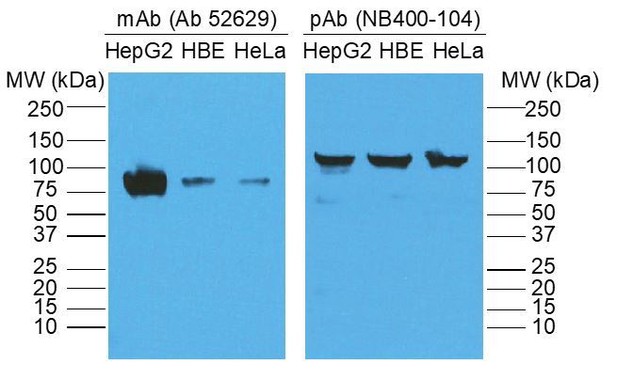
Identification of scavenger receptor B1 as the airway microfold cell receptor for Mycobacterium tuberculosis
Figures
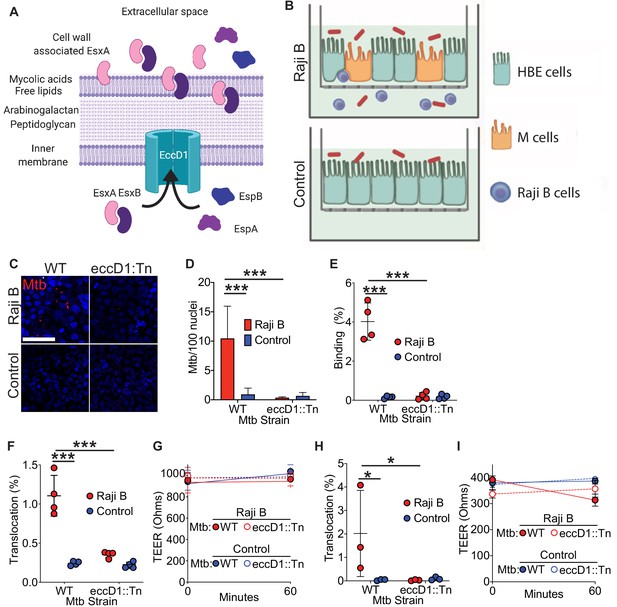
Mtb T7SS is necessary to mediate binding and translocation across M cells.
(A) Model of Mtb T7SS. (B) Schematic of human airway M cell transwell model. (C,D) Control and HBE/RajiB transwells were incubated with Mtb strains at 4°C for 1 hr and binding was analyzed by confocal microscopy (C) with quantification of bacterial number (D). Scale bar, 20 µm. (E) Control and HBE/RajiB transwells were incubated with Mtb strains at 4°C for 1 hr and lysed to determine binding by quantifying bacterial CFU and comparing with the initial inoculum. (F) Control and HBE/RajiB transwells were incubated with Mtb strains at 37°C for 1 hr and bacterial translocation was determined by quantifying bacterial CFU from the basal compartment and comparing with the inoculum. (G) TEER measurements from transwells from (F). (H) Caco-2/Raji B transwells were infected as described in F. (I) TEER measurements from transwells from (H). Experiments shown are representative of at least three independent experiments. *p<0.05, ***p<0.0005 as determined by one-way ANOVA. Where not shown, comparisons were not significant.
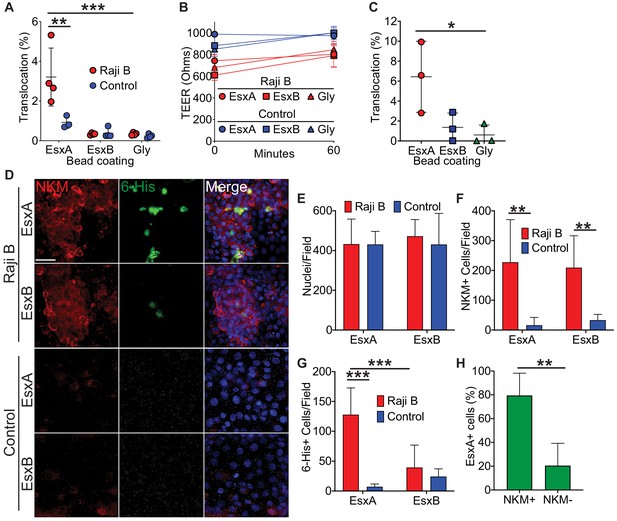
Mtb EsxA is sufficient to mediate binding and translocation across M cells.
(A) Control and HBE/RajiB transwells were incubated with fluorescent beads coated with EsxA, EsxB, or glycine. Translocation was determined by comparing the number of beads in the basal compartment with the inoculum. (B) TEER measurements from transwells from (A). (C) Caco-2/Raji B transwells were treated as described in A. (D) Control and HBE/RajiB transwells were incubated with recombinant EsxA or EsxB and stained with NKM 16-2-4 (red) and an anti-6-His antibody (green). Scale bar, 30 µm. (E–G) Quantification of nuclei (E), NKM 16-2-4+ cells (F), and EsxA+ (G) cells from the transwells described in (D). (H) Quantification of NKM 16-2-4 staining on EsxA+ cells from the HBE/Raji B transwells described in (D). Experiments shown are representative of at least three independent experiments. *p<0.05, **p<0.005, ***p<0.0005 as determined by one-way ANOVA. Where not shown, comparisons were not significant.
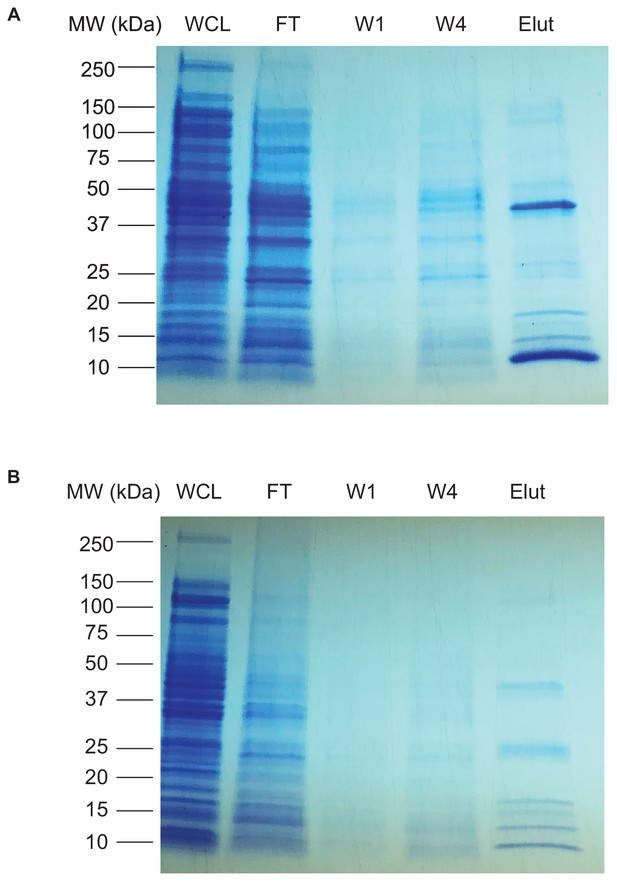
Coommassie stain of EsxA and EsxB purification.
(A,B) Whole cell lysate (WCL), flow-through (FT), wash 1 (W1), wash 4 (W4), and elution fractions (Elut) were obtained during EsxA (A) and EsxB (B) purification. Fractions were analyzed using SDS-PAGE followed by Coommassie Blue staining.
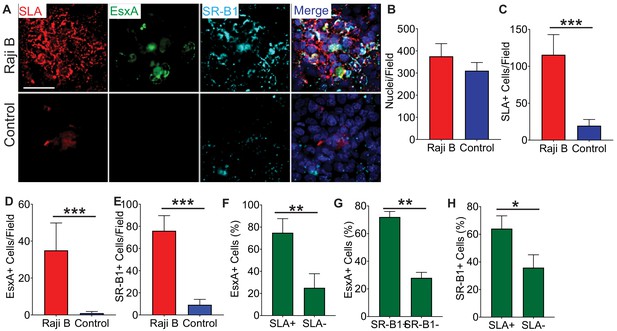
Recombinant EsxA and SR-B1 colocalize with the M cell marker Sialyl LewisA on Raji B-treated transwells.
(A) HBE/Raji B or control transwells were incubated with biotinylated EsxA and stained with anti-Sialyl LewisA (red), anti-SR-B1 (cyan), and Alexa Fluor 488 conjugated streptavidin (green). Scale bar, 30 µm. (B–E) Multiple images of the transwells described in (A) were taken and the number of nuclei (B), SLA+ (C), EsxA+ (D), and SR-B1+ (E) cells was determined using ImageJ. (F–H) Expression of SLA on EsxA+ cells (F), of SR-B1 on EsxA+ cells (G), and the expression of SLA on SR-B1+ cells (H) on HBE/Raji B transwells was determined by ImageJ. *p<0.05, **p<0.005, ***p<0.0005 as determined by one-way ANOVA. Where not shown, comparisons were not significant.
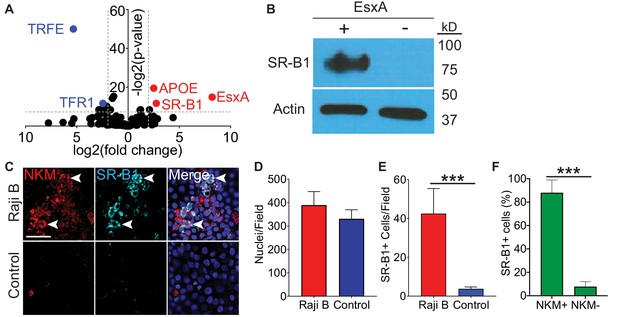
SR-B1 is the M cell EsxA receptor.
(A) Volcano plot displaying peptides enriched when Caco-2 cells were treated either with transferrin (blue dots on left) or EsxA (red dots on right). Results are of a single experiment with three biologic replicates per condition. (B) Western blot using an anti-SR-B1 antibody (top) or an anti-actin antibody (bottom) of proteins enriched after HBE cells were incubated with biotinylated EsxA or control. (C) Control and HBE/RajiB transwells were stained with NKM 16-2-4 and an anti-SR-B1 antibody and analyzed by confocal microscopy. Arrows denote examples of double positive cells. Scale bar, 40 µm. (D,E) Quantification of the number of nuclei (D) or SR-B1+ cells (E) from the transwells described in (C). (F) Quantification of NKM 16-2-4 staining on SR-B1+ cells from the HBE/Raji B transwells described in (C). Experiments shown are representative of at least three independent experiments. ***p<0.0005 as determined by one-way ANOVA. Where not shown, comparisons were not significant.
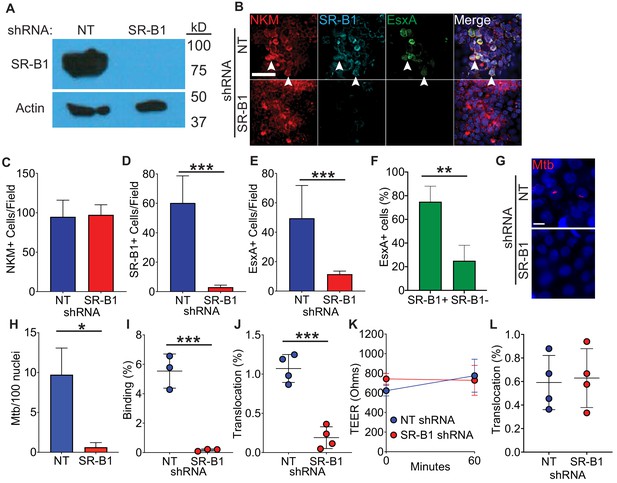
Loss of SR-B1 reduces EsxA binding and Mtb translocation through M cells.
(A) Western blot of SR-B1 (top) or beta-actin (bottom) of shRNA expressing HBE cells. (B) HBE/RajiB transwells with shRNA expressing HBE cells were incubated with biotinylated EsxA and stained with NKM 16-2-4 (red), anti-SR-B1 (cyan), and Alexa Fluor 488 conjugated streptavidin (green). Arrows denote examples of triple positive cells. Scale bar, 40 µm. (C–E) Quantification of the number of NKM+ (C), SR-B1+ cells (D) and EsxA+ cells (E) on transwells described from (B). (F) Quantification of SR-B1 staining on EsxA+ cells from HBE NT shRNA/Raji B transwells described in (B). (G,H) HBE/RajiB transwells with shRNA expressing HBE cells were incubated with mCherry Mtb and Mtb binding was analyzed by confocal microscopy (G) with quantification of bacterial number (H). Scale bar, 10 µm. (I) HBE/RajiB transwells with shRNA expressing HBE cells were incubated with Mtb strains at 4°C and lysed to determine binding by quantifying bacterial CFU and comparing with the initial inoculum. (J) HBE/RajiB transwells with shRNA expressing HBE cells were incubated with Mtb strains at 37°C and bacterial translocation was determined by quantifying bacterial CFU from the basal compartment and comparing with the inoculum. (K) TEER of the transwells from (J). (L) HBE/RajiB transwells with shRNA expressing HBE cells were incubated with Pseudomonas aeruginosa at 37°C and bacterial translocation was determined by quantifying bacterial CFU from the basal compartment and comparing with the inoculum. Experiments shown are representative of at least three independent experiments. *p<0.05, **p<0.005, ***p<0.0005 as determined by one-way ANOVA. Where not shown, comparisons were not significant.
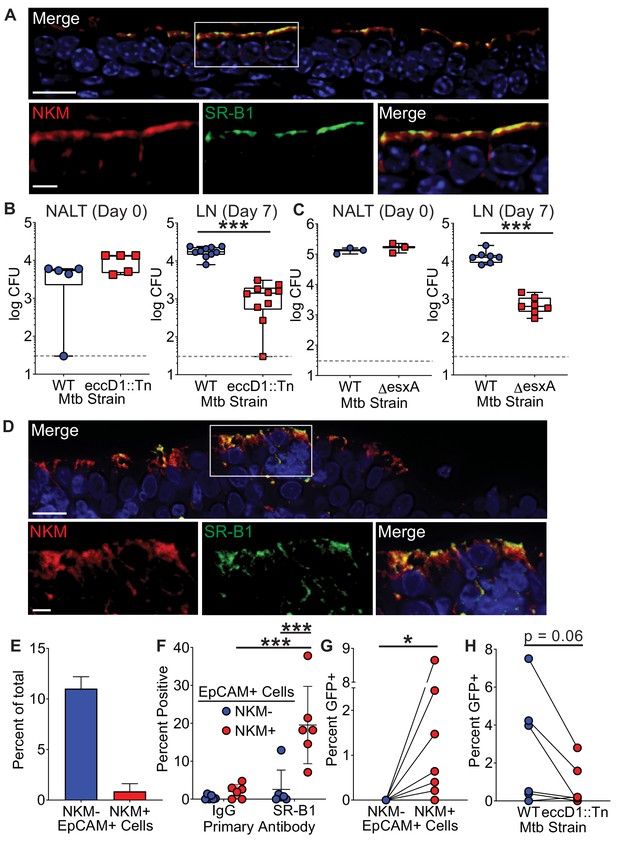
The Mtb type VII secretion system is necessary for Mtb entry in mice and humans.
(A) Mouse NALT sections were stained with NKM 16-2-4 and anti-SR-B1 antibodies and analyzed by confocal microscopy. Scale bar, top, 15 µm, bottom, 5 µm. (B,C) Mice were intranasally infected with either WT Mtb, Mtb eccD1::Tn (B), or MtbΔesxA (C). CFU was determined in the NALT on day 0 (left) or in the cervical lymph nodes on day 7 (right). Symbols represent CFU from individual animals (n = 8–10 per strain). ***p<0.0005 compared to WT by Mann-Whitney U test. (D) Human adenoid sections were stained with NKM 16-2-4 and anti-SR-B1 antibodies and analyzed by confocal microscopy. Scale bar, top, 15 µm, bottom, 5 µm. (E) Human adenoids were disaggregated, stained with NKM 16-2-4 and anti-EpCAM antibodies, and analyzed by flow cytometry. (F) Human adenoids were treated as in (E), stained with anti-SR-B1 or control IgG antibodies and analyzed by flow cytometry. Symbols represent adenoids from individual donor (F–H). **p<0.005, Wilcoxon matched pairs signed rank test. (G) Human adenoids were infected with GFP+ Mtb, disaggregated, immunostained and analyzed by flow cytometry to determine the proportion of GFP+ Mtb containing NKM+/EpCAM+ and NKM-/EpCAM+ cells. *p<0.05, Wilcoxon matched pairs signed rank test. (H) Human adenoids were infected with GFP+ Mtb or GFP+ Mtb eccD1::Tn. The percentage of GFP+ Mtb containing NKM+/EpCAM+ double positive cells was determined by flow cytometry. The Wilcoxon matched pairs signed rank test was used for comparison.
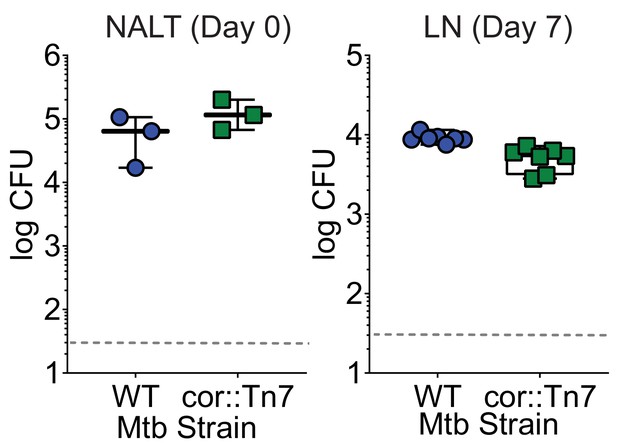
Mtbcor::Tn7 does not display a translocation defect following a mouse intranasal infection Mice were intranasally infected with either WT Mtb or Mtbcor::Tn7.
CFU was determined in the NALT on day 0 (left) or in the cervical lymph nodes on day 7 (right). Symbols represent CFU from individual animals (n = 8 per strain).
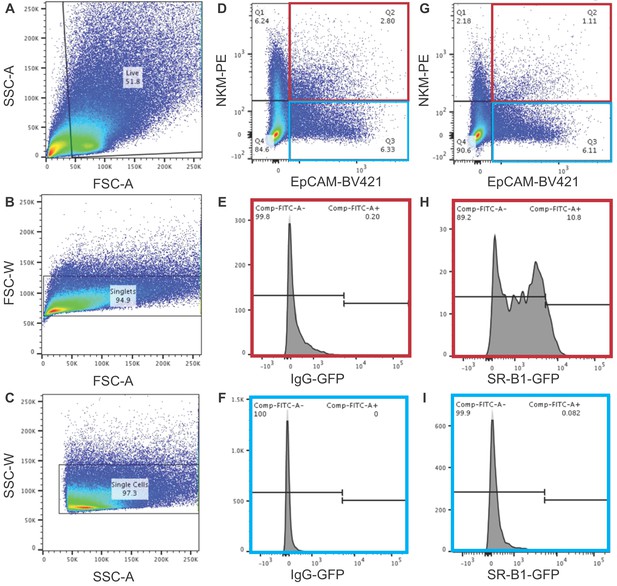
Adenoid gating strategy to determine SR-B1 positive cells.
(A–C) Gating strategy for unstained cells. Debris was excluded using FSC-A and SSC-A to yield the live cell population (A). Single cells were identified using the FSC-A and FSC-W (B) and singlets were identified using SSC-A and SSC-W (C). Gates for stained samples were established using unstained samples. (D–F) Adenoids were stained with mouse BV421 conjugated anti-EpCAM, mouse PE conjugated NKM 16-2-4, and a rabbit IgG followed by a donkey-anti-rabbit 488 conjugated antibody. Using the gating strategy described in (A–C), NKM+/EpCAM+ cells (highlighted in red) and NKM-/EpCAM+ cells (highlighted in blue) were analyzed for fluorescence in the green channel. (G–I) Adenoids were stained with mouse BV421 conjugated anti-EpCAM, mouse PE conjugated NKM 16-2-4, and rabbit anti-SR-B1 followed by a donkey-anti-rabbit 488 conjugated antibody. Using the gating strategy described in (A–C), NKM+/EpCAM+ cells (highlighted in red) and NKM-/EpCAM+ cells (highlighted in blue) were analyzed for fluorescence in the green channel.
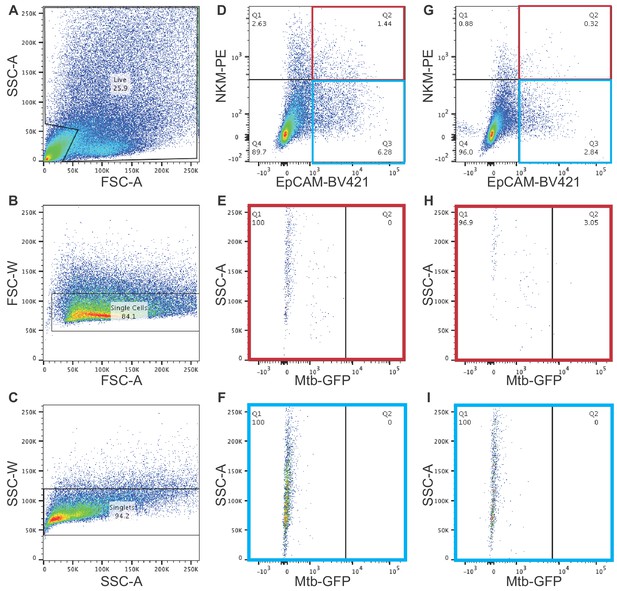
Adenoid gating strategy to determine GFP+ Mtb containing cells.
(A–C) Gating strategy for unstained cells. Debris was excluded using FSC-A and SSC-A to yield the live cell population (A). Single cells were identified using the FSC-A and FSC-W (B) and singlets were identified using SSC-A and SSC-W (C). Gates for stained samples were established using unstained samples. (D–F) Adenoids were infected with a vehicle control, disaggregated, and stained with mouse BV421 conjugated anti-EpCAM and mouse PE conjugated NKM 16-2-4. Using the gating strategy described in (A–C), NKM+/EpCAM+ cells (highlighted in red) and NKM-/EpCAM+ cells (highlighted in blue) were analyzed for fluorescence in the green channel. (G–I) Adenoids were infected with GFP Mtb, disaggregated, and stained with mouse BV421 conjugated anti-EpCAM and mouse PE conjugated NKM 16-2-4. Using the gating strategy described in (A–C), NKM+/EpCAM+ cells (highlighted in red) and NKM-/EpCAM+ cells (highlighted in blue) were analyzed for fluorescence in the green channel.
Additional files
-
Supplementary file 1
Variant table for WT Mtb, Mtb eccD1::Tn and MtbΔesxA.
Genomic DNA from the indicated strains was sequenced and variants identified by comparison to the Mtb Erdman reference genome.
- https://cdn.elifesciences.org/articles/52551/elife-52551-supp1-v3.xls
-
Supplementary file 2
Mass spectrometry peptide analysis of transferrin or EsxA cell surface binding.
TriCEPS-labeled transferrin or EsxA were affinity purified after binding to the surface of Caco-2 cells, and bound proteins analyzed by mass spectrometry.
- https://cdn.elifesciences.org/articles/52551/elife-52551-supp2-v3.xls
-
Supplementary file 3
Key resources table.
- https://cdn.elifesciences.org/articles/52551/elife-52551-supp3-v3.xls
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52551/elife-52551-transrepform-v3.docx