Rationally derived inhibitors of hepatitis C virus (HCV) p7 channel activity reveal prospect for bimodal antiviral therapy
Figures
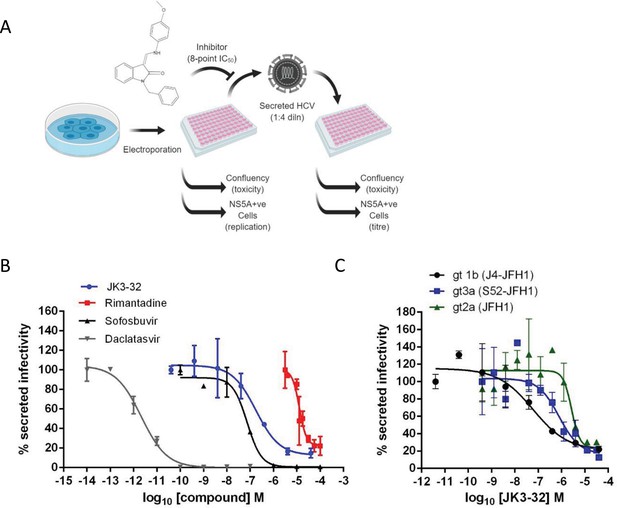
Activity of JK3/32 against HCV particle secretion.
(A) Diagram of workflow for rapid throughput assay for secreted infectivity (generated using ‘Biorender’, https://app.biorender.com). (B) Comparison of JK3/32 potency vs. virion secretion of J4/JFH-1 with licensed HCV DAAs sofosbuvir and daclatasvir, as well as the prototypic adamantane viroporin inhibitor, rimantadine. Curves are representative of at least four experimental repeats for JK3/32, multiple for sofosbuvir and daclatasvir, and two for rimantadine, where each condition is carried out in quadruplicate and error bars represent standard deviations. (C) Comparative EC50 curves for JK3/32 effects upon GT1b, 2a and 3a chimaeric HCV (J4, JFH-1, S52/JFH-1) secreted infectivity post-electroporation. Curves are again representative of multiple experiments and error bars show standard deviations between quadruplicate repeats.
-
Figure 1—source data 1
Raw data from EC50 curves.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig1-data1-v4.zip
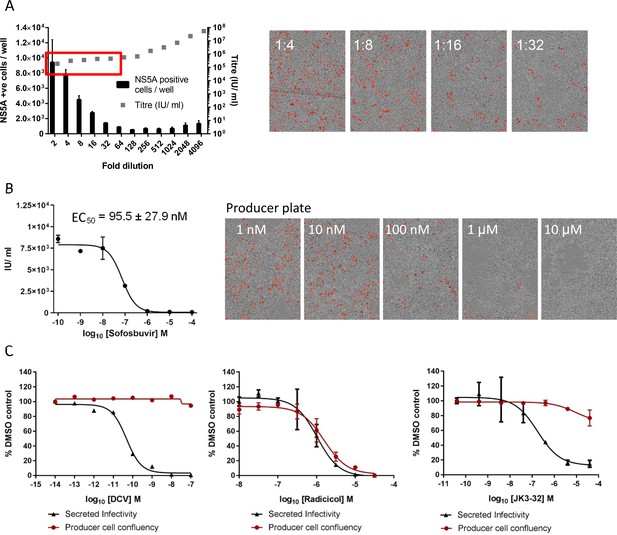
Optimised quantitation of secreted HCV infectivity using the IncuCyte Zoom.
(A) Dilutions of infectious supernatants were assessed for optimal signal/noise whilst remaining within the linear range of quantitation (red box). Appropriate dilutions for untreated virus were repeated alongside every inhibitor EC50 experiment to ensure that titres could be back-calculated accurately; each condition repeated in quadruplicate across multiple experiments, error bars show standard deviation. Example IncuCyte images are shown for comparison. (B) EC50 and corresponding IncuCyte images of sofosbuvir (SOF) treated producer cells stained for NS5A, correlating secreted infectivity with producer cell replication. (C) Determination of cytotoxic effects by producer cell confluency. In each case, infectivity (black line) was assessed alongside producer cell confluency (red line) as an indicator of viability (MTT assays were also conducted for compounds showing effects in initial screens). Examples shown for a control DAA daclatasvir (DCV), the Hsp90 inhibitor radicicol as a Huh7 cytotoxic agent, and the lead p7i JK3-32. Error bars show standard deviation for quadruplicate samples in each representative experiment.
-
Figure 1—figure supplement 1—source data 1
Raw data for EC50 titrations.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig1-figsupp1-data1-v4.zip
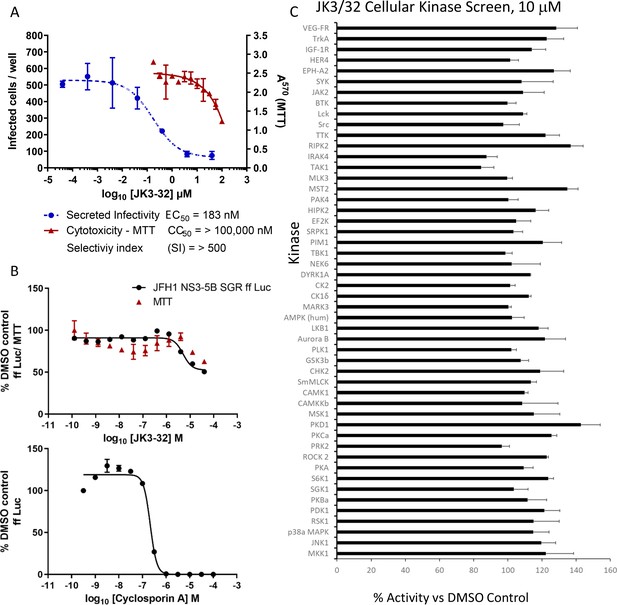
Screening for potential JK3/32 off-target effects.
(A) Summary data establishing the JK3/32 selectivity index with respect to cytotoxicity measured by MTT assay. Each condition in quadruplicate across multiple experiments (see Figure 2—figure supplement 1—source data 1), error bars show standard deviation. (B) Top - Lack of JK3/32 activity against replication (firefly luciferase activity) of HCV subgenomic replicon (JFH-1), bottom – efficient blockade of replicon replication using cyclosporine A (CsA). (C) MRC PPU cellular kinase screen (see Materials and methods) testing 10 μM JK3/32,~20 x the EC50 concentration versus particle secretion. Error bars represent standard deviations for two experimental repeats.
-
Figure 1—figure supplement 2—source data 1
Raw data for replicon and cyclosporine validations.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig1-figsupp2-data1-v4.zip
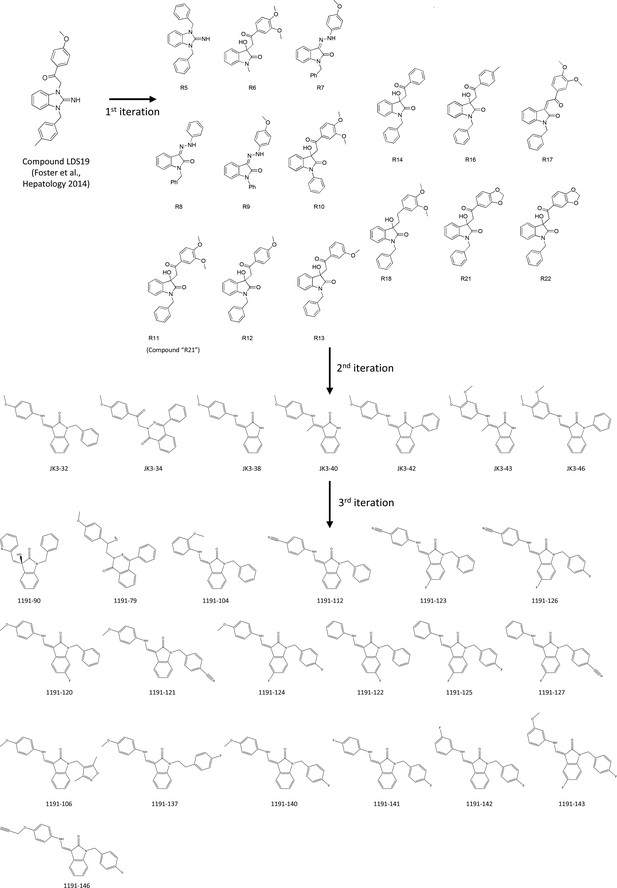
Evolution of LDS19 compound series including JK3/32.
LDS19 was identified from an in silico screen using a 3ZD0-based channel structure model and demonstrated to have potent anti-p7 activity in vitro and in cell culture, as described previously (Foster et al., 2011). Three subsequent iterations of compound structure variations were undertaken to establish a comprehensive SAR and JK3/32 was established as the series lead. Compound RS11 (aka R21) was used as a negative control in cell entry assays due to a lack of activity versus genotype 1b and 3a HCV.
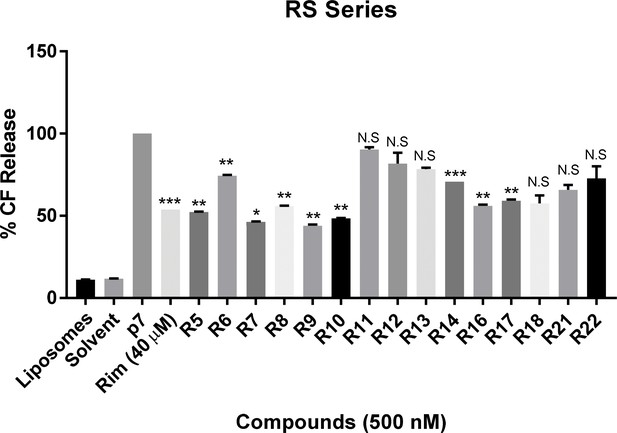
In vitro dye release assay screen for activity of RS compounds versus genotype 1b p7.
RS series compounds were tested at a concentration of 500 nM versus recombinant J4 strain genotype 1b p7, alongside a positive control for inhibition (40 μM rimantadine). End-point fluorescence values from two experimental repeats were normalised against protein in the presence of no drug (100%). Error bars represent standard deviations (***p≤0.0001, **p≤0.001, *p≤0.05, Student's t-test, n = 2).
-
Figure 1—figure supplement 4—source data 1
Raw data for RS series iteration dye release assays.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig1-figsupp4-data1-v4.zip
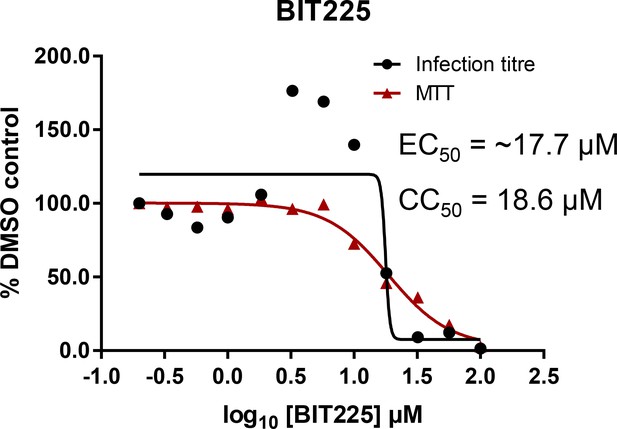
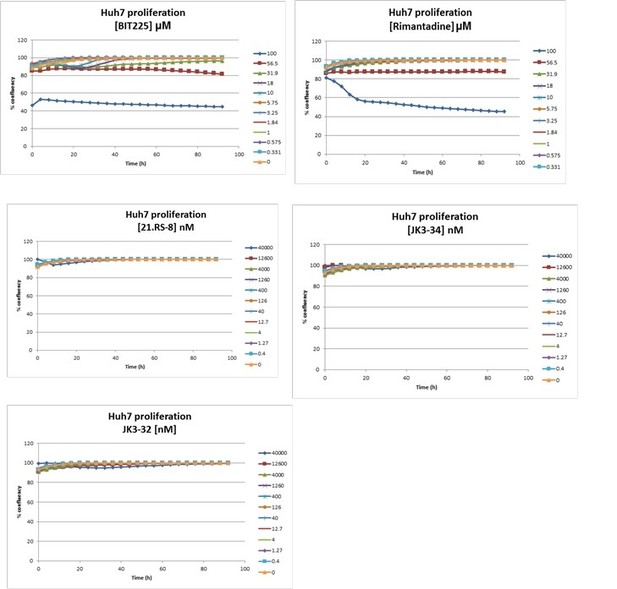
Effects of clinically advanced viroporin inhibitor, BIT225, versus particle secretion.
The amiloride derivative BIT225 was tested for effects against secreted HCV infectivity (J4/JFH-1 genotype 1b chimaera) using the optimised IncuCyte protocol. Conditions in quadruplicate, error bars show standard deviation. Calculated EC50 was ~17.7 μM, whereas CC50 was near identical at 18.6 μM.
-
Figure 1—figure supplement 5—source data 1
Raw data for BIT225 EC50 curve.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig1-figsupp5-data1-v4.zip
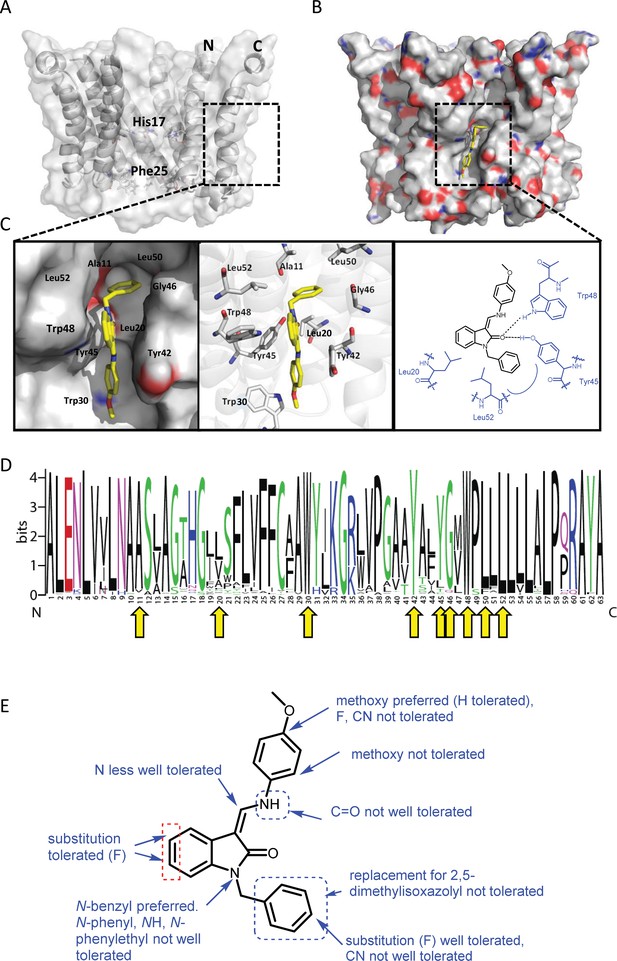
Predicted interactions of JK3/32 with genotype 1b p7 heptamer complexes.
(A) Cutaway image of PDB: 3ZD0-based heptamer showing orientation of N- and C-terminal helices, predicted gating residue (Phe25) and proton sensor (His17). Box shows approximate region corresponding to peripheral drug-binding site. (B) Space-filling model of PDB: 3ZD0-based heptamer showing basic (blue) and acidic (red) charge distribution and positioning of JK3/32 (yellow) within peripheral binding site (box). (C) Zoomed images showing peripheral drug-binding site and predicted energetically preferred binding pose (in Glide) for JK3/32 (yellow) within membrane-exposed binding site as space fill (left), side chains (middle) and key interactions, including with Tyr45 and Trp48 (right). (D) Amino acid conservation within p7 across ~1500 sequences from the EU HCV database. Height corresponds to relative conservation of individual residues (quantified in Table 1). Yellow arrows indicate residues predicted to form direct interactions with JK3/32. (E) Summary of the structure-activity relationships for the JK3/32 series of inhibitors (Table 1) for their effects upon chimaeric GT1b HCV (J4/JFH-1) secreted infectivity following electroporation.
-
Figure 2—source data 1
Jalview analysis data for EU HCV database p7 sequences.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig2-data1-v4.zip
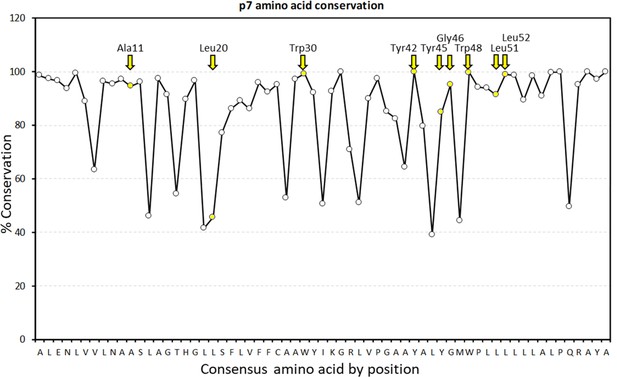
Graphical representation of percentage conservation for consensus p7 sequence (see Figure 2—figure supplement 1—source data 1 for numerical summary).
JK3/32 peripheral binding site loci are highlighted in yellow and by yellow named arrows for residues present within the J4 p7 sequence.
-
Figure 2—figure supplement 1—source data 1
Raw data for p7 sequence alignments.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig2-figsupp1-data1-v4.zip
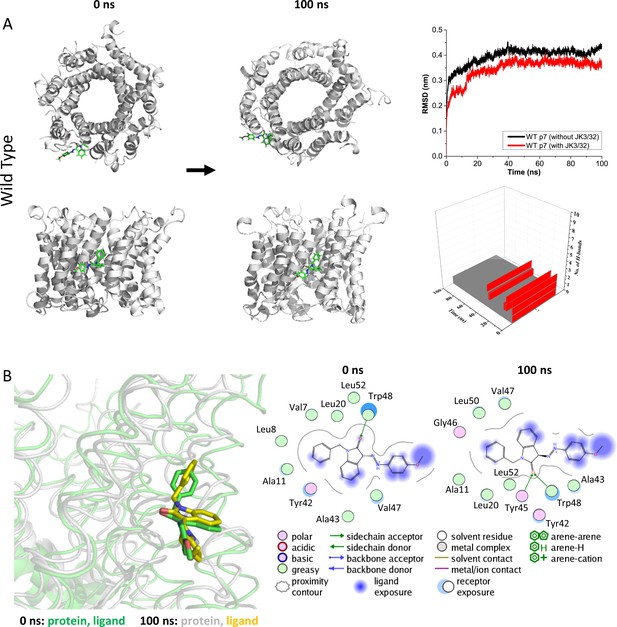
Atomistic simulations of JK3/32 interaction with PDB: 3ZD0-based heptamers.
(A) 100 ns atomistic molecular dynamics simulation of p7 complexes bound to JK3/32, starting from a minimised pose in a hydrated lipid bilayer environment. Top-down and side views of complexes at the start and end of simulation are shown with a single molecule of JK3/32 bound at the membrane-exposed binding site. Graphs on the right show the root mean-squared variation (RMSD) over time (top) in the presence (red) or absence (black) of ligand, and the number of H-bonds formed throughout the simulation (bottom). (B) Overlay of p7 protein structures and JK3/32 orientation at 0 ns and 100 ns (left) alongside molecular interactions at the two time points (right); note bifurcation of H-bond interaction between indole carboxyl and Trp48 (0 ns) to both Trp48 and Tyr45 (100 ns).
-
Figure 3—source data 1
Output data for MD simulations for Jk3/32 with wild type/L20F p7.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig3-data1-v4.zip
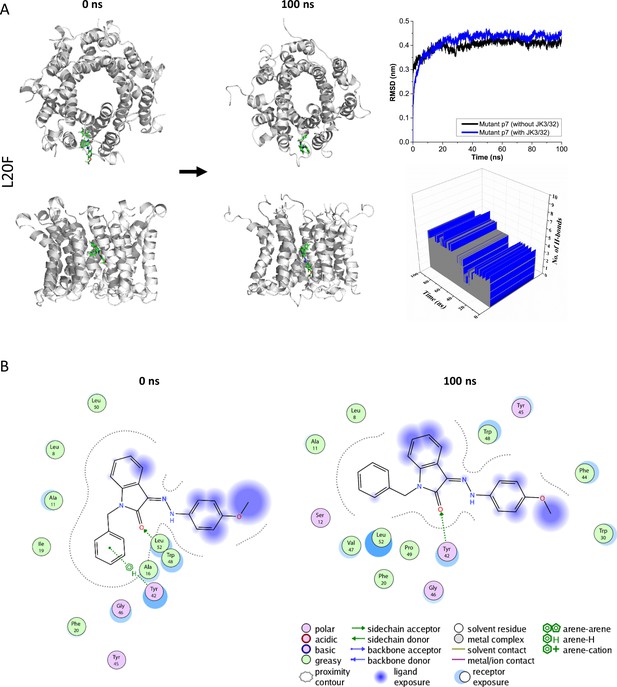
Atomistic simulations of JK3/32 interaction with PDB: 3ZD0-based heptamers containing Leu20Phe polymorphism.
(A) 100 ns atomistic molecular dynamics simulation of p7-L20F complexes bound to JK3/32, starting from a minimised pose in a hydrated lipid bilayer environment. Top-down and side views of complexes at the start and end of simulation are shown with a single molecule of JK3/32 bound at the membrane-exposed binding site. Graphs on the right show the root mean-squared variation (RMSD) over time (top) in the presence (red) or absence (black) of ligand, and the number of H-bonds formed throughout the simulation (bottom). (B) Molecular interactions at the two time points (right); note, crowding of the binding pocket leads to higher stable RMSD and an increased number of H-bonds formed over time compared with the wild type protein (see main Figure 3).
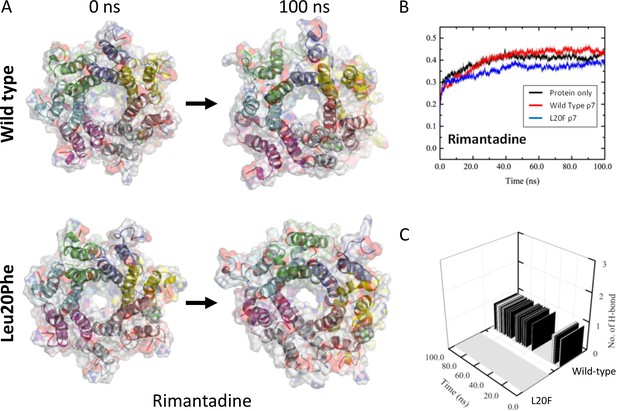
Molecular Dynamic Simulation of rimantadine with heptameric 3ZD0-derived p7 and resistant Leu20Phe complexes.
An L20F polymorphism was introduced into the genotype 1b p7 sequence in the context of the 3ZD0 heptamer, then subjected to energy minimisation within a hydrated lipid bilayer. (A) Location of rimantadine into the peripheral binding pocket led to stable, but weak interactions with wild type p7, whereas the Leu20Phe mutation caused a loss of binding interactions and migration of Rimantadine away from the pocket. This was also evident from RMSD traces (B), as well as the loss of H-bonds between Rimantadine and the protein (C).
-
Figure 3—figure supplement 2—source data 1
Output data for rimantadine MD simulation.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig3-figsupp2-data1-v4.zip
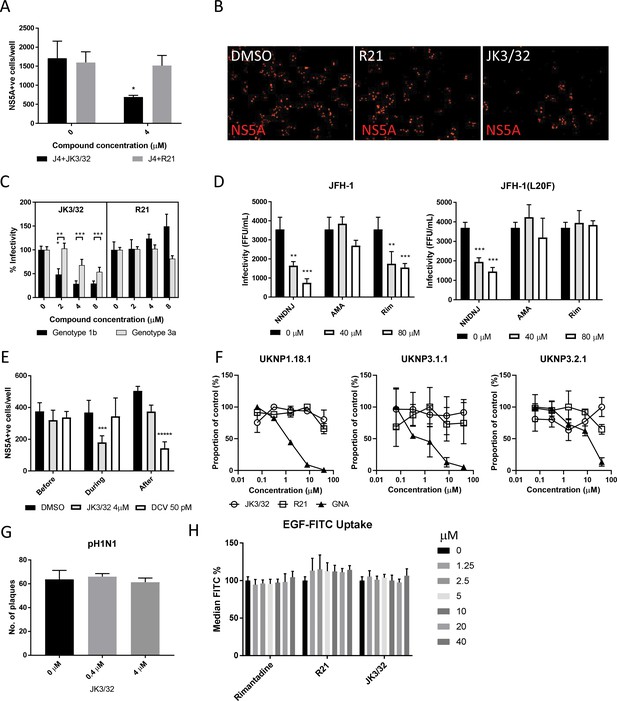
Characterisation of JK3/32 effects upon HCV entry.
(A) Infectivity following application of JK3/32, or the inactive R21 analogue, during entry of GT1b chimaeric HCV into Huh7 cells. Virus innoculae were pre-treated with either compounds or DMSO for 20 min at room temperature prior to application to Huh7 cells overnight. Cells were washed extensively and assessed for infectivity 48 hr post infection by NS5A immunofluorescence, quantified using the Incucyte Zoom. (*p≤0.05, Student's t-test, n = 2). (B) Representative images from Incucyte analysis of NS5A-stained Huh7 cells in A for comparison. (C) Titrated JK3/32 concentrations were assessed during entry as described in A, comparing chimaeric GT1b (J4/JFH-1) and GT3a (S52/JFH-1) viruses (***p≤0.001, Student's t-test, n = 2). (D) Effects of prototypic p7 channel blockers against wild type and rimantadine-resistant GT2a HCV (JFH-1) during entry into Huh7 cells (**p≤0.01, ***p≤0.001, Student's t-test, n = 2). (E) JK3/32 effects upon entry when added prior (2 hr), during (4 hr) or post-infection (48 hr) with chimaeric GT1b HCV (J4/JFH-1), compared to Daclatasvir NS5Ai (***p≤0.001, *****p≤0.00001, Student's t-test, n = 2). (F) Lack of effect of JK3/32 or R21 upon entry of Lentiviral HCV pseudotypes into Huh7 cells compared to GNA positive control. HCV envelopes were derived from genotype 1b (UKNP1.18.1) or 3a (UKNP3.1.1 and UKNP3.2.1) patients. Results are representative of two experiments. (G) Lack of effect for JK3/32 upon IAV entry into MDCK cells. (H) Lack of effect for JK3/32, R21 or rimantadine upon clathrin-mediated endocytosis, measured by fluorescent EGF uptake.
-
Figure 4—source data 1
Raw data for virus entry experiments.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig4-data1-v4.zip
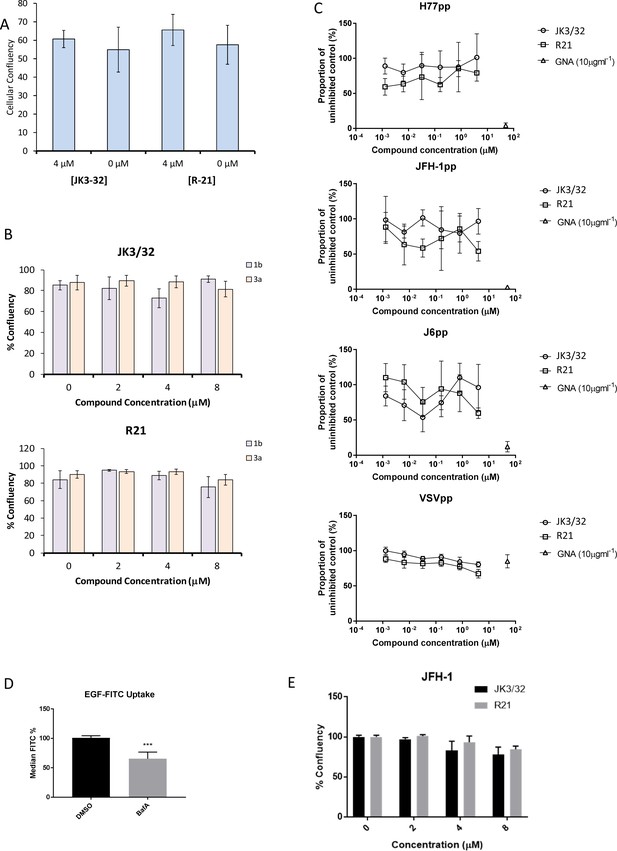
Additional control experiments for JK3/32 effects during HCV entry.
(A, B) Cell viability (Incucyte cell confluency) during treatment of Huh7 cells with JK3/32 or R21 during virus entry experiments shown in Figure 4a–c. (C) Additional Lentiviral pseudotype assays using HCV envelopes derived from standard laboratory strains (H77, J6 and JFH-1), as well as a VSV-G control. Treatment with JK3/32, R21 and GNA. (D) Positive control for EGF uptake assays using 1 μM Bafilomycin A. E. Cell viability controls as for A, B, for cells infected with JFH-1 (see Figure 5b).
-
Figure 4—figure supplement 1—source data 1
Raw data for virus entry control experiments.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig4-figsupp1-data1-v4.zip
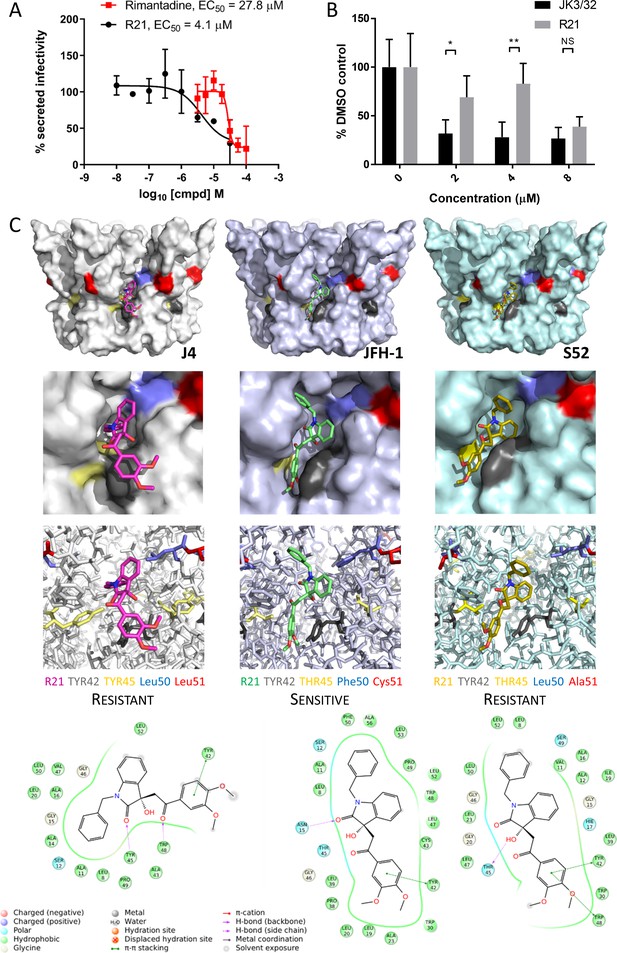
Susceptibility of genotype 2a JFH-1 HCV to R21 further supports a role during entry as well as peripheral site specificity.
(A) Comparison of R21 potency vs. virion secretion of JFH-1 with rimantadine. Curves are representative of two experimental repeats, where each condition was carried out in quadruplicate and error bars represent standard deviations. (B) Infectivity following application of JK3/32 or R21 during JFH-1 entry into Huh7 cells. Virus innoculae were pre-treated with compounds or DMSO for 20 min at room temperature prior to application to Huh7 cells overnight. Cells were washed extensively and assessed for infectivity 48 hr post infection by NS5A immunofluorescence, then quantified using the Incucyte Zoom. (*p≤0.05, **p≤0.01 Student's t-test, n = 2). (C) Predicted energetically preferred binding poses and residue interactions for R21 within the peripheral binding pocket for genotype 1b (J4), 2a (JFH-1) and 3a (S52) HCV p7 structure models. Key residues are colour coded as shown, including positions 45 (yellow) and 50 (blue), where variants show a strong predicted influence upon R21 binding.
-
Figure 5—source data 1
Raw data for R21 docking interactions.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig5-data1-v4.zip
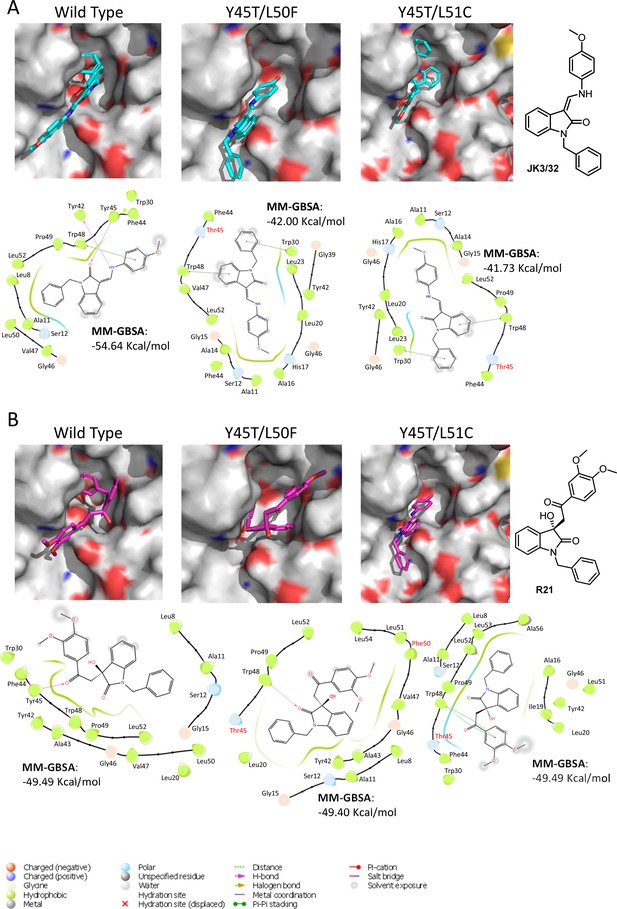
Predicted binding of JK3/32 and R21 to mutated J4 p7 channel complexes.
Structure homology models of the genotype 1b J4 p7 channel complex containing either Y45T/L50F or Y45T/L51C double mutations, were generated and energy-minimised using Maestro (Schrödinger). (A) JK3/32 was re-docked into the peripheral binding site for the wild type or mutated channel complexes using Glide (see Materials and methods), identifying interactions within the binding site. Predicted binding energies were calculated using MM-GBSA. (B) As for A, but for the R21 ligand.
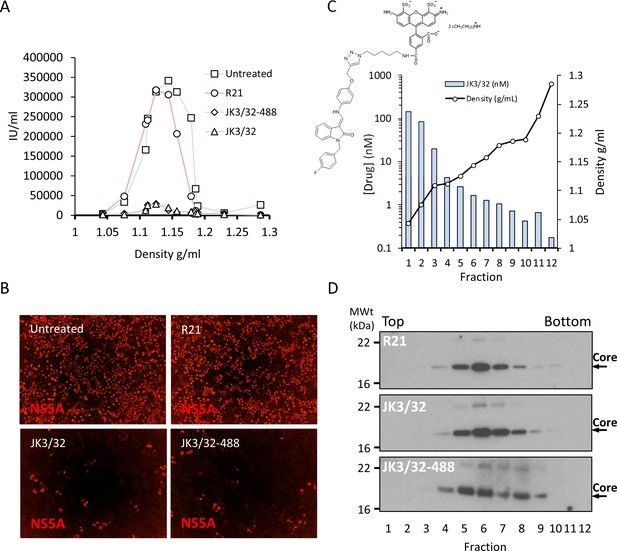
Direct JK3/32 virion dosing reduces HCV infectivity with minimal exposure of compound to cells.
Concentrated, purified high titre chimaeric GT1b HCV (J4/JFH-1) was incubated with DMSO, active JK3/32 (-488) or control compounds (R21) at 10 μM for 20 min prior to separation on a continuous 10–40% iodixinol/PBS density gradient followed by fractionation. (A) IncuCyte quantitation of NS5A immunofluorescence staining within each fraction expressed as infectious units (IU) per mL. (B) Immunofluorescence staining of HCV NS5A protein within naïve Huh7 cells 48 hr post-infection with 10 μL of fraction 6, corresponding to peak infectivity for untreated control gradients. (C) Calculated concentration of JK3/32-488 (chemical structure shown in inset) based upon fluorimetry within each of 12 fractions taken from the top of the gradient and corresponding fraction densities. (D) Western blot analysis of drug treated gradient fractions, probing for HCV core protein.
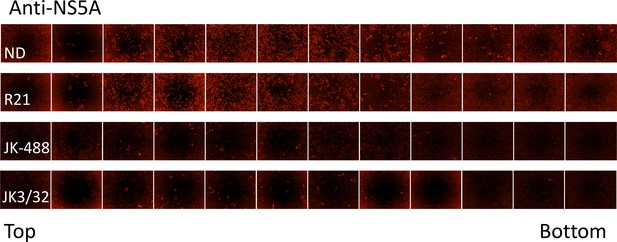
IncuCyte image set from iodixinol gradients shown in Figure 7.
10 μL of each gradient fraction was added to 10 000 naïve Huh7 cells within each well of a black-walled 96-well plate. 48 hr post-infection, cells were immunofluorescence stained for HCV NS5A protein and infectious units counted using the IncuCyte.
-
Figure 7—figure supplement 1—source data 1
Raw image data for gradient immunofluorescence.
- https://cdn.elifesciences.org/articles/52555/elife-52555-fig7-figsupp1-data1-v4.zip
Tables
SAR table for JK3/32 series showing compounds contributing directly.
Core oxindole scaffold for JK3/32 shown, indicating six R-groups (R1-R6) subjected to modification, as well as a position within the core structure (A).
| Activity vs gt1b p7 (J4-JFH1 HCV), 72 hr treatment | ||||||||
|---|---|---|---|---|---|---|---|---|
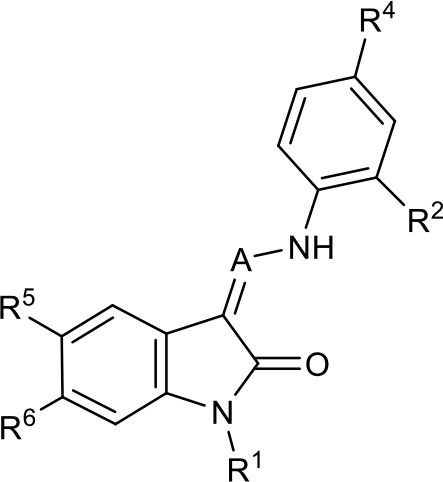 | ||||||||
| Compound | R1 | R2 | R4 | R5 | R6 | A | EC50 (µM) | CC50 (µM) |
| JK3-32 | Bzl | H | OMe | H | H | CH | 0.184 ± 0.089 (n = 4) | >100 |
| JK3-42 | Ph | H | OMe | H | H | CH | 0.932 ± 0.812 (n = 2) | >4 |
| JK3-38 | H | H | OMe | H | H | CH | >4 (n = 2) | >4 |
| 21-RS-7 | Bzl | H | OMe | H | H | N | 1.34 ± 0.37 (n = 3) | >4 |
| 21-RS-8 | Bzl | H | H | H | H | N | 0.775 ± 0.700 (n = 2) | >40 |
| 21-RS-9 | Ph | H | OMe | H | H | N | >40 (n = 1) | >40 |
| 1191–104 | Bzl | OMe | H | H | H | CH | 11.33 (n = 1) | >40 |
| 1191–112 | Bzl | H | CN | H | H | CH | >4 (n = 1) | 13.3 ± 12.4 (n = 2) |
| 1191–121 | Bzl | H | OMe | H | F | CH | 0.40 ± 0.12 (n = 2) | 12.8 |
| 1191–120 | 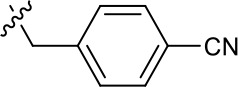 | H | OMe | H | H | CH | 1.42 ± 1.38 (n = 2) | 5.4 ± 4.4 (n = 2) |
| 1191–124 | 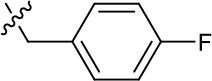 | H | OMe | F | H | CH | 0.32 ± 0.35 (n = 2) | 8.3 ± 3.9 (n = 2) |
| 1191–106 | 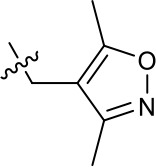 | H | OMe | H | H | CH | 11.58 (n = 1) | >40 |
| 1191–137 | 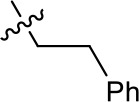 | H | OMe | H | H | CH | 2.30 (n = 1) | 12.5 |
| 1191–140 | 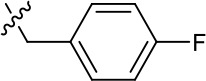 | H | OMe | H | H | CH | 0.461 (n = 1) | 10.4 |
| 1191–141 | 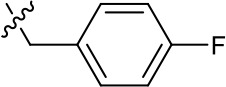 | H | F | H | H | CH | >1.26 | >1.26 |
| 1191–146 | 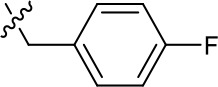 | H | 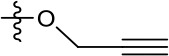 | H | H | CH | 2.48 (n = 1) | >12.6 |
| 1191–125 | 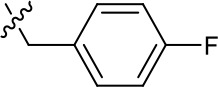 | H | H | F | H | CH | 0.209 (n = 1) | 12.9 |
| 1191–126 | 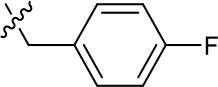 | H | CN | F | H | CH | 3.76 (n = 1) | >40 |
| Additional compounds: | ||||||||
| Compound | Structure | EC50 (µM) | CC50 (µM) | |||||
| 21-RS-17 | 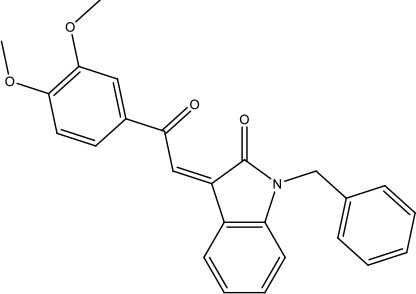 | 1.73 ± 1.68 (n = 2) | >4 | |||||
| R21 | 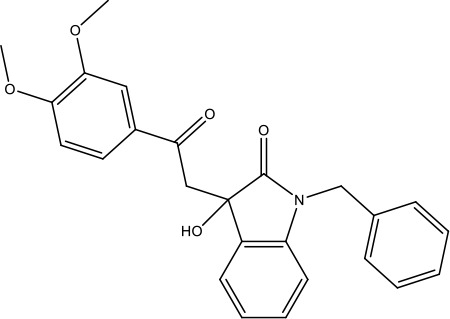 | >40 (n = 6) | >40 | |||||
-
Table 1—source data 1
JK series compound structures.
- https://cdn.elifesciences.org/articles/52555/elife-52555-table1-data1-v4.docx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (include species and sex here) | Hepatitis C virus full length infectious cell culture clone: JFH-1 | Wakita et al., 2005. (via Apath) under MTA | GenBank: AB047639.1 | |
| Strain, strain background (include species and sex here) | Hepatitis C virus full length infectious (chimaeric) cell culture clone: J4/JFH-1 | PMID:21177811, direct from Prof Bukh under MTA | GenBank: JF343781.2 | |
| Strain, strain background (include species and sex here) | Hepatitis C virus full length infectious (chimaeric) cell culture clone: S52/JFH-1 | PMID:21177811, direct from Prof Bukh under MTA | GenBank: JF343784.2 | |
| Strain, strain background (include species and sex here) | Influenza A/England/195/2009 (E195) | Prof Wendy Barclay, Imperial College | NCBI:txid645582 | Virus stock |
| Genetic reagent (include species here) | Hepatitis C virus full length infectious (chimaeric) cell culture clone: JFH-1 (L20F) | PMID:21520195 | Modified GenBank: AB047639.1 | Adamantane resistant p7, JFH-1 genotype 2a HCV |
| Cell line (include species here) | Huh7 (human hepatocellular carcinoma) | JCRB, via Prof John McLauchlan, CVR Glasgow | JCRB0403 | Mycoplasma tested and STR profile in-house |
| Cell line (include species here) | Madin-Derby Canine Kidney (MDCK) cells | ATCC | ATCC CCL34 | As provided |
| Cell line (include species here) | Human Embryonic Kidney (HEK) 293 T cells | ATCC | ATCC CRL-3216 | Adenovirus transformed, expressing SV40 large T antigen (G418r), as provided |
| Transfected construct (include species here) | (MLV) based Lentiviral luciferase reporter vector (pTG126) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | MLV Gag-Pol expressing plasmid (phCMV-5349) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | vesicular stomatitis virus glycoprotein (VSV-G) expressing plasmid | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | pcDNA3.1D-E1/E2 encoding HCV envelope from genotype 1a (H77) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | pcDNA3.1D-E1/E2 encoding HCV envelope from genotype 2a (JFH-1) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | pcDNA3.1D-E1/E2 encoding HCV envelope from genotype 2a (J6) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | pcDNA3.1D-E1/E2 encoding HCV envelope from patient isolate UKNP1.18.1 (genotype 1b) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | pcDNA3.1D-E1/E2 encoding HCV envelope from patient isolate UKNP3.2.1 (genotype 3a) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Transfected construct (include species here) | pcDNA3.1D-E1/E2 encoding HCV envelope from patient isolate UKNP3.1.1 (genotype 3a) | DOI: 10.1128/JVI.02700-15 | MLV Lentiviral reporter pseudotyped with HCV envelope glycoproteins. | |
| Antibody | (Include host species and clonality) Sheep anti-NS5A, polyclonal serum | DOI: 10.1074/jbc.M210900200 | (include dilution) 1. diluted 1/2500 for immunofluorescence | |
| Antibody | Monoclonal Mouse anti-HCV core, C7-50 | Thermo Fisher | catalogue # MA1-080 | Diluted 1/1000 for western blot |
| Antibody | AlexaFluor 594 nm Donkey anti-Sheep antibody | Thermo Fisher | catalogue # A-11016 | Diluted 1/500 |
| Antibody | HRP conjugated Goat anti Rabbit secondary | Thermo Fisher | cat # G-21234 | Diluted 1/5000 |
| Peptide, recombinant protein | Hepatitis C virus genotype 1b, J4 isolate, FLAG-p7 (N-terminal tag) | DOI: 10.1002/hep.26685 DOI: 10.1074/jbc.M602434200 | In-house expression. Cleaved from GST, HPLC purified | |
| Commercial assay or kit | T7 Ribomax In vitro transcription kit | Promega | cat # P1320 | |
| Commercial assay or kit | Firefly luciferase assay system | Promega | cat# E1500 | |
| Chemical compound, drug | Rimantadine hydrochloride | SIGMA | 1604508 | |
| Chemical compound, drug | Amantadine hydrochloride | SIGMA | A1260 | |
| Chemical compound, drug | NN-DNJ | Toronto Biochemicals | N650300 | |
| Chemical compound, drug | JK3/32 series | This paper. Synthesised in-house, available upon request, subject to MTA and stock availability | ||
| Chemical compound, drug | BIT225 | This paper. Synthesised in-house, available upon request, subject to MTA and stock availability | ||
| Software, algorithm | Maestro with the OPLS3 force field and solvation model VSGB | Schrödinger | RRID:SCR_016748 | |
| Protein Preparation Wizard (Version 12.3.13) | Schrödinger | RRID:SCR_016748 | ||
| ‘Ligand Docking’ tool | Schrödinger | RRID:SCR_016748 | ||
| Prime MM-GBSA tool in Schrödinger Maestro | Schrödinger | RRID:SCR_016748 | ||
| MOE software (Version 2015:1001) | http://www.chemcomp.com | RRID:SCR_014882 | ||
| AMBER 94 force field | J. Am. Chem. Soc. 1995, 117, 5179 | RRID:SCR_018497 | ||
| MMFF94 force field | RRID:SCR_015986 | |||
| Amber ff99SB-ILDN force field (ff) | RRID:SCR_018497 | |||
| Gromacs 4.5.5 | RRID:SCR_014565 | |||
| Jalview | http://www.jalview.org | RRID:SCR_006459 |