G1 release as in Figure 1D.
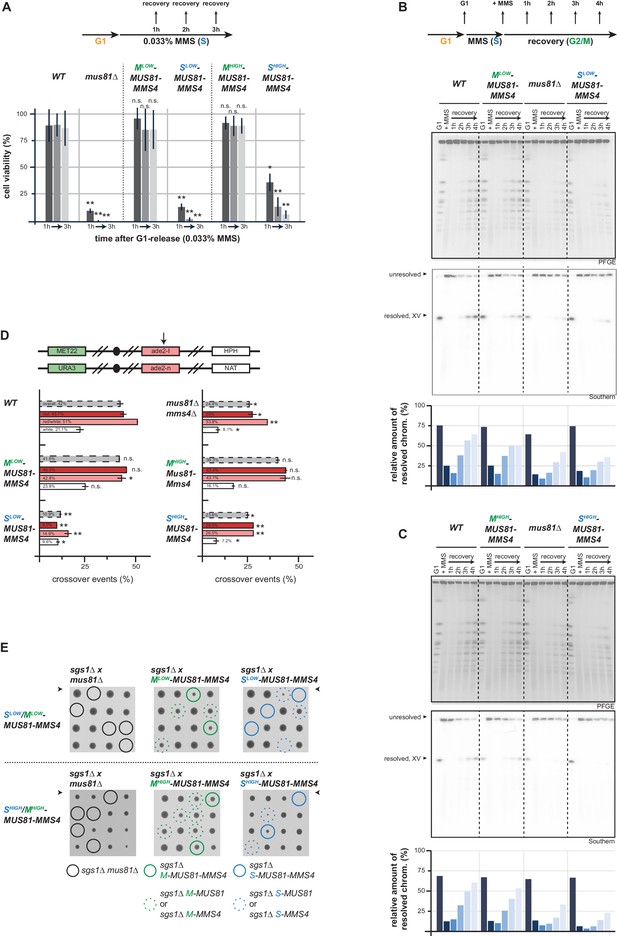
Mus81-Mms4 act as a post-replicative resolvase.
(A) Viability after a pulse of MMS in S phase and subsequent replication fork stalling depends on the M phase function of Mus81-Mms4. Viability assay scoring survivors after pulses of MMS in S phase for one to three hours (upper). Cell viability (%) was determined by colony forming units normalized to untreated cells (0 hr) and is depicted as mean of biological replicates (n = 3) with error bars indicating standard deviation. Significance: n.s. p>0.05, *p<0.05, **p<0.005 as calculated by an unpaired Student´s T-test (see Figure 4—source data 1 for underlying values and exact p-values). (B/C) Resolution of replication/repair intermediates arising in response to replication stalling in S phase requires mitotic Mus81-Mms4 function. PFGE analysis of cells recovering (1–4 hr in Nocodazole) from a pulse of MMS (0.033%, 1 hr) in S phase (see upper panel for experimental setup). PFGE gels were stained with EtBr or subjected to southern blot hybridization with a probe against the ADE2 locus located on chromosome XV. The relative number of resolved chromosomes XV from the southern blots was quantified using ImageJ and is depicted below. (B) PFGE/southern analysis of Mlow-Mus81-Mms4/Slow-Mus81-Mms4 cells. (C) PFGE/southern analysis of Mhigh-Mus81-Mms4/Shigh-Mus81-Mms4 cells. (D) HR repair resulting in crossovers depends on the mitotic function of Mus81. I-SceI induced recombination assay between heterologous ade2 alleles in diploid cells as described in Ho et al., 2010. Upper panel: arrangement of marker genes on chromosomes IV used for classifying the genetic outcomes of DSB repair. The arrow indicates the I-SceI site. Bottom panel: genetic outcome of repair, with overall crossover events (grey) and crossovers among individual classes (red, red/white, white) that differ in conversion tract length. Depicted are mean values from two independent experiments each scoring 400–600 cells with the standard deviation as error bars. Significance: n.s. p>0.05, *p<0.05, **p<0.005 compared to WT cells by unpaired Student´s T-test (see Figure 4—source data 2 for underlying values and exact p-values). (E) The essential requirement of Mus81 in the absence of SGS1-dependent dissolution occurs during M phase. Tetrad analysis of yeast diploid cells with indicated genotypes reveals synthetic lethality between sgs1∆ and Slow-/Shigh-Mus81-Mms4 while Mlow-/Mhigh-Mus81-Mms4 shows no discernible effect on cell growth in the background of sgs1∆ (see Figure 4—figure supplement 2 for a growth analysis of the individual spores of the tetrad analysis with Shigh-/Mhigh-Mus81-Mms4).
-
Figure 4—source data 1
Average, stdv and p-values of normalized colony numbers from replicates 1–3 depicted in Figure 4A.
- https://cdn.elifesciences.org/articles/52459/elife-52459-fig4-data1-v3.xlsx
-
Figure 4—source data 2
Average, stdv and p-values of CO rates from two independent experiments depicted in Figure 4D.
- https://cdn.elifesciences.org/articles/52459/elife-52459-fig4-data2-v3.xlsx
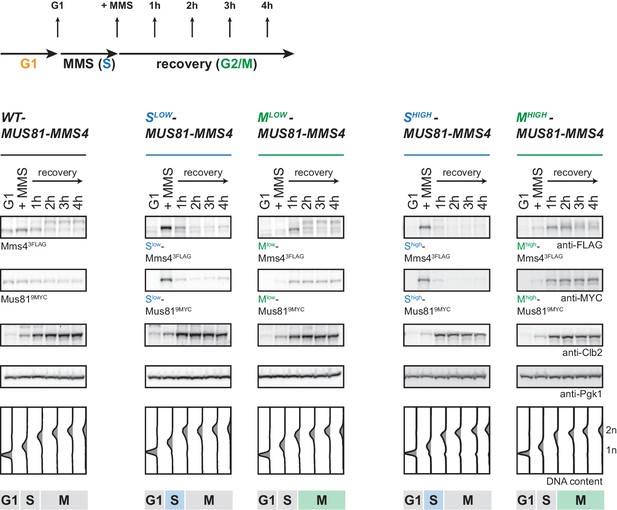
Cell cycle tags restrict efficiently to S or M phase also after DNA damage treatment.
Western blot analysis of WT cells (left), of cells expressing Slow-Mus81-Mms4 or Mlow-Mus81-Mms4 (middle) and of cells expressing Shigh-Mus81-Mms4 or Mhigh-Mus81-Mms4 (right) S- and M-tagged Mus81-Mms4 recovering from a pulse of MMS (0.033%, 1 hr) as in Figure 4B/C. Rise in Clb2 levels are coincidental with entry into M phase. Pgk1 serves as control.

Mus81 function during M phase is required in the absence of Sgs1 function.
Growth assay of yeast haploid cells with the indicated genotypes derived from the tetrad analysis of Shigh-/Mhigh-Mus81-Mms4 in Figure 4E including very slow growing Shigh-/Mhigh-Mus81-Mms4 sgs1∆ haploids. Four strains of indicated genotypes were spotted in 5-fold serial dilutions and growth was analysed after incubation of 1.5 days at 30°C on YPD plates.
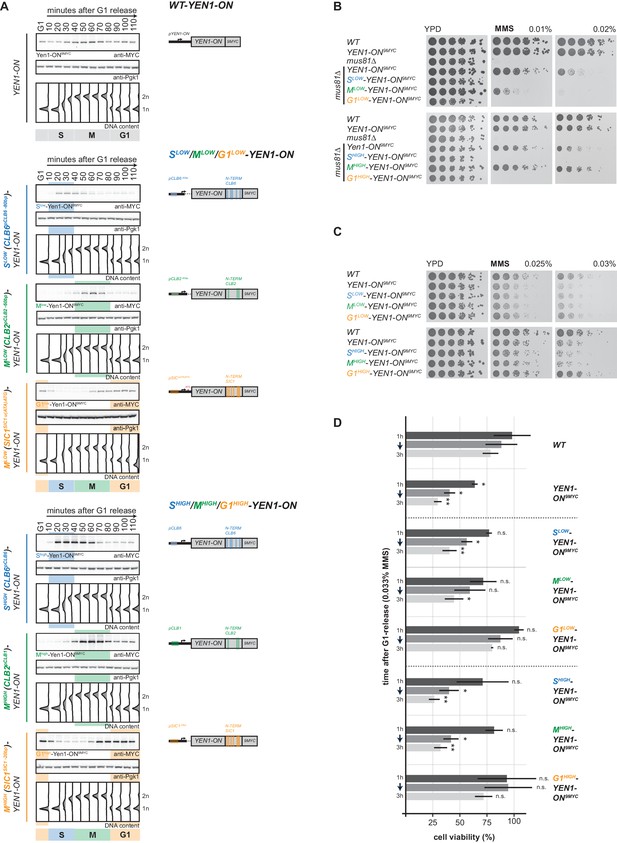
Premature activation of Yen1 in S or early M phase interferes with the response to replication stalling lesions.
(A) Cell cycle-tagged Yen1-ON-9MYC constructs restrict expression of constitutively active Yen1-ON to S, M or G1 phase. (Left) Western blot analysis of strains expressing WT, S (Slow (Clb6pClb6 -80bp)-Yen1-ON9MYC/Shigh (Clb6pClb6)-Yen1-ON9MYC), M (Mlow (Clb2pClb2 -60bp)-Yen1-ON9MYC/Mhigh (Clb2pClb1)-Yen1-ON9MYC) and G1 (G1low (Sic1pSic1 u(ATA)ATG)-Yen1-ON9MYC/G1high (Sic1pSic1 -20bp)-Yen1-ON9MYC) phase-restricted Yen1-ON during synchronous cell cycle progression as in Figure 2A. (see Figure 5—figure supplement 2 for quantification of peak expression levels for individual constructs). (Right) schematic representation of endogenously expressed and cell cycle-tagged Yen1-ON constructs. Blue, green and orange bars indicate location of cell cycle regulatory elements in the promoter and N-terminal degron sequence. (B), The M phase function of Yen1-ON is able to bypass Mus81 requirement after MMS induced replication fork stalling. Strains with indicated genotypes were chronically exposed to MMS as in Figure 3A (note the mus81Δ background). (C–D), Viability after MMS induced replication fork stalling decreases when Yen1-ON is restricted to S or early M phase. (C) Survival of indicated strains after chronic MMS exposure as in (B). (D) Viability assay after a single pulse of MMS in S phase was measured for indicated strains as in Figure 4A (see Figure 5—source data 1 for underlying values and exact p-values).
-
Figure 5—source data 1
average, stdv and p-values of normalized colony numbers from replicates 1–3 depicted in Figure 5D.
- https://cdn.elifesciences.org/articles/52459/elife-52459-fig5-data1-v3.xlsx
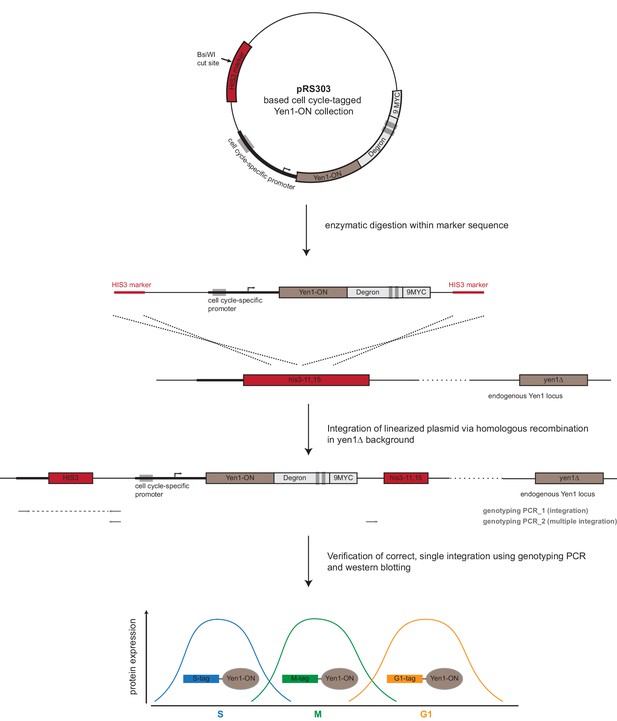
Strategy for C-terminal cell cycle tagging of Yen1-ON.
In order to generate Yen1-ON alleles expressed under cell cycle-regulated promoters and with C-terminal degrons, Yen1-ON constructs were generated in pRS303 integrative vectors. Cell cycle-tagged Yen1-ON constructs were linearized and integrated at the his3-11,15 locus via homologous recombination. A detailed description of the workflow can be found in Materials and methods (construction of cell cycle-tagged strains).
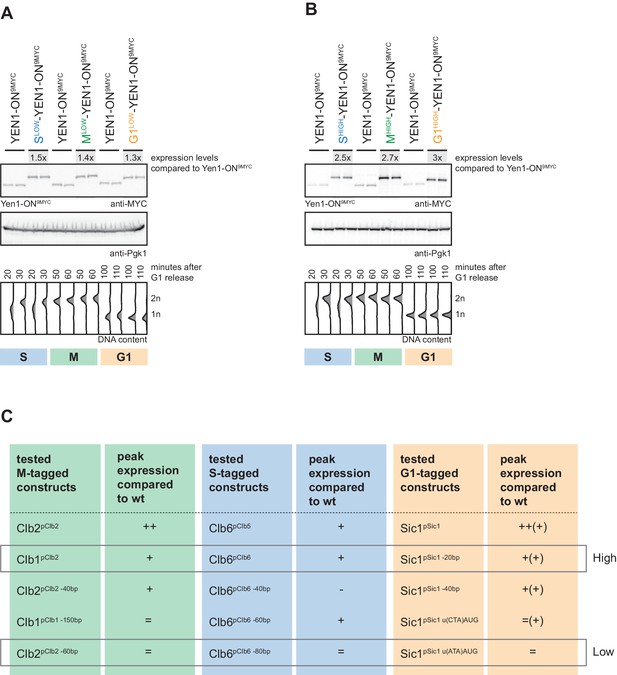
Analysis of ‘low’ and ‘high’ S-, M- and G1-tagged Yen1-ON peak expression levels.
(A/B) Sets of Slow (Clb6pClb6 -80bp)-, Mlow (Clb2pClb2 -60bp)- and G1low (Sic1pSic1 u(ATA)ATG)-Yen1-ON (A) and Shigh (Clb6pClb6)-, Mhigh (Clb2pClb1)- and G1high (Sic1pSic1 -20bp)-Yen1-ON (B) display similar peak expression levels in S, M and G1 phase, respectively. Western blot analysis of peak expression levels of cell cycle-tagged Yen1-ON-9MYC variants at individual time points after G1 release (20,30 min = S; 50,60 min = M; 100,110 = G1) as controlled by DNA content measurement by flow cytometry (lower). Peak expression levels were quantified using Image-J and signals of the individual constructs were divided by the corresponding Pgk1 sample to normalize to overall protein levels. Normalized peak expression levels were compared to WT expression at the corresponding time point (fold difference) and the two analysed time points were averaged for each construct to result in a mean fold difference that is indicated above the western blots. (C) Summary of peak expression levels of various tested cell cycle-tagged Yen1-ON constructs. Depicted are relations of peak expression values in comparison to WT, which lead to selection of ‘high’ and ‘low’ expressing constructs.
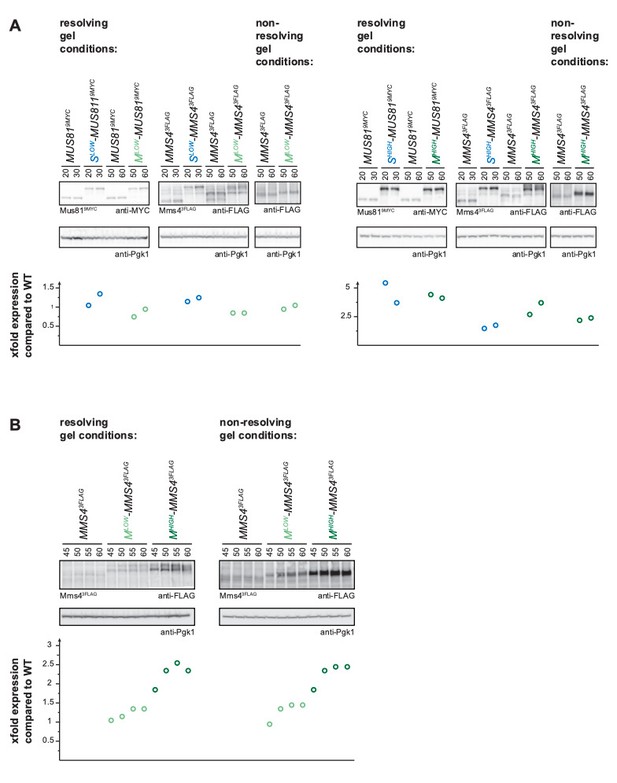
Quantification of Mus81 and Mms4 levels after resolving and non-resolving gel running conditions.
Expression levels were quantified using Image-J and signals at the individual timepoints were divided by the corresponding Pgk1 sample to normalize to overall protein levels. Normalized protein levels were compared to WT expression at the corresponding timepoint (fold difference) and the resulting fold difference is depicted in the graphs below the western blots. For the mitotic Mms4 samples (Mlow-Mms4 and Mhigh-Mms4) the analysis was done both after resolving (4-12% gel, 90min) and after non-resolving (12% gel, 25 min) gel conditions to show the validity of quantifying phosphorylated Mms4 signals. The Mus81 and S phase Mms4 samples were run with resolving gel conditions only. A, Quantification of peak expression levels from Figure 2—figure supplement 1, extended by an analysis with non-resolving gel conditions for the mitotic Mms4 samples. B, Quantification of peak expression levels from samples of the experiment shown in Figure 2C with resolving and non-resolving gel conditions.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-FLAG (rabbit-polyclonal) | Sigma | Cat# F7425, RRID:AB_439687 | WB (1:2000) |
| Antibody | anti-FLAG M2-Peroxidase (mouse-monoclonal) | Sigma | Cat# A8592, RRID:AB_439702 | WB (1:3000) |
| Antibody | anti-MYC (mouse-monoclonal) | Millipore | Cat# 05–724, clone 4A6, RRID:AB_11211891 | WB (1:2000) |
| Antibody | anti-Clb2 (rabbit-polyclonal) | Santa Cruz | Cat# sc-9071, RRID:AB_667962 | WB (1:500) |
| Antibody | anti-Hsp70 (mouse-polyclonal) | Enzo Life Sciences | Cat# ADI-SPA-822, RRID:AB_10615940 | WB (1:10000) |
| Antibody | anti-H4 (rabbit-polyclonal) | Abcam | Cat# ab10158, RRID:AB_296888 | WB (1:2000) |
| Antibody | anti-Nsp1 (mouse-monoclonal) | Abcam | Cat# ab4641, RRID:AB_304549 | WB (1:10000) |
| Strain, strain background (S. cerevisiae) | strain background, W303 | Rothstein, 1983 | ||
| Strain, strain background (S. cerevisiae) | yeast strains | This study | Supplementary File 1 | |
| Recombinant DNA reagent | plasmid backbone, pYM-N31 | Janke et al., 2004 Euroscarf | P30284 | |
| Recombinant DNA reagent | plasmid backbone, pRS303 | ATCC | Cat# 77138 | |
| Recombinant DNA reagent | plasmids | This study | Supplementary file 1 | |
| Sequence-based reagent | S1 | Janke et al., 2004 | PCR primers | cgtacgctgcaggtcgac |
| Sequence-based reagent | S4 | Janke et al., 2004 | PCR primers | catcgatgaattctctgtcg |
| Sequence-based reagent | yeGFP S1 | This study | PCR primers | attgtaatacgactcact atagggcgaattggag ctccaccgcggtggcg gccgccgtacgctgca ggtcgac |
| Sequence-based reagent | yeGFP S4 | This study | PCR primers | ctaattcaaccaaaattg ggacaacaccagtgaa taattcttcacctttagaca tcatcgatgaattctctgtcg |
| Sequence-based reagent | Mus81 S1 | This study | PCR primers | caaagtttcaaaggatt gatacgaacacacattc ctagcatgaaagcatgc gtacgctgcaggtcgac |
| Sequence-based reagent | Mus81 S4 | This study | PCR primers | caactaattcttgtaaccatt caatatataggtcttttaagtt tgatgagagttccatcgatg aattctctgtcg |
| Sequence-based reagent | Mms4 S1 | This study | PCR primers | acaatgtatggattatgg tatagaataatagtagtc acatattgcagctagttaa cgtacgctgcaggtcgac |
| Sequence-based reagent | Mms4 S4 | This study | PCR primers | gaatactggcatcgtttct tgaatctttgtcctcaaca aaatcaacgatctggctc atcgatgaattctctgtcg |
| Commercial assay or kit | In-Fusion HD Cloning | Clontech | 639648 | |
| Chemical compound, drug | Sytox-green | Invitrogen | S7020 | |
| Chemical compound, drug | RNaseA | Sigma Aldrich | R4875 | |
| Chemical compound, drug | ProteinaseK | Sigma Aldrich | P2308 | |
| Chemical compound, drug | Pierce ECL Western Blotting Substrate | Thermo Fisher | 2106 | |
| Chemical compound, drug | Nocodazol | Sigma Aldrich | M1404 | |
| Chemical compound, drug | Hydroxyurea | Sigma Aldrich | H8627 | |
| Chemical compound, drug | MMS | Sigma Aldrich | 129925 | |
| Chemical compound, drug | Camptothecin | Sigma Aldrich | C9911 | |
| Peptide, recombinant protein | alpha-Factor | MPIB core facility | ||
| Other | Zymolyase Z100T | Roth | 9329.2 | |
| Other | Pulsed Field certified Agarose | BioRad | 1620138 | |
| Other | Amersham Protran Premium 0.45 um Nitrocellulose membrane | GE Healthcare | 10600003 | |
| Other | Amersham Hybond N+ Nylon membrane | GE Healthcare | RPN203B | |
| Other | NuPAGE Novex, 4–12% BIS-TRIS gels | Invitrogen | NP0323 | |
| Software, algorithm | T-Test calculator | GraphPad | GraphPad, RRID:SCR_000306 | http://www.graphpad.com/quickcalcs/ttest2 |
Additional files
-
Supplementary file 1
Yeast strains and Plasmids.
Table 1. Yeast strains used in this study. Table 2. Cell cycle tagging plasmids used in this study Table 3. Other plasmids used in this study
- https://cdn.elifesciences.org/articles/52459/elife-52459-supp1-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52459/elife-52459-transrepform-v3.docx





