The hippocampus encodes delay and value information during delay-discounting decision making
Figures
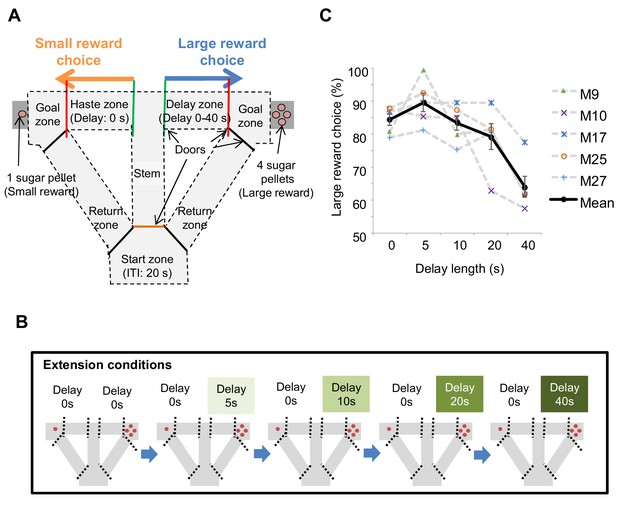
Task design of the delay-based decision making in the T-maze.
(A) Schematic diagram for the experimental setup. Mice can choose the right or left arms assigned to obtain the small reward without delay or the large reward with delay, respectively. (B) Flow of the extension conditions. The delay lengths were extended sequentially. Red circles indicate the number of sugar pellets. (C) Percentages of large-reward choices as a function of delay length. Error bars indicate the standard error of the means (SEM).
-
Figure 1—source data 1
Source Data File for Figure 1C.
- https://cdn.elifesciences.org/articles/52466/elife-52466-fig1-data1-v2.xlsx
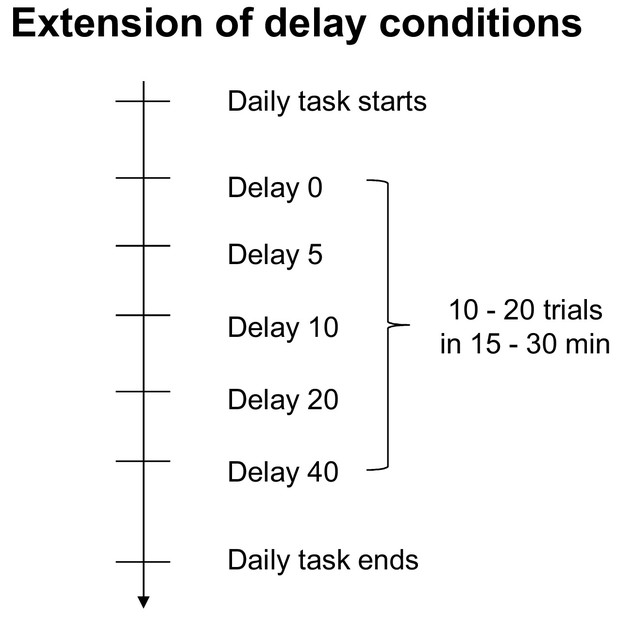
Daily timeline of the extension of delay conditions.
The arrow indicates time flow. Mice individually had 10–20 trials for each delay condition (0, 5, 10, 20 and 40 s) in the right or left arm.

Schedule of running experimental conditions.
All the experimental conditions were performed in different weekly sessions.
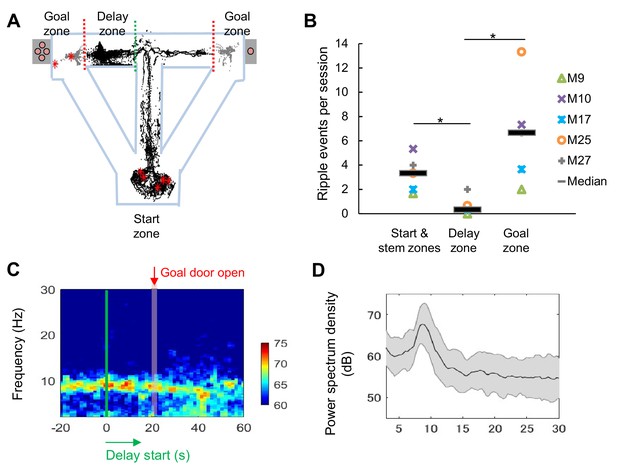
LFP signals during the long delay were characterized by strong theta power and lack of SWRs.
(A) Sharp-wave/ripple events (SWRs) rarely occurred during the delay in the task (data from one session of delay 20 s extension conditions). Red asterisks indicate the locations of SWRs. Black and gray dots indicate the path of animal movements before (black) and after arriving at the goal (gray), respectively. (B) SWRs per session in specific experimental zones. The total number of SWRs for each zone was counted and color-coded according to individual animals (the average number of events acquired from 3 sessions of delay 20 s extension conditions). *, p<0.05, Mann-Whitney’s U-test. (C) Spectrogram of the hippocampal CA1 region during the peri-delay period (averaged from three mice, total six sessions of delay 20 s extension conditions) Green line: delay-onset; red line: estimated goal-onset. (D) Power spectrum density during 2 s at the beginning of the delay. Shaded area indicates ± SD.
-
Figure 2—source data 1
Source Data File for Figure 2B.
- https://cdn.elifesciences.org/articles/52466/elife-52466-fig2-data1-v2.xlsx
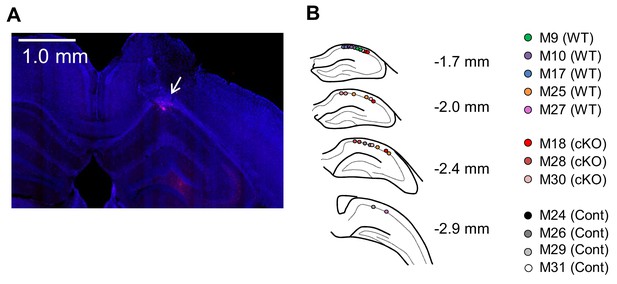
Histological verification of recording sites in the CA1.
(A) An example of a hippocampal slice with DiI pasted at the tip of the electrode and DAPI staining. White arrows indicate inserted points. (B) Electrode tracks are represented by colored dots corresponding to the subjects. The recording electrodes for M9, M10, M18, M24, and M26 were silicon probes (A4-2tet-5mm-150-200-121-H32, NeuroNexus), and M17, M25, M28, M29, M30 and M31 were nichrome tetrodes. All of the electrodes were set on the microdrives and adjusted to the targeting cites.
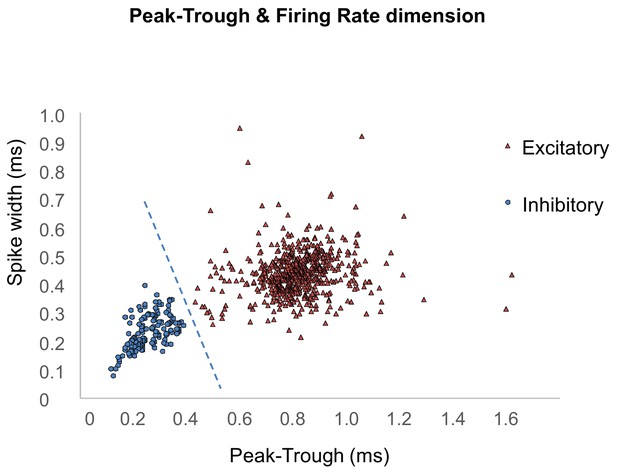
Cell-type classification by plot in peak-trough and spike width.
Red triangles indicate excitatory neurons, while blue circles indicate inhibitory neurons. The dotted line indicates classification criteria for excitatory and inhibitory determinations.
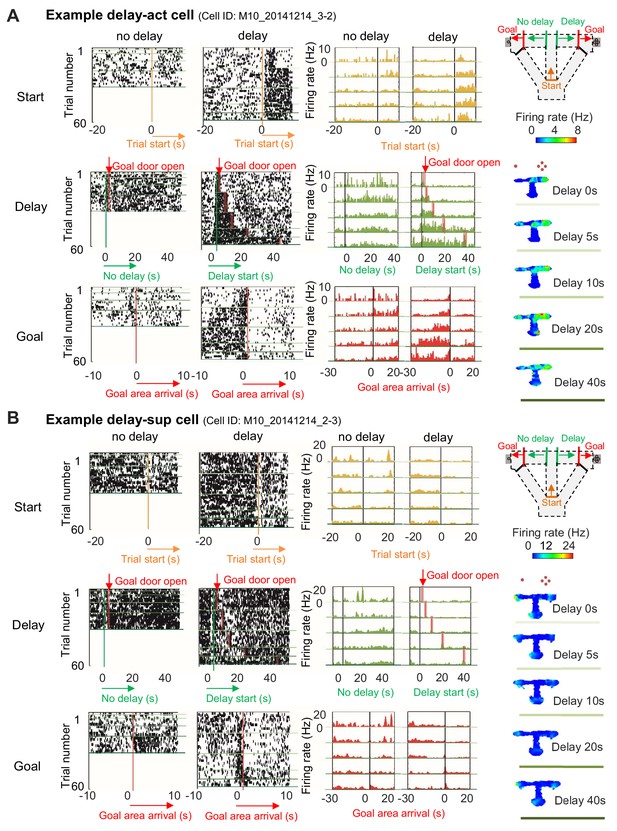
Increased or decreased neuronal activity of CA1 cells during delay.
(A) An example of CA1 delay-active (delay-act) cells, which showed an increment in the firing rate as a function of delay length. Left, raster plots of the firing activity of the cells aligned with start-onset (top), delay-onset (middle) and goal-onset (bottom). Orange lines indicate start-onset. Green lines indicate delay-onset. Pale red lines indicate expected delay-offset. Red lines indicate goal-onset. Center, peristimulus time histograms (PSTHs) of the firing activity of the cells aligned with start-onset (top), delay-onset (middle) and goal-onset (bottom). Right, color-coded rate maps. The delayed arm was assigned to the right side with a large reward for this recording session. Red dots indicate the number of sugar pellets. (B) An example of CA1 delay-suppressed cells, which showed a decrement in the firing rate during delay. The delayed arm was assigned to the right side with a large reward for this recording session.
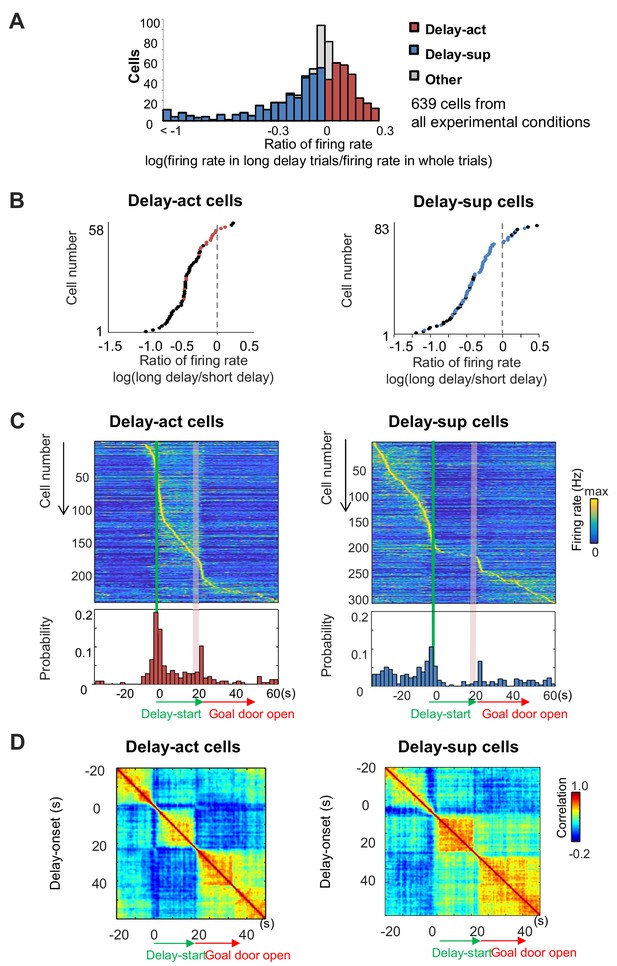
Delay-dependent firing patterns of CA1 delay-active and delay-suppressed cells.
(A) The distribution of delay-active and delay-suppressed cells aligned with the ratio of firing rate in the long delay period and in whole trials. (B) The ratio of mean firing rate during long delays (20 or 40 s) to that during short delay (5 s) for all neurons (base-10 log-transformed). Each dot indicates an individual neuron. Black dots indicate neurons that had a statistically significant difference in firing rate between short and long delay conditions (p<0.001). (C) Temporal patterns of firing rates in CA1 delay-active and delay-suppressed cells during delay. Top, color-coded temporal firing patterns. Neurons were ordered by the time of their peak firing rates. Bottom, temporal distribution of the peak firing rates of the neurons. Green lines indicate delay-onset. Pale red lines indicate expected delay-offset. (D) Correlation matrix of population vectors as a function of time for CA1 delay-active and delay-suppressed cells.
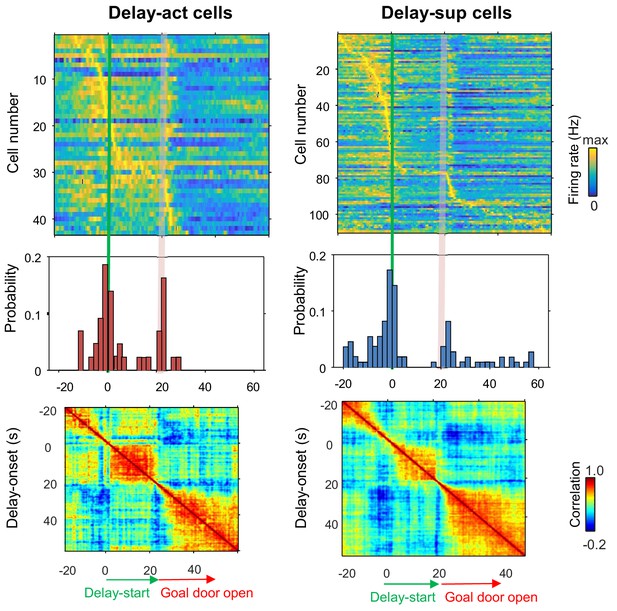
Population coding of long delay in the inhibitory CA1 cells.
Temporal patterns of firing rates in inhibitory CA1 delay-active and delay-suppressed cells during delay. Top, color-coded temporal firing patterns. Neurons were ordered by the time of their peak firing rates. Middle, temporal distribution of the peak firing rates of the neurons. Green lines indicate delay-onset. Pale red lines indicate expected delay-offset. Bottom, correlation matrix of population vectors as a function of time for CA1 delay-active and delay-suppressed cells.
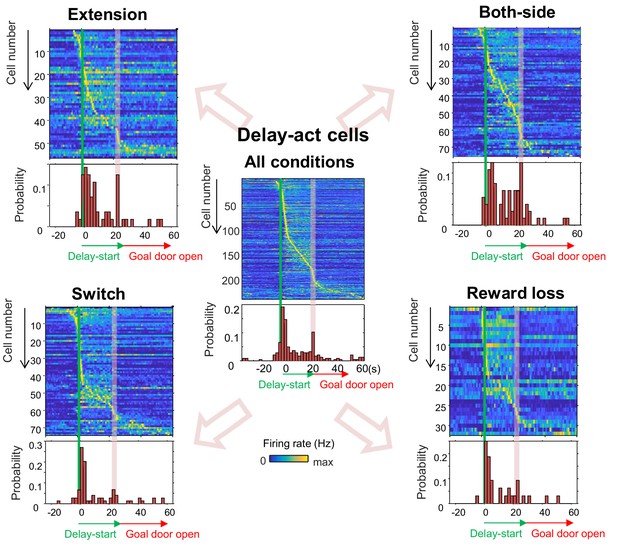
Population coding of long delay in CA1 delay-active cells in different experimental conditions.
Temporal patterns of firing rates in CA1 delay-active cells from all conditions (center), extension (upper left), switch (lower left), both-side (upper right), and reward loss (lower right) conditions. A relatively uniform distribution of peak firing rate was found in both-side conditions (one sample kstest, p=0.46) compared to the other conditions (p<0.001, for extension; p<0.001, for switch; and p=0.01, for Reward lose). Top, color-coded temporal firing patterns. Neurons were ordered by the time of their peak firing rates. Bottom, temporal distribution of the peak firing rates of the neurons. Green lines indicate delay-onset. Pale red lines indicate expected delay-offset.
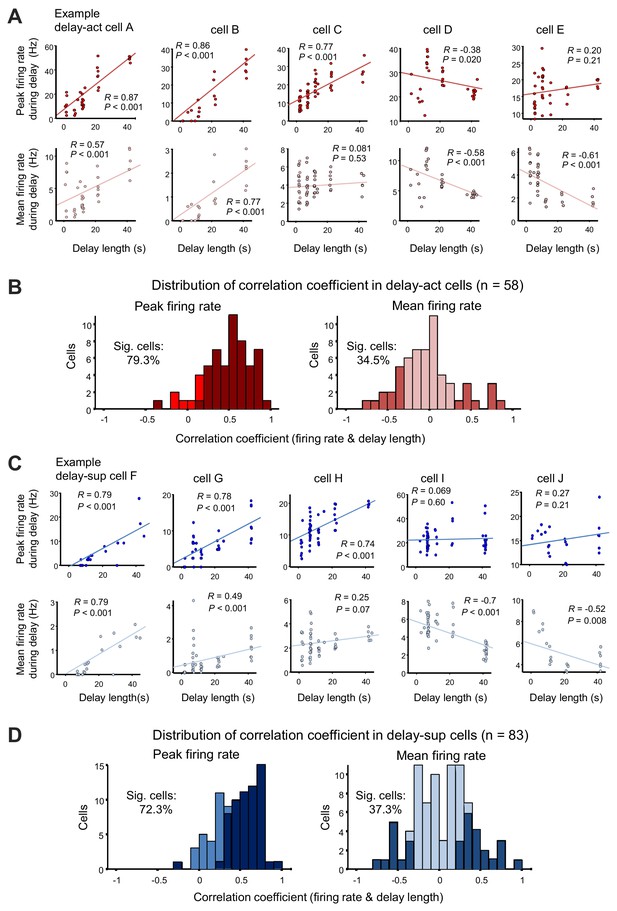
Delay-dependent firing patterns of CA1 delay-active and delay-suppressed cells.
(A) Scatter plots show correlations between firing rate during delay (upper, peak firing rate; lower, mean firing rate) and the delay length of five representative delay-act cells. Cells A and B show positive correlations in both peak and mean firing rate; on the other hand, cell C shows either. Cells D and E show negative correlations. (B) Distribution of correlation coefficients between firing rate (left, peak firing rate; right, mean firing rate) and delay length in delay-act cells. Dark color bars indicate statistically significant neurons (p<0.05), whereas bright color bars indicate neurons do not reach statistical significance. (C) Scatter plots show correlations between firing rate during delay (upper, peak firing rate; lower, mean firing rate) and delay length for five representative delay-suppressed cells. (D) Distribution of correlation coefficients between firing rate (left, peak firing rate; right, mean firing rate) and delay length in delay-suppressed cells. Dark color bars indicate statistically significant neurons (p<0.05), whereas bright color bars indicate that neurons do not reach statistical significance.
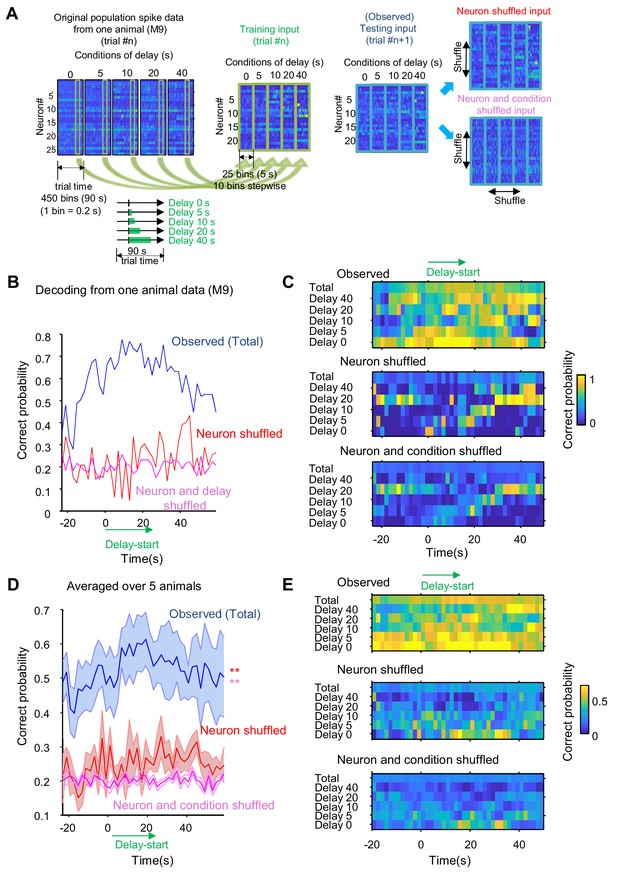
Decoding of delay length from population spike activity.
(A) Schematic of the SVM decoding of delay length. Spiking trains of five delay conditions (0, 5, 10, 20 and 40 s) are transformed into normalized firing rates as training datasets. The population rate estimates are binned (200 ms) and those of 5 s (25 bins) were coupled and given in the multiclass models for SVM as training datasets. Testing datasets are produced with the spike activity of the next trial. Two control testing datasets are created by shuffling the labels of conditions and/or neurons. (B) Representative result of delay length decoding computed from the spike activity of 28 neuron from one animal (M9). The blue line indicates the probability of correct prediction (predicted from observed but untrained testing data). Red and magenta lines indicate the result of shuffled testing inputs (red: neuron shuffled; magenta: neuron and condition shuffled). (C) Color-coded correction rate map for the five delay conditions (0, 5, 10, 20 and 40 s) and total average. Top, result for observed inputs; middle, result for neuron-shuffled inputs; bottom, result for neuron- and condition-shuffled inputs. (D) Averaged decoding results from population spikes of five animals (M9, 28 neurons; M10, 43 neurons; M17, 22 neurons; M25, 34 neurons; and M27, 23 neurons). Data are shown as mean ± SEM. Red **: observed vs neuron shuffled, p<0.001; magenta **, observed vs neuron and condition shuffled, p<0.001; group effect, F2=882.1, p<0.001, two-way ANOVA. (E) The decoding results of individual delay length. Color-coded rate maps for the five delay conditions (0, 5, 10, 20 and 40 s) and total average. Top, result for observed inputs; middle, result for neuron-shuffled inputs; bottom, result for neuron- and condition-shuffled inputs. Data from five mouse were used.
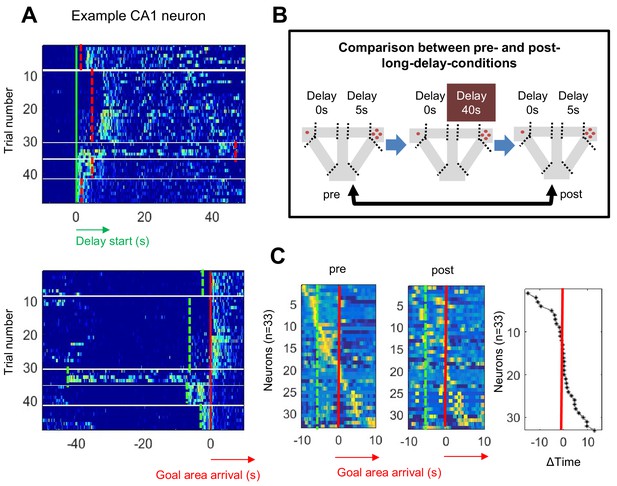
Temporal patterns of the CA1 neuron shift as sessions progressed.
(A) Firing gradually shifted backward. (B) Task schedule for comparison of firing patterns between pre- and post-long delay-session(s). (C) Normalized firing rate sorted by peak firing rate time in pre- (left) and post-long-delay-sessions (center). The order of the cells in the pre- and post-long-delay-sessions was identical. Difference in peak firing time between pre- and post-long-delay-session (sorted by time; right).
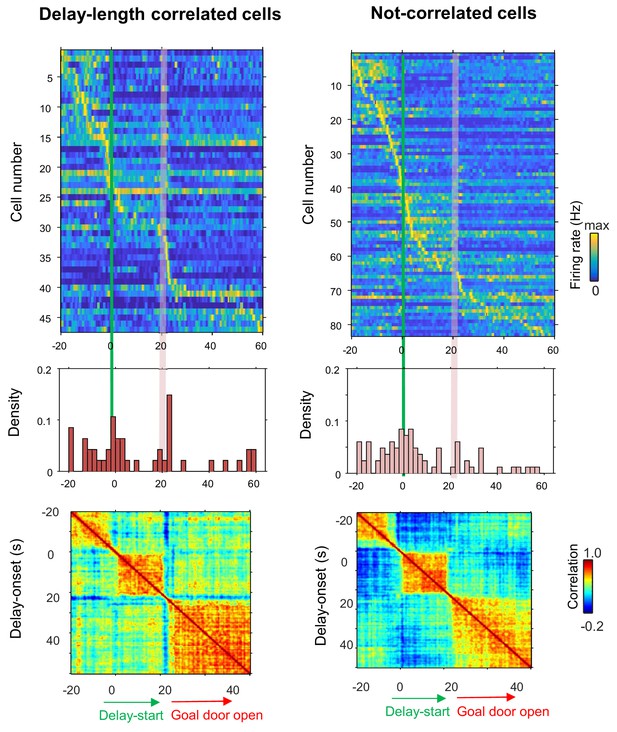
Population coding of long delay in the delay-length-correlated and non-correlated CA1 cells.
Temporal patterns of firing rates in CA1 delay-length-correlated and non-correlated cells during delay. Top, color-coded temporal firing patterns. Neurons were ordered by the time of their peak firing rates. Middle, temporal distribution of the peak firing rates of the neurons. Green lines indicate delay-onset. Pale red lines indicate expected delay-offset. Bottom, correlation matrix of population vectors as a function of time for CA1 delay-active and delay-suppressed cells.
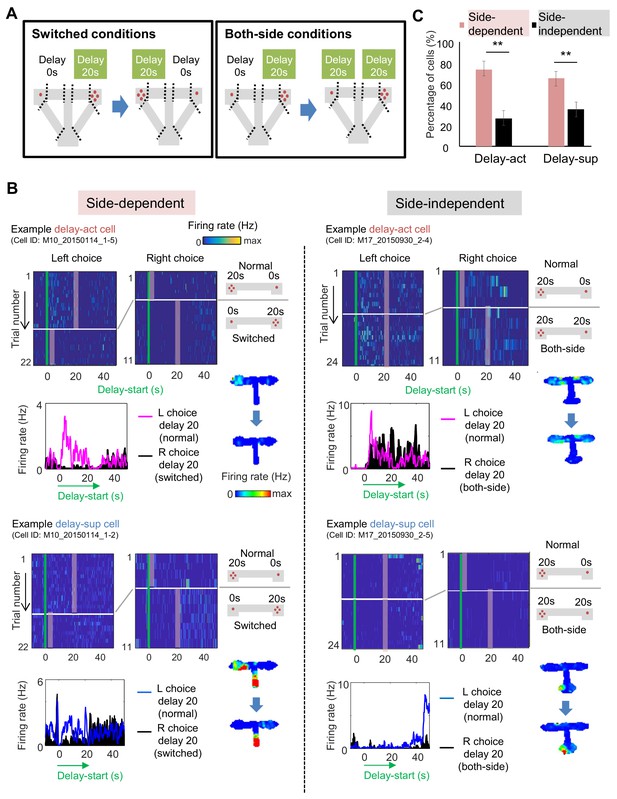
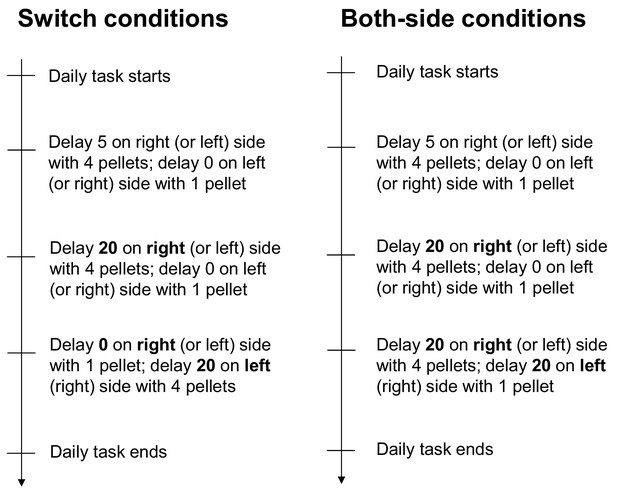
Spatial-selective delay coding in CA neurons.
(A) Experimental conditions to investigate the location selectivity in delay-active neurons. The location of the delay zone was switched to the other side (switched conditions) or doubled to both sides (both-side conditions). (B) Example CA1 delay-active and delay-suppressed cells. Side-dependent and side-independent neurons are shown as left and right rows, respectively. Top left, colored raster plots expressing relative firing rates. Green lines indicate delay-onset. Pale red lines indicate expected delay-offset. Top right, information of conditions corresponded to the raster plots on the left. Red dots indicate the number of sugar pellets. Bottom left, Peri-event time histograms showing the averaged firing rates. Magenta lines indicate the firing rate of the left choice with a 20 s delay. Black-filled histograms indicate the firing rate of the right choice with a 20 s delay. Bottom right, color-coded rate maps for the two conditions (normal delay and switch or both-side conditions). (C) Percentage of place-dependent and -independent CA1 delay-active and delay-suppressed neurons. Error bars indicate 95% Clopper-Pearson’s confidence intervals. **: p<0.01, Mann-Whitney’s U-test.
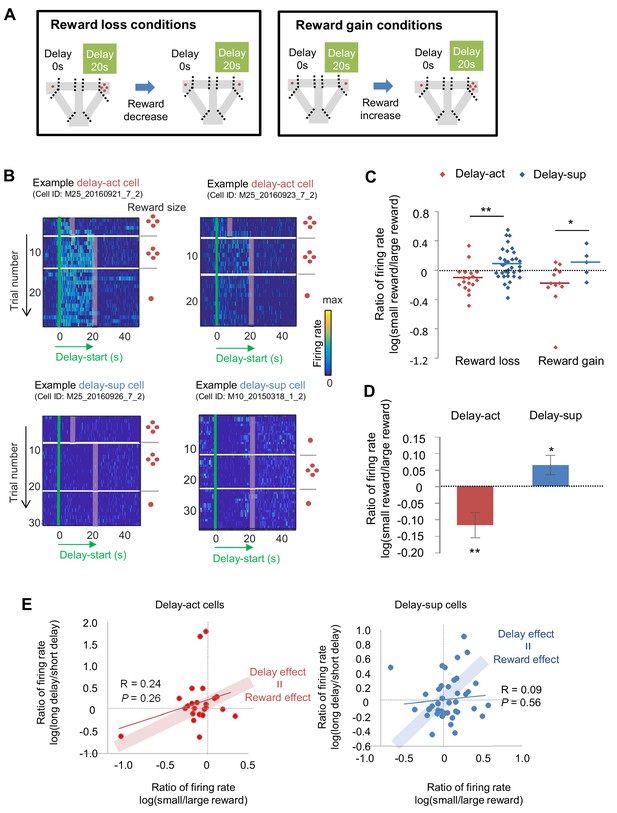
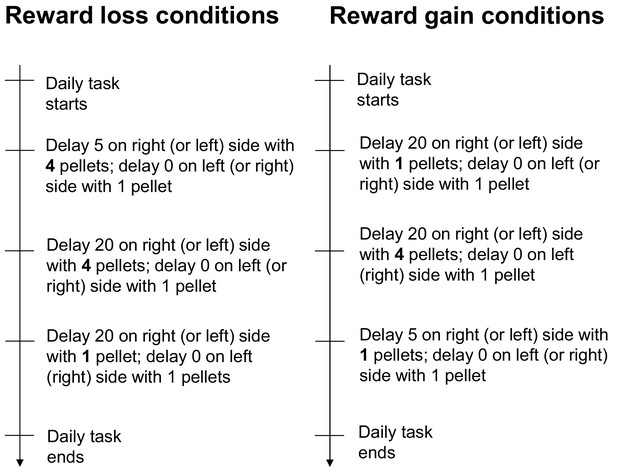
The firing of CA1 delay-active and delay-suppressed cells is distinctly changed by reward size manipulations.
(A) Left, experimental reward loss conditions: the reward size was changed from 4 to 1 (or 0) pellets. Right, experimental reward gain conditions: the reward size was changed from 1 (or 0) to 4 pellets. (B) Example CA1 delay-active (top) and delay-suppressed cells (bottom) fired during delay in reward loss conditions. Green lines indicate delay-onset. Red lines indicate expected delay-offset. Red dots indicate the number of sugar pellets. (C) Ratio of firing rates of delay-active and -suppressed cells in reward loss and gain conditions. Dots indicate individual data for delay-active cells (red) and delay-suppressed cells (blue). Central bars indicate the medians. *, p<0.05; **, p<0.01, Mann-Whitney’s U-test. (D) Ratio of firing rates of delay-active and -suppressed cells in mixed population. Error bars indicate SEM. *, p<0.05; **, p<0.01, One-sample t-test. (E) Scatter plots of firing rate ratios between small/large reward conditions and between long delay/short delay conditions. The computed correlation coefficient R and p value are indicated.
-
Figure 7—source data 1
Source Data File for Figure 7C and D.
- https://cdn.elifesciences.org/articles/52466/elife-52466-fig7-data1-v2.xlsx
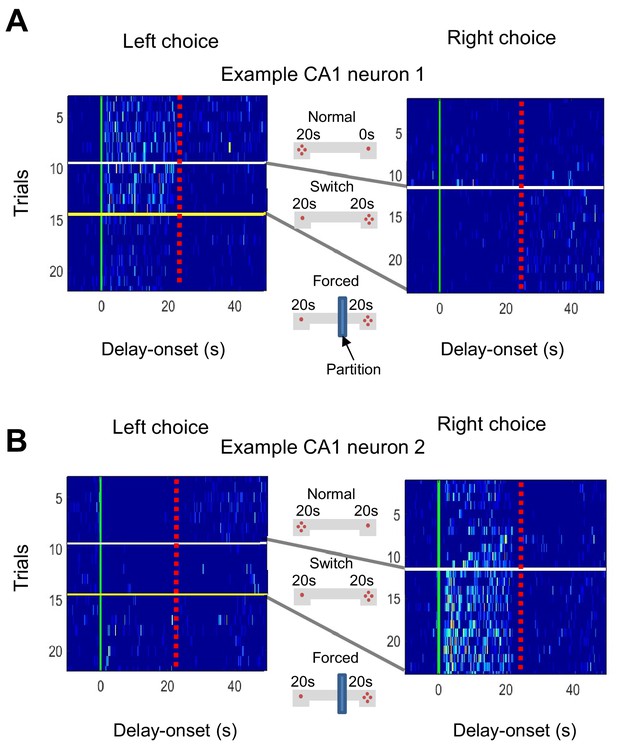
Firing in CA1 delay-active cells depended on animal preference.
Forced conditions in which the animals were forced to choose one side by a partition (here, left side). The yellow line indicates the trial converted to forced conditions. (A) The firing in the left side disappeared when the preference to the left side was extinguished. (B) Simultaneously, other neurons showed increased firing when the preference recovered to the other side (here, the right side).
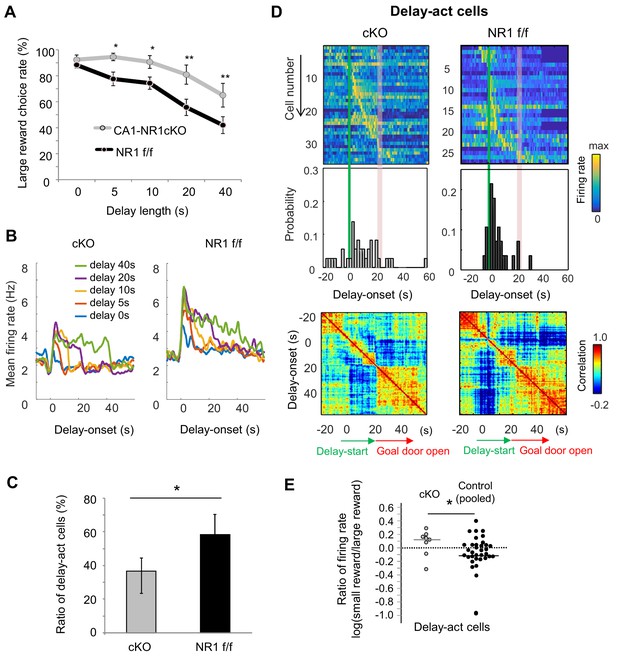
NMDAR-dependent mechanism for delay-discounting.
(A) Impaired delay-discounting in CA1-NR1 cKO mice. *, p<0.05; **, p<0.01; post-hoc Scheffe’s test. Error bars indicate SEM. (B) NMDAR deficiency disrupted the delay tuning in the CA1 activity. Average firing patterns of the CA1 delay-active cells from cKO and control mice for different delay lengths (0, 5, 10, 20, and 40 s). (C) Abnormal delay-active and –suppressed cell proportion in cKO mice. Ratio of delay-act cells to delay-sup cells for cKO and control mice. Error bars indicate 95% Clopper-Pearson’s confidence intervals. *, p<0.05; Mann–Whitney’s U test. (D) NMDAR deficiency disrupted the populational activity in CA1. Top, color-coded temporal firing patterns of the CA1 delay-active cells in cKO and control mice. Neurons were ordered by the time of their peak firing rates. Middle, temporal distribution of neurons. Green lines indicate delay-onset. Red lines indicate expected delay-offset. Bottom, correlation matrix of population vectors as a function of time for CA1 delay-act cells in cKO and control mice. (E) NMDAR deficiency disrupted the negative skew in the firing rate ratio of delay-active cells. Ratio of firing rates of delay-active cells in CA1 of cKO and WT mice. Dots indicate individual data for cKO (gray) and control (black) mice. The central bar indicates the median. *, p<0.05; Mann–Whitney’ U test.
-
Figure 8—source data 1
Source Data File for Figure 8A and E.
- https://cdn.elifesciences.org/articles/52466/elife-52466-fig8-data1-v2.xlsx
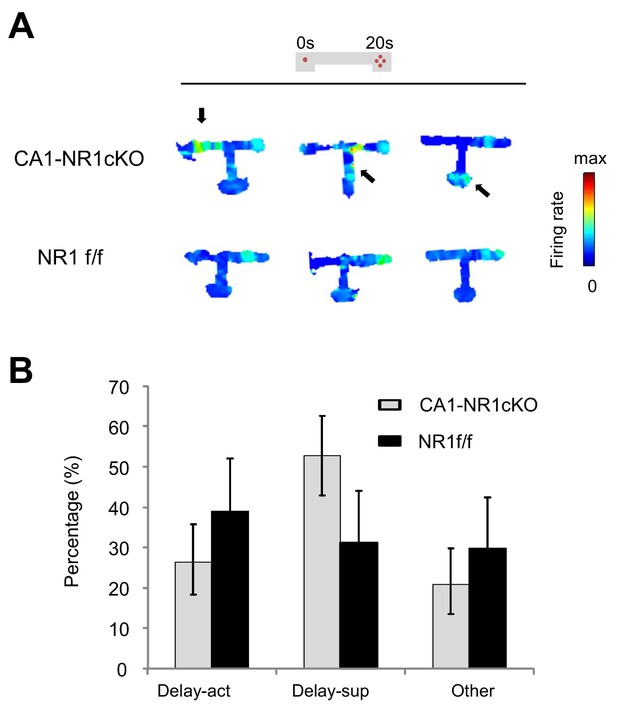
Mutant mice (CA1-NR1cKO) exhibiting impaired delay discounting showed less specific spatial representation in place cell activities and less in the number of delay-active cells.
(A) Spatial representation of example neurons from delay-act in CA1-NR1cKO (upper) and control mice (lower). Arrows indicate areas with non-specific firing. (B) Percentages of delay-act and delay-sup cells in mutant and control mice. Error bars indicate 95% Clopper-Pearson’s confidence intervals.

Moving patterns and speed during delay.
Left: Moving traces of single trial on delay 5, 10, 20 and 40 s conditions. Right: Moving speed corresponding to the trials shown in the left traces. Initial high speed movements (around 20-24 cm/s) occurred by starting a trial are followed by constant intermitted speed movements (around 16 cm/s). Note that there is no obvious difference in moving speed among the delay 5, 10, 20 and 40 s conditions. Data from mouse M9.
Tables
The number of delay-active and delay-suppressed CA1 excitatory and inhibitory neurons recorded from all sessions.
| Cell type | Delay-active | Delay-suppressed | Other | Total |
|---|---|---|---|---|
| Excitatory neurons | 243 | 313 | 83 | 639 |
| Inhibitory neurons | 43 | 100 | 26 | 169 |
Full distribution of CA1 excitatory neurons for all of the tested conditions.
| Test conditions | Delay responsiveness | Neuron number |
|---|---|---|
| Extension | Delay-active | 58 |
| Delay-suppressed | 83 | |
| Other | 36 | |
| Switched or both-side | Delay-active | 155 |
| Delay-suppressed | 191 | |
| Other | 34 | |
| Reward loss or gain | Delay-active | 30 |
| Delay-suppressed | 39 | |
| Other | 13 |
Distribution of side-dependent and side-independent, delay-active and delay-suppressed, CA1 excitatory and inhibitory neurons.
| Delay responsibility | Side-dependency | Cell types | N | % |
|---|---|---|---|---|
| Delay-active | Side-dependent | Excitatory neuron | 114 | 73.5 |
| Inhibitory neuron | 12 | 60.0 | ||
| Side-independent | Excitatory neuron | 41 | 26.5 | |
| Inhibitory neuron | 8 | 40.0 | ||
| Delay-suppressed | Side-dependent | Excitatory neuron | 124 | 64.9 |
| Inhibitory neuron | 45 | 71.4 | ||
| Side-independent | Excitatory neuron | 67 | 35.1 | |
| Inhibitory neuron | 18 | 28.6 |
Full distribution of CA1 excitatory neurons for the NMDAR mutant study.
The numbers in parentheses are cells from the wildtype.
| Test conditions | Delay responsiveness | Neurons | |
|---|---|---|---|
| cKO | Control | ||
| Extension | Delay-active | 28 | 25 |
| Delay-suppressed | 56 | 20 | |
| Other | 22 | 19 | |
| Reward loss and gain | Delay-active | 8 | 33 (30) |
| Delay-suppressed | 6 | 0 | |
| Other | 3 | 2 | |
Ages of CA1-NMDAR cKO mutant and control mice used for the electrophysiological study.
| Genotype | Animal ID | Age at surgery | Age at experiments ended |
|---|---|---|---|
| CA1-NR1 cKO (CaMK2-Cre; NR1-flox/flox) | M18 | 2 months | 3 months |
| M28 | 3 months | 3 months | |
| M30 | 3 months | 4 months | |
| Control (NR1-flox/flox) | M24 | 5 months | 5 months |
| M26 | 3 months | 4 months | |
| M29 | 3 months | 4 months | |
| M31 | 4 month | 5 months |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6J | RIKEN Bio Resource Center | RRID: IMSR_JAX:000664 | Wild-type mouse |
| Strain, strain background (Mus musculus) | NR1flox | PMID: 8980237 | 005246 (Jackson Laboratory) | Targeted mutation line |
| Strain, strain background (Mus musculus) | Tg(Camk2a-cre)T29-1Stl/J | PMID: 8980237 | 005359 (Jackson Laboratory) | Cre transgenic line |
| Strain, strain background (Mus musculus) | Tg(Camk2a-cre)T29-1Stl/J, NR1flox/flox | PMID: 8980237 | RRID: MGI:3581524 | Conditional knockout line |
| Commercial assay or kit | T-maze | O’hara and Co., Ltd. | RRID: SCR_018016 | Automatic operant test |
| Other | Neural probes | NeuroNexus | A4 × 2-tet-5mm-150-200-312 | 32-ch electrode |
| Other | nDrive | NeuroNexus | RRID: SCR_018019 | Micro driver to control movement of electrode |
| Other | Amplipex: KJE-1001 | Amplipex | RRID: SCR_018017 | Recording system for neural signals |
| Software, algorithm | MATLAB_R2018a | Mathworks | RRID: SCR_001622 | |
| Software, algorithm | Klusters | PMID: 16580733 | RRID:SCR_015533 | |
| Software, algorithm | NDmanager | PMID: 16580733 | RRID:SCR_015533 | |
| Software, algorithm | Neuroscope | PMID: 16580733 | RRID:SCR_015533 | |
| Software, algorithm | KlustaKwik2 | PMID: 25149694 | RRID:SCR_014480 | |
| Sequence-based reagent | Cre_F | PMID: 28244984 | PCR primers | ACC TGA TGG ACA TGT TCA GGG ATC G |
| Sequenced-based reagent | Cre_R | PMID: 28244984 | PCR primers | TCC GGT TAT TCA ACT TGC ACC ATG C |
| Sequenced-based reagent | NR1flox-F | PMID: 28244984 | PCR primers | TGT GCT GGG TGT GAG GGT TG |
| Sequenced-based reagent | NR1flox-R | PMID: 28244984 | PCR primers | GTG AGC TGC ACT TCC AGA AG |
| Other | DAPI stain | ThermoFisher | Thermo Fisher Scientific Cat# D1306 | (1 µg/mL) |
| Other | DiI stain | ThermoFisher | Thermo Fisher Scientific Cat# D3911 | (200 µg/mL) |