Piezo1/2 mediate mechanotransduction essential for bone formation through concerted activation of NFAT-YAP1-ß-catenin
Figures
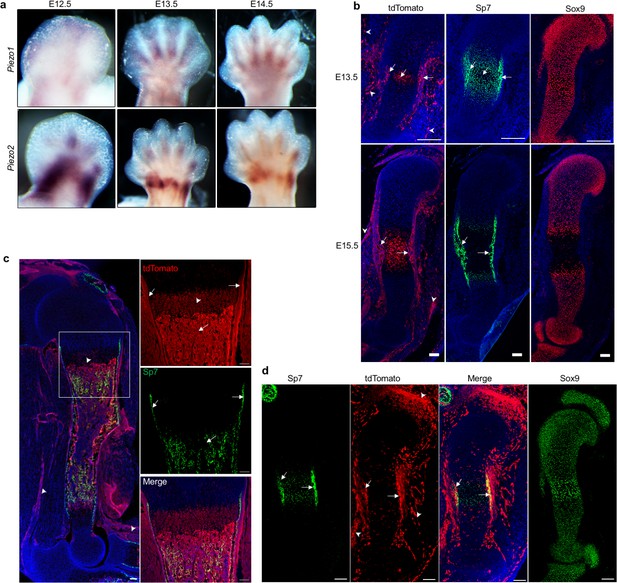
Characterization of Piezo1 and Piezo2 expression in the developing long bones.
(a) Expression of Piezo1 and Piezo2 examined by whole-mount in situ hybridization of embryonic limb buds at the indicated stages. (b) Immunostaining for tdTomato, Sp7 and Sox9 in consecutive humerus sections of the Piezo1-tdTomato forelimb buds. Arrows: Membranous tdTomato and nuclear Sp7 were found in the differentiating osteoblasts and hypertrophic chondrocytes in the same regions. Arrowheads: Extra-skeletal Piezo1-tdTomato expression. (c) Representative image of GFP and tdTomato fluorescent staining of humerus sections from the Piezo1-tdTomato; Sp7-Cre::GFP mouse at P0. The boxed region was shown in higher magnification in the right panel. Arrows: colocalization of tdTomato and GFP in the differentiating osteoblasts. Arrowheads: Hypertrophic chondrocyte or extra-skeletal expression of Piezo1-tdTomato. (d) Representative images of tdTomato costained with Sp7 or Sox9 on consecutive humerus sections from the E13.5 Piezo2Cre; Rosa26-tdTomato limb bud. Arrows: TdTomato costained with Sp7 in the differentiating osteoblasts. Arrowheads: Joint or extra-skeletal Piezo1-tdTomato expression. All scale bars, 100 μm. DAPI (blue) stain the nucleus.
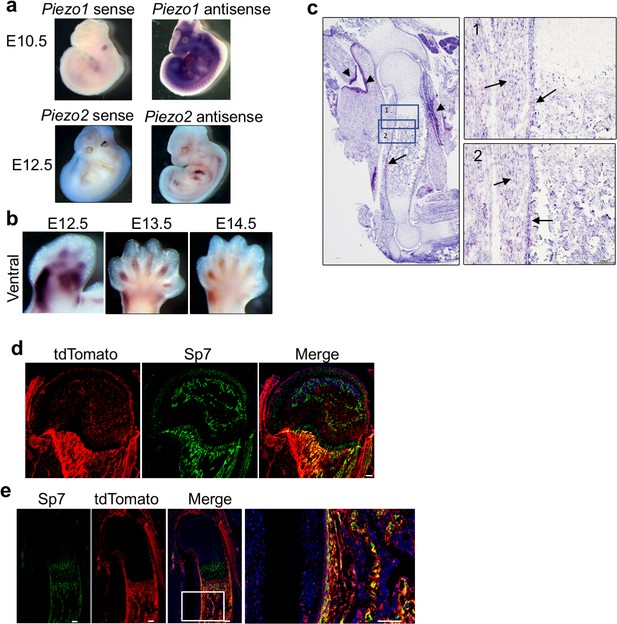
Mouse Piezo1 and Piezo2 expression.
(a, b) Expression of Piezo1 and Piezo2 examined by whole mount in situ hybridization of mouse embryos at E10.5 and E12.5 with Piezo1 and Piezo2 sense or antisense probes. (b) Ventral view of Piezo2 expression in the limb bud at the indicated stages. (c) Section in situ hybridization by RNAscope to detect Piezo1 mRNA expression in E16.5 humerus. Arrows: Piezo1 expression in the differentiating osteoblasts and muscle. Arrowheads: stronger expression of Piezo1 in the non-muscle connective tissues. Boxed regions are shown in higher magnification on the right panel. Scale bar, 100 μm. (d and e) Fluorescent images of tdTomato with costained Sp7 in humerus of the Piezo2Cre; Rosa26-tdTomato mouse at P14 (d) and E17.5 (e). Boxed region in (e) is shown on the right side. Some of the Piezo 2 lineage cells expressed Sp7. Scale bars, 100 μm. DAPI stains the nucleus.
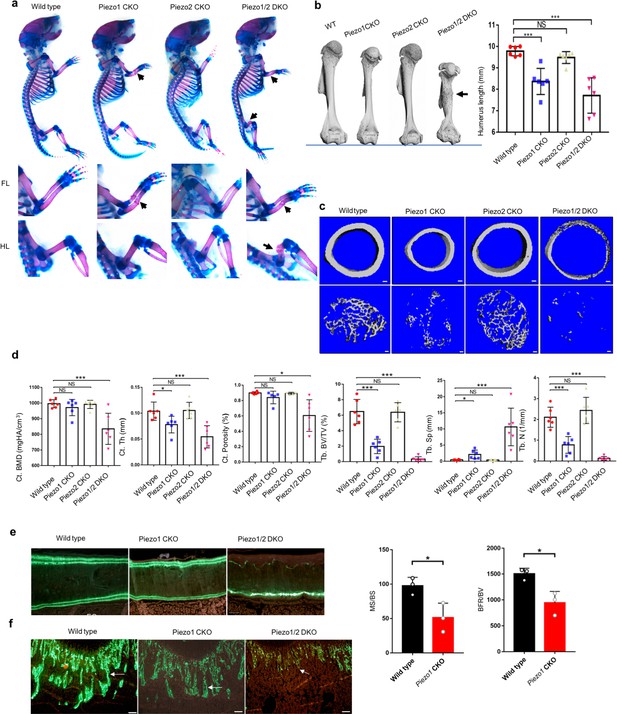
Loss of Piezo1 or both Piezo1/2 in embryonic limb mesenchyme led to reduced bone formation and spontaneous bone fractures in neonatal mice.
(a) Whole mount alizarin red and alcian blue staining of P0 mouse pups from the same litter. Bone fractures were indicated by arrows. The forelimb (FL) and hindlimb (HL) were taken out and shown in the lower panel. (b) Representative three dimensional μCT images of humerus from 3 weeks old littermate mice with the indicated genotypes. The humerus length was quantified (n = 6, mean ± SD). Bone fracture is indicated by an arrow. (c) Representative cross section μCT images of the cortical and trabecular femur bones from 3 weeks old littermate mice. (d) Quantified analysis of μCT data. Data are shown as means ± SD. (e, f) Histomorphometric analysis of distal femurs from wild type and Prrx-Cre driven Piezo1 CKO and Piezo1/2 DKO mutant mice. Representative images of double Calcein labeling in the cortical (e) and trabecular (f) bones were showed, and the dynamic bone formation parameters were only quantifiable in the wild type and Piezo1 CKO group, due to the severely reduced cortical bone formation in the Piezo1/2 DKO and little bone formation in the secondary spongiosa of the distal metaphysis in both Piezo1 CKO and Piezo1/2 DKO mutant mice. PO: periosteum; EO; endosteum. Scale bars: 50 μm (e); 100 μm (f). *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Tukey’s multiple comparisons tests. In (e), p value is calculated by two-tailed unpaired Student’s t-test (Source data 1).
-
Figure 2—source data 1
Loss of Piezo1 or both Piezo1/2 in embryonic limb mesenchyme led to reduced cartilage growth and osteoblast differentiation.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig2-data1-v2.xls
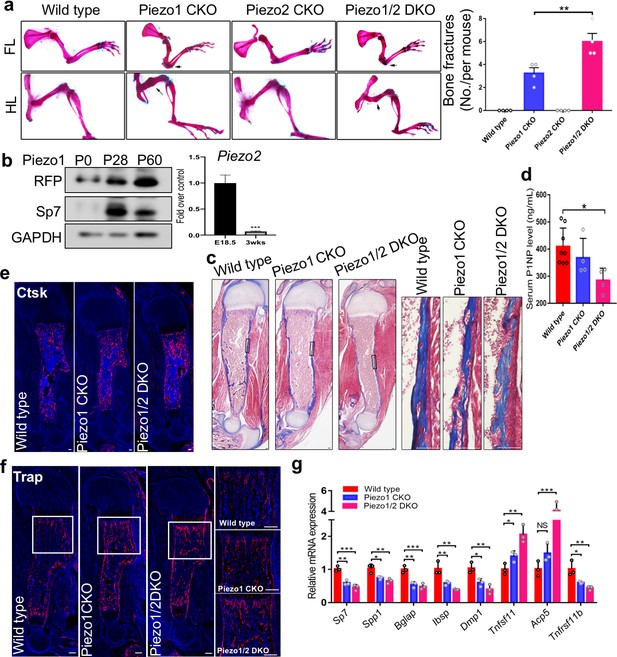
Gross characterization of Prrx1-Cre driven Piezo1 and Piezo2 mutant phenotypes at postnatal stages.
(a) Representative images of alizarin red and alcian blue stained limbs from 3 weeks old mice with the indicated genotypes. Bone fractures are indicated with arrows and quantification (n = 4) is shown on the right side. (b) Expression of Pieozo 1 and 2. Western blotting analysis of the humerus bone devoid marrow from the Piezo1-Tdtomato mice of the indicated ages. Piezo1 protein levels were shown by RFP immunoblotting. Piezo2 protein was too low to be detected. RT-qPCR analysis of Piezo2 expression of cortical bones from E18.5 and 3 weeks old mice was performed. n = 5–6. (c) Masson’s Trichrome staining of humerus sections from P5 mouse pups. Reduction in collagen content (blue staining) was observed in the Piezo1 CKO or Piezo1/2 DKO mutants. Boxed regions are shown in higher magnification in the right panel. (d) Detection of serum PINP levels in the 4 weeks old mice with the indicated genotypes by ELISA. Bone formation was most significantly reduced in the Piezo1/2 DKO mutant. (n = 4–7, means ± SD). (e, f) Immunostaining of Ctsk (e) and Trap1 (f) in humerus sections from P0 mouse pups. Boxed regions were shown in higher magnification in the right panel. Osteoclast differentiation was increased in both Piezo1 CKO and Piezo1/2 DKO mutants. (g) qPCR analysis of osteoblast and osteoclast gene expression from bone tissues of P0 pups with indicated genotypes (n = 3, means ± SD). *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t-test or one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant (a, d and g) (Figure 2—figure supplement 1—source data 1).
-
Figure 2—figure supplement 1—source data 1
Original numbers used for quantification and Western blots.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig2-figsupp1-data1-v2.xls
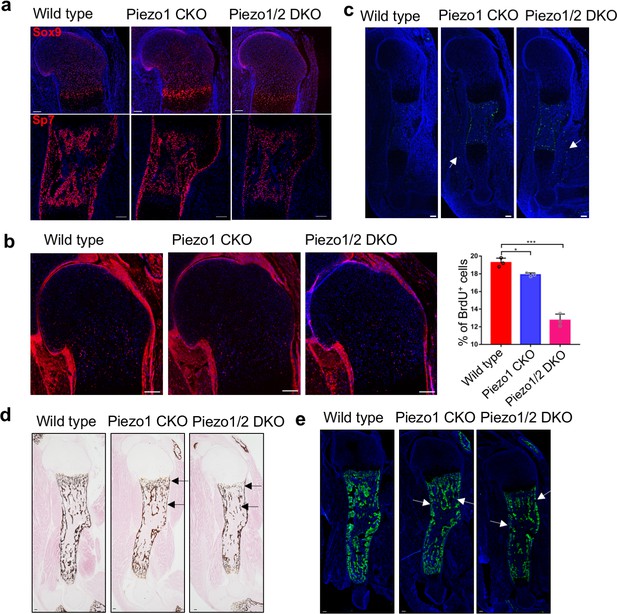
Loss of Piezo1 or both Piezo1/2 in embryonic limb mesenchyme led to reduced cartilage growth and osteoblast differentiation.
(a) Immunofluorescent images of Sox9 and Sp7 expression in the humerus sections of E16.5 embryos. (b) Representative BrdU of humerus sections of E16.5 embryos. The percentage of BrdU+ cells in the growth plate was quantified (n = 3, means ± SD). *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Tukey’s multiple comparisons tests. Proliferative cartilage regions were indicated by double headed arrows (Figure 3—source data 1). (c) Representative TUNEL staining of humerus sections of E16.5 embryos. TUNEL signals in the muscle were indicated by arrows. (d) von Kossa staining and (e) Spp1 immunostaining of humerus sections from P0 littermate pups. Reduced staining was indicated by arrows. All scale bars, 100 μm. DAPI (blue) stain the nucleus.
-
Figure 3—source data 1
Quantification of cell proliferation.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig3-data1-v2.xls
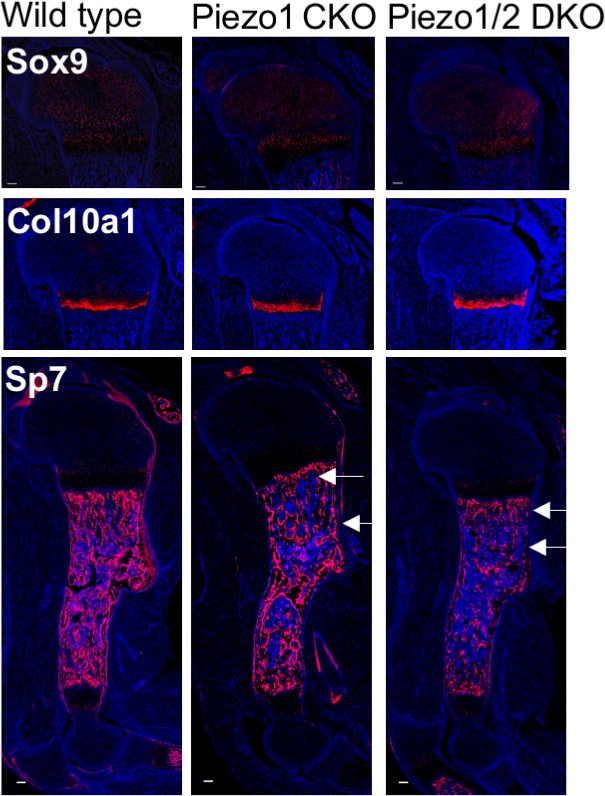
Immunostaining of Sox9, Col10 and Sp7 in sections of humerus cartilage from P0 pups.
All scale bars: 100 μm. DAPI stains the nucleus.
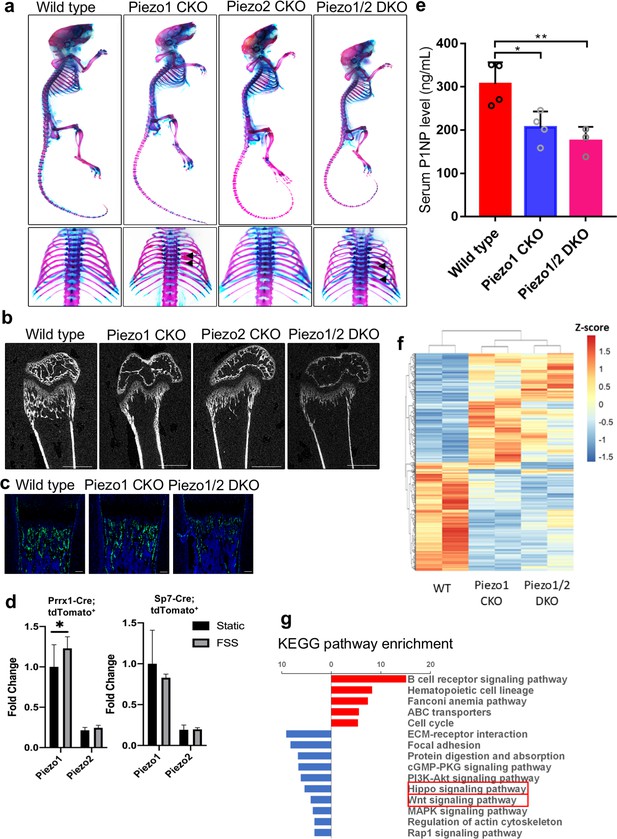
Loss of Piezo1 or Piezo1/2 in osteoblastic cells driven by the Sp7-GFP::Cre shows reduced bone mass and rib fractures.
(a) Whole mount alizarin red and alcian blue staining of P21 mice with indicated genotypes. Dorsal view of the ribcages was shown in the lower panel. Arrows indicate bone fractures in the ribs. (b) Representative 2D μCT images of the femurs from P21 mice with indicated genotypes. The quantified parameters are shown in Supplementary file 1. (c) Expression of Sp7-Cre::GFP in humerus sections of P0 pups with indicated genotypes. (d) tdTomato+ cells were sorted from long bones of the Prrx1-Cre;tdTomatofl/+ and Sp7-Cre;tdTomatofl/+ mice and cultured under osteogenic differentiation under static or fluid shear stress (FSS) condition. Piezo1 and Piezo2 expression were analyzed using RT-qPCR. (e) Serum PINP levels in 6 weeks old mice of indicated genotypes detected by ELISA. As all bones were affected by the Sp7-GFP::Cre driver, PINP levels were significantly reduced in both Piezo1 or Piezo1/2 mutants (n = 3–4, means ± SD). (f) RNA samples from the P0 humerus bone tissues of indicated Prrx1-Cre-driven mutants were subject to RNA seq. Heat-map analysis of differentially expressed genes with fold change increase >2.82 fold and reduction to <0.35 fold. Piezo1 CKO or Piezo1/2 DKO mutants showed similar alteration of gene expression genome wide. (g) KEGG pathway analysis of differentially expressed genes. Prominent reductions in Hippo and Wnt signaling were identified (boxed). *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant (Figure 4—source data 1).
-
Figure 4—source data 1
Differentially expressed genes and orginal numbers for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig4-data1-v2.xls
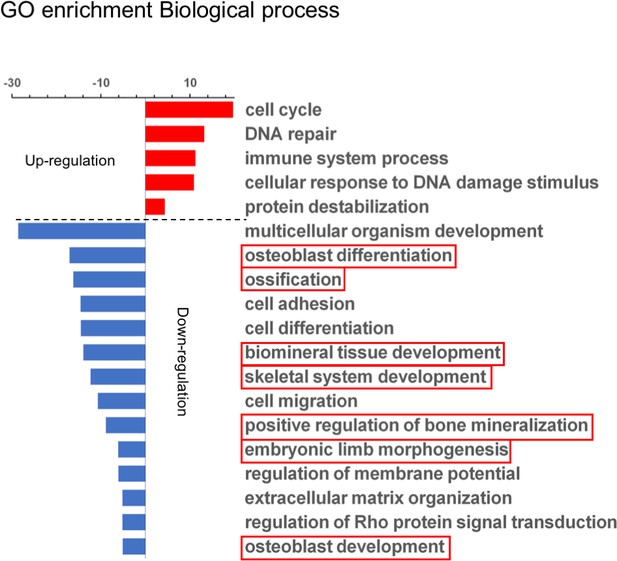
Gene ontology (GO) biological process analysis of differentially expressed genes.
Downregulations in osteoblast differentiation, ossification and mineralization were enriched (boxed).
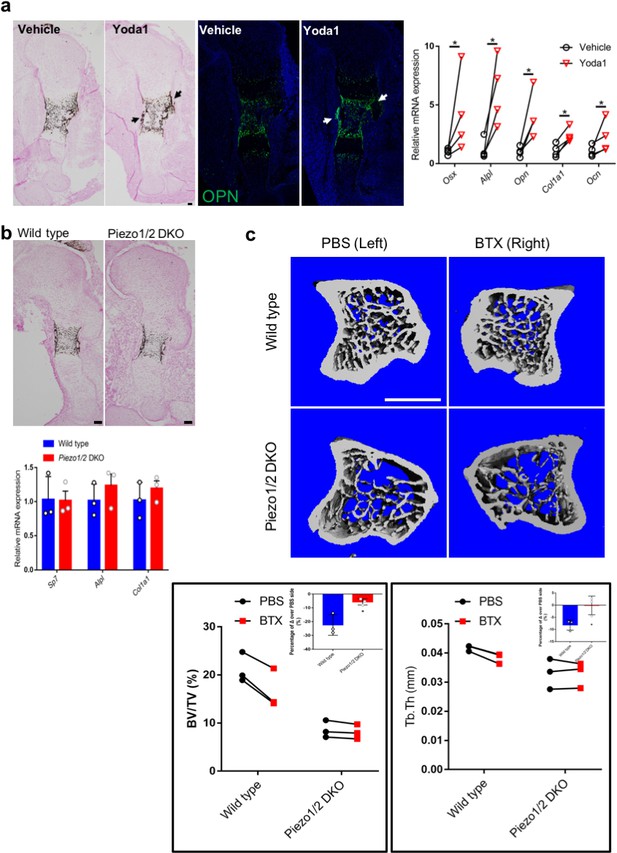
Loss of Piezo1/2 diminished reduction in bone formation caused by unloading in both embryonic development and adult life.
(a, b) Sections of the embryonic humerus from the limb bud dissected from E13.5 embryos and cultured for 4 days in BGJb medium under static conditions. Osteoblast differentiation was further determined by qRT-PCR anlysis of osteoblastic markers. (a) The right limb bud was treated with Yoda1 (400 nM), and the contralateral left limb bud was treated with equal volume of vehicle as control. Representative images of von Kossa or SPP1 staining of wild-type humerus from the cultured limbs. Scale bar: 100 μm. difference in osteoblast gene expression between limb bud pairs from the same embryo was shown on the right side. *p<0.05, according to paired ratio t test. (b) Representative images of von Kossa stained wild type or Sp7-Cre-driven Piezo1/2 DKO sections of the cultured humerus. Gene expression was shown below. No significant difference was found between the cultured wild type and Piezo1/2 DKO limbs. (c) Representative μCT images of the tibia metaphysis in an unloading model of BTX-induced muscle paralysis. The BTX-injected right leg and the PBS-injected contralateral left leg from male mice are shown. Scale Bar: 1 mm. The BV/TV and trabecular bone thickness of each inject mouse were analyzed and compared. *p<0.05, according to paired ratio t test. The percentage of difference between BTX and PBS injected side over the PBS injection side was calculated and shown in the inserts. *p<0.05, according to unpaired student t test (Figure 5—source data 1).
-
Figure 5—source data 1
Original numbers collected for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig5-data1-v2.xls
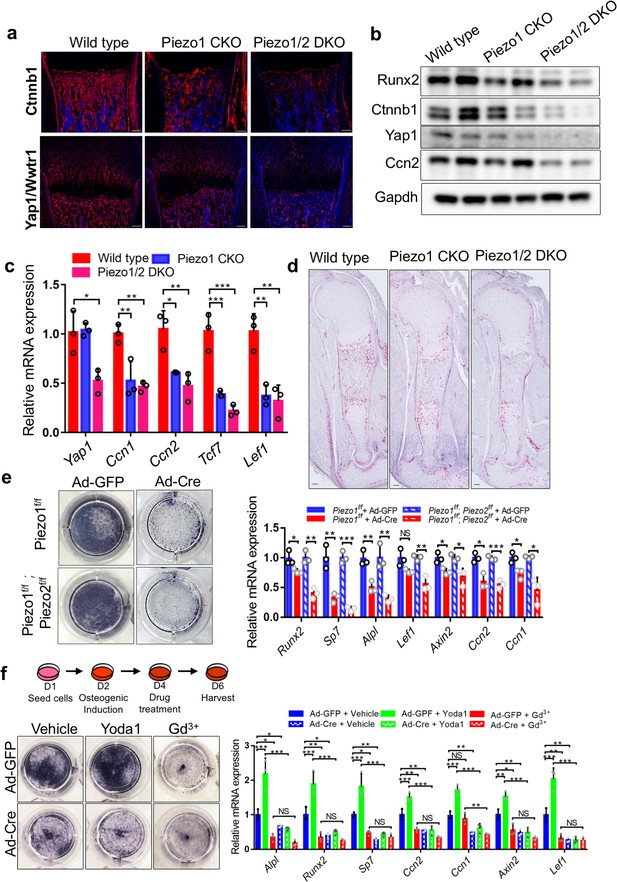
Loss of Piezo1/2 reduced Yap1 and Wnt/Ctnnb1 signaling activities and osteoblast differentiation in vivo and in vitro.
(a) Immunofluorescent staining of Ctnnb1 and Yap1/Wwtr1 in the humerus sections from the P0 Prrx1-Cre driven Piezo1 and Piezo1/2 mutants and littermate controls. (b) Western blot analyses of P0 femur bone tissue lysates from P0 pups. (c) qPCR analyses of gene expression from femur bone tissues of P0 pups (n = 3, means ± SD). (d) Ccn2 expression by RNAScope analysis of humerus sections of E16.5 embryos. (e) Alkaline phosphatase staining (left) and qPCR analyses (right, n = 3, means ± SD) in the indicated BMSCs infected with Ad-GFP or Ad-Cre 5 days after osteogenic induction. (f) Alkaline phosphatase staining (left) and qPCR analyses (right, n = 3, means ± SD) in Piezo1f/f;Piezo2f/f BMSCs treated with Piezo1 agonist (Yoda1) and antagonist (Gd3+) 6 days after osteogenic induction. The schematics of the induction process is shown on the top. All scale bars: 100 μm. DAPI (blue) stained the nucleus. *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t-test (e) or one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant (c and f) (Figure 6—source data 1).
-
Figure 6—source data 1
Original numbers and Western blots.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig6-data1-v2.xls
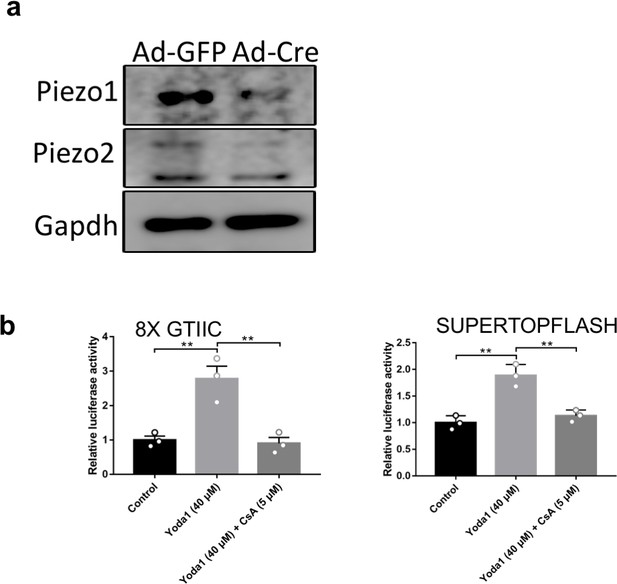
Regulation of NFAT and CTNNB1 signaling by Piezo.
(a) Western Blotting analyses of Piezo1/2 in lysates of Ad-GFP or Ad-Cre infected Piezo1f/f; Piezo2f/f BMSCs. Piezo1 is expressed at higher levels than Piezo2 and Ad-Cre infection efficiently reduced Piezo1/2 protein levels. (b) Luciferase reporter assay of Hippo and Wnt pathway activities after Yoda1 and CsA treatment of HEK293T cells. The effects of Yoda1 treatment were largely cancelled by CsA treatment. N=3, data are shown as means ± SD (Figure 6—figure supplement 1—source data 1).
-
Figure 6—figure supplement 1—source data 1
Original numbers and Western blots.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig6-figsupp1-data1-v2.xls
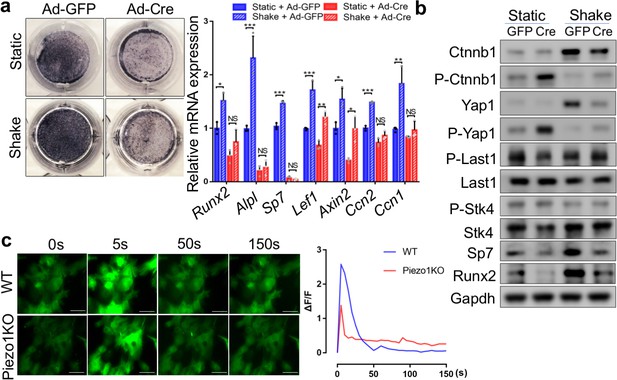
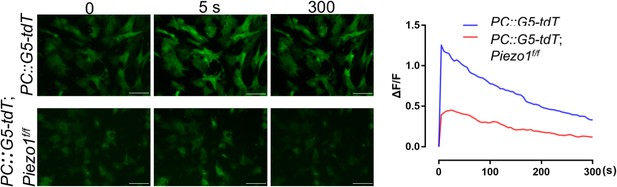
Piezo1 was required to sense mechanical forces generated by FSS and upregulate Yap1 and Ctnnb1 activities in primary mouse BMSCs.
(a) Alkaline phosphatase staining (left) and qPCR analyses (right) in BMSCs cultured with or without shaking during osteogenic induction (n = 3, means ± SD). *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Tukey’s multiple comparisons tests (Figure 7—source data 1). (b) Western blotting analyses of cell lysates from BMSCs cultured under static or shaking conditions during osteogenic induction. (c) Representative images (left) of BMSCs with fluorescent GCaMP5G reporter at different time points after FSS. Scale bars: 80 μm. The fluorescent intensities of GCaMP5G were quantified (right) every 5 s (Figure 7—source data 1).
-
Figure 7—source data 1
Original Western blots and numbers collected for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig7-data1-v2.xls
Representative GFP images of the GCaMP5G Ca2+ reporter in Ad-Cre infected BMSCs at different time points after Yoda1 treatment.
Ad-Cre infection of the PC::G5-tdT; Piezo1f/f BMSCs-induced GCaMP5G reporter expression and simultaneous deletion of Piezo1. The fluorescent intensity of GCaMP5G was quantified (Right). Scale bar: 130 μm (Figure 7—figure supplement 1—source data 1).
-
Figure 7—figure supplement 1—source data 1
Original numbers collected for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig7-figsupp1-data1-v2.xls
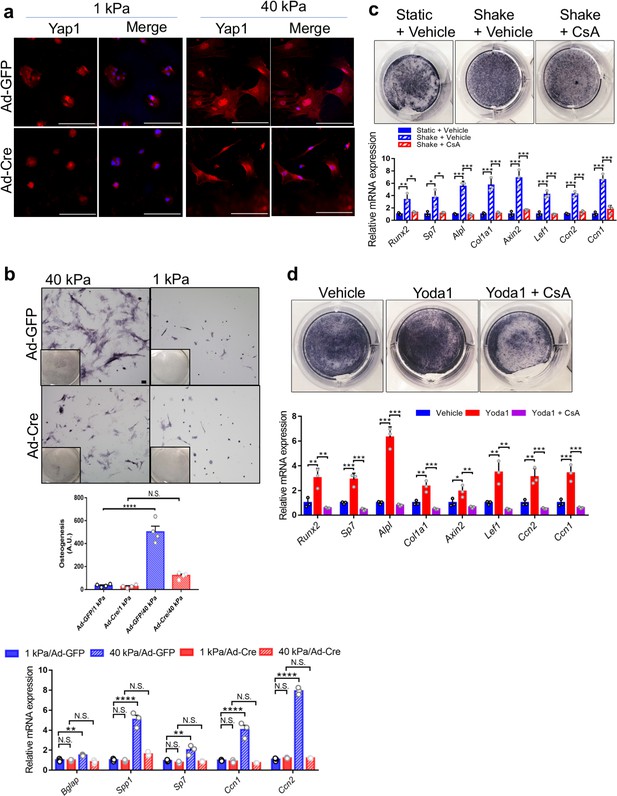
Piezo1 was required to sense ECM stiffness and upregulate Yap1 and Ctnnb1 activities in primary mouse BMSCs.
(a) Yap1 immunostaining of Piezo1f/f;Piezo2f/f BMSCs seeded on matrices with the indicated stiffness after Ad-GFP or Ad-Cre infection. Scale bars: 100 μm. (b) BMSC were infected with the indicated Ad-virus, plated on stiff (40 kPa) or soft (1 kPa) substrates and induced to differentiate into osteoblasts for 6 days. Representative alkaline phosphatase stainings images were shown. Scale bar: 100 mm. As shown previously (Dupont et al., 2011). Osteogenic differentiation was quantified by the alkaline-phosphatase-positive area determined with ImageJ as the number of blue pixels across the picture. This value was normalized to the number of cells (Hoechst/nuclei) for each picture (arbitrary units). RT-PCR anlaysis of BMSCs grown on the indicated hydrogels was shown below. (c) Alkaline phosphatase staining and qPCR analyses of primary mouse BMSCs treated with shaking with or without a Ppp3ca inhibitor CsA (n = 3, means ± SD). (d) Alkaline phosphatase staining and qPCR analyses of BMSCs treated with Yoda1 and CsA (n = 3, means ± SD). *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Tukey’s multiple comparisons tests (b–d) (Figure 8—source data 1).
-
Figure 8—source data 1
Original numbers for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig8-data1-v2.xls
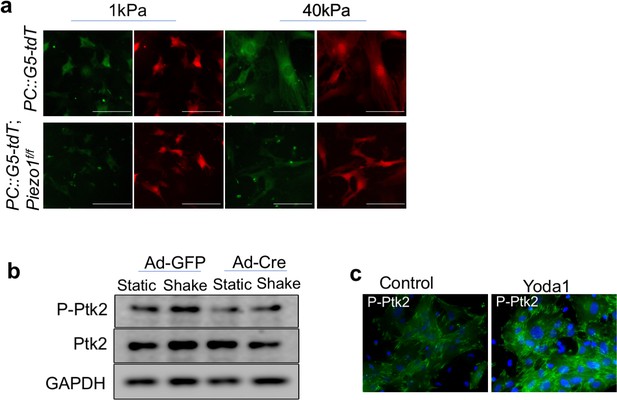
Piezo mutant primary BMSCs were defective in mechanotransduction.
(a) Representative images of the GCaMP5G reporter and the internal tdTomato fluorescent control in BMSCs of the indicated genotypes after Ad-Cre infection. The BMSCs were cultured on matrices with the indicated stiffness. While there was no difference in the tdTomato fluorescent intensity, Ca2+ signaling activity indicated by GFP intensity was increased in cells on stiffer matrix and Piezo1-deficient BMSCs were less spreading with weaker GFP signals on both soft and stiff matrix. Scale bar: 100 μm. (b) Ptk2 (Fak) phosphorylation in the Ad-Cre-induced Piezo1/2 deficient BMSCs was reduced compared to the Ad-GFP infected BMSCs, suggesting focal adhesion was reduced in the absence of Piezo1/2. (c) Fluorescent immunostaining of pPtk2 in mouse primary BMSCs. Activation of Piezo1 by Yoda1 treatment increased pPtk2 staining (Figure 8—figure supplement 1—source data 1).
-
Figure 8—figure supplement 1—source data 1
Original Western blots.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig8-figsupp1-data1-v2.xls
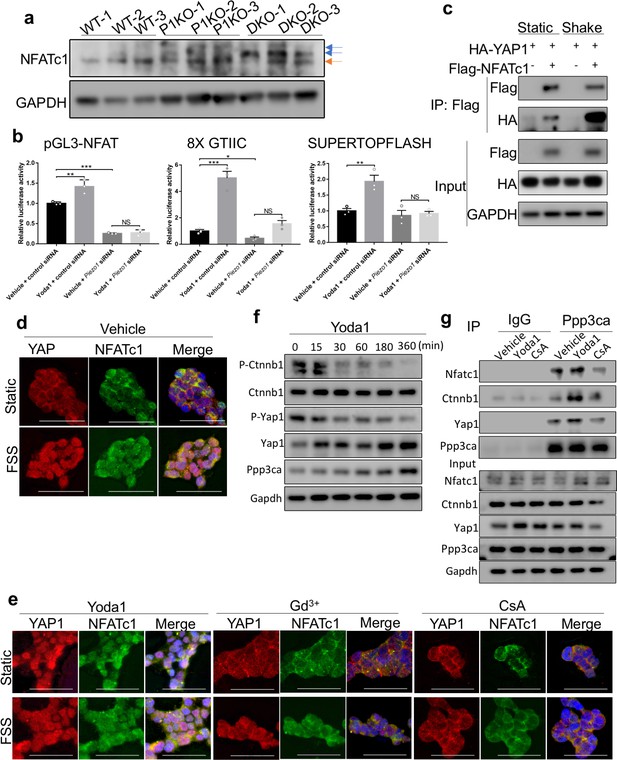
Concerted activation of NFATc1, Yap1 and Ctnnb1 by Piezo channel activation.
(a) Western blotting analysis of NFATc1 expression in bone tissue lysates from P0 pups. The slower migrating phosphorylated NFATc1 forms were indicated by blue arrows. (b) Luciferase reporter assays of NFATc1 (left), YAP1 (middle) and Ctnnb1 (right) activities in HEK293T cells (n = 3, means ± SD). *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t-test and one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant (Figure 9—source data 1). Yoda1 treatment promoted transcription activities of NFATc1, YAP1 and Ctnnb1, which were abolished by PIEZO1 knocking down. (c) Immunoprecipitation (IP) assays of HEK293T cell lysates with the indicated expression constructs. Shaking promoted NFATc1 and YAP1 binding. (d, e) Immunostaining of YAP1 and NFATc1 in HEK293 cells with indicated treatments. Activation of PIEZO1 with Yoda1 or FSS through shaking promoted nuclear localization of both YAP1 and NFATc1, which was blocked by Gd3+ or CsA treatment. Scale bars, 100 μm. (f) Western blotting analyses of Yoda1 treated primary mouse BMSCs. Phosphorylated Ctnnb1 and Yap1 were quickly reduced. (g) IP assays of Ppp3ca binding to NFATc1, Yap1 and Ctnnb1 in mouse primary BMSCs. IP with IgG was a negative control. Yoda1 treatment promoted CnA binding to NFATc1, Yap1 and Ctnnb1, which was inhibited by CsA.
-
Figure 9—source data 1
Original Western blots and data for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig9-data1-v2.xls
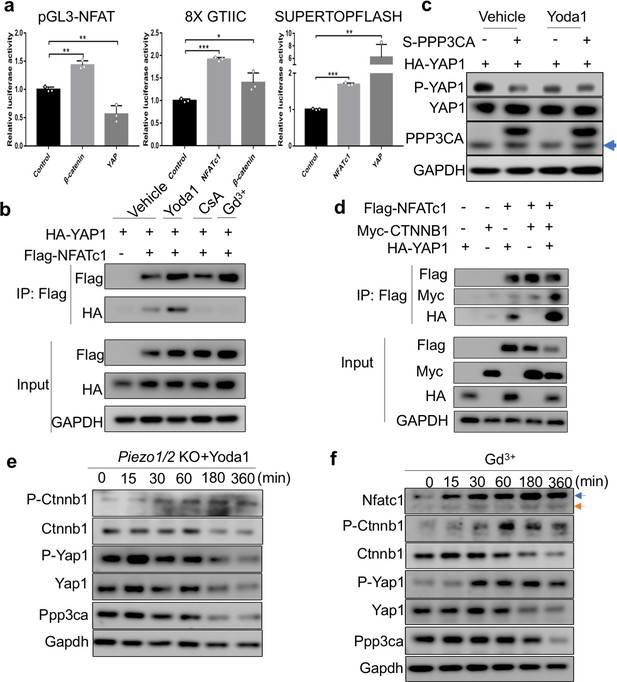
PIEZO1 activation led to concerted activation of NFATc1, YAP1 and CTNNB1.
(a) Luciferase reporter assays of NFATc1 (pGL3-NFAT), YAP1 (8XGTIIC) and CTNNB1 (SUPERTOPFLASH) transcription activities in HEK293T cells. N = 3, data are shown as means ± SD. *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t-test and one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant (Figure 9—figure supplement 1—source data 1). Expression one of NFATc1, YAP1 and CTNNB1 led to activation of the other two. The only exception is that YAP1 overexpression reducd NFATc1 activties. (b, d) Immunoprecipitation (IP) assays of HEK293T cell lysates with the indicated expression constructs. (b) Yoda1 treatment (40 μM, 4 hr) promoted complex formation of NFATc1 and YAP1, which was inhibited by CsA or Gd3+ treatment. (c) Western blotting assay of HEK 293 T cells transfected with HA-tagged YAP1 and S-tagged PPP3CA as indicated. DMSO (vehicle) or Yoda1 (40 μM) treatment was performed 24 hr after transfection for 4 hr. Arrow indicates endogenous PPP3CA. (d) Expression of NFATc1, YAP1 and CTNNB1 together promoted their complex formation. (e) Western blotting analyses of Piezo1/2 deficient BMSCs treated with Yoda1. Yoda1 treatment failed to reduce phosphorylation of Ctnnb1 and Yap1. Ppp3ca: Catalytic subunit of CaN. (f) Western blotting analyses of wild-type primary BMSCs treated by Gd3+. Increased Nfatc1 phosphorylation (blue arrow), as well as increased phosphorylation of Yap1 and Ctnnb1 was observed (Figure 9—figure supplement 1—source data 1).
-
Figure 9—figure supplement 1—source data 1
Original Western blots.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig9-figsupp1-data1-v2.xls
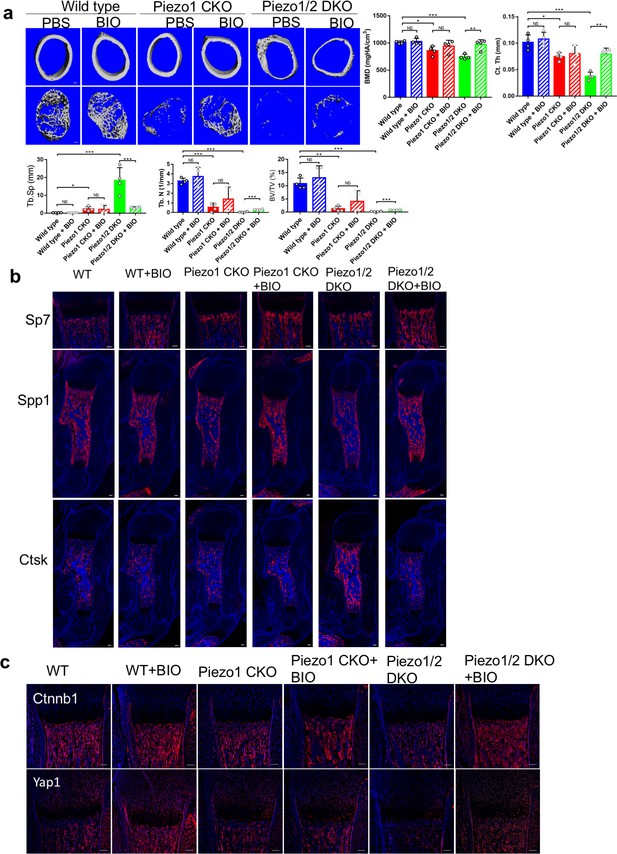
GSK3 inhibitor BIO partially rescued the bone development defects in the Piezo1/2 mutants.
(a) Representative cross section images and quantification of μCT scans of the femurs from P21 mice. N = 4. *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t-test and one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant. Data are shown as means ± SD (Figure 10—source data 1). (b) Immunostaining of humerus sections from P0 mouse pups. (c) Representative immunofluorecent images of Yap1 and Ctnnb1 staining of humerus sections from P0 mouse pups with the indicated genotypes. All scale bars: 100 μm. DAPI (blue) stained the nucleus.
-
Figure 10—source data 1
Original data for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig10-data1-v2.xls
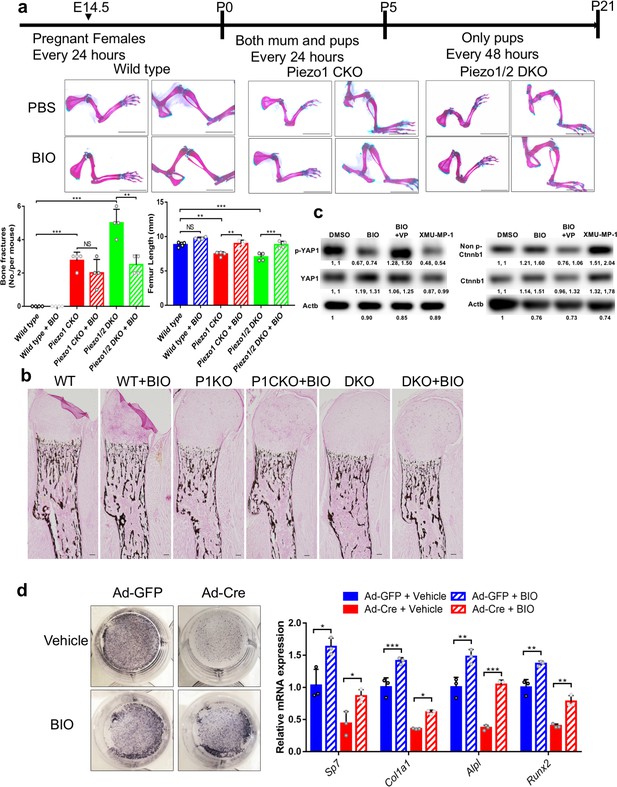
BIO injection partially rescued the reduced bone mass phenotypes in Piezo mutants.
(a) Alizarin red and alcine blue staining of the limbs from P21 mice with the indicated genotypes after PBS or BIO injection. The injection procedure is schematically shown on the top. The bone fracture numbers and femur length were quantified (n = 4). Scale bar: 1 cm. (b) von Kossa staining of humerus sections from P0 mice. Scale bar, 100 μm. BIO treatment increased mineralization of Piezo1/2 mutants. (c) Western blot analyses of lysates from primary BMSC cultured with indicated treatment. BIO (500 nM), Verteporfin (VP) (200 μg/μL), and XMU-MP-1 (2 μM), Cells were collected 3 hr after treatment. Protein bands in the blot were quantified by densitometry and analyzed using ImageJ. The first number indicated relative grayscale values to DMSO treated samples. The second number indicated the ratio of each protein level to β-actin. (d) Alkaline phosphatase staining and qPCR analyses of Ad-GFP or Ad-Cre infected Piezo1/2 conditional mutant BMSCs that had been treated with PBS or BIO in vitro. N = 3, data are shown as means ± SD. *p<0.05, **p<0.01, ***p<0.001, two-tailed unpaired Student’s t-test and one-way ANOVA followed by Tukey’s multiple comparisons tests when ANOVA was significant (a, b, c, d; Source data 1).
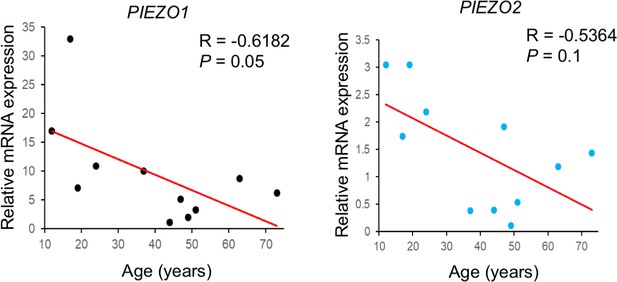
Expression of POEZO1/2 in human BMSCs.
QRT-PCR results using the cDNA of human BMSCs from 11 male subjects, between 12 to 75 years old. PIEZO1/2 Expression levels were normalized to GAPDH expression. The correlation between the expression of PIEZO1 or PIEZO2 and age was then calculated by Spearman’s correlation in Microsoft Excel (Figure 11—source data 1).
-
Figure 11—source data 1
Original numbers collected for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-fig11-data1-v2.xlsx
Videos
Pseudovideo recording of the GFP signals of the intracellular Ca2+ sensor GCaMP5G after FSS stimulation.
Images were continuously taken at time intervals of 5 s, and this pseudovideo was made from the first parallel 30 images. Left: Ad-Cre virus infected primary BMSCs from the control PC::G5-tdT mice; Right: Ad-Cre virus infected primary BMSCs from the Piezo1fl/fl; PC::G5-tdT mice.
Pseudovideo recording of the GFP signals of the intracellular Ca2+ sensor GCaMP5G after Yoda1 treatment.
Images were continuously taken at time intervals of 5 s, and this pseudovideo was made from the first parallel 60 images. Left: Ad-Cre virus infected primary BMSCs from the control PC::G5-tdT mice; Right: Ad-Cre virus infected primary BMSCs from the Piezo1fl/fl; PC::G5-tdT mice.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. musculus) | Piezo1 | Gene ID: 234839 | ||
| Gene (M. musculus) | Piezo2 | Gene ID: 667742 | ||
| Strain, strain background (C57BL/6J, Female and male) | C57BL/6J Wild type mice | Jackson laboratory | Cat# JAX:000664, RRID:IMSR_JAX:000664 | |
| Strain, strain background (C57BL/6J, Female and male) | Piezo1-tdTomato mice | Jackson laboratory | Cat# JAX:029214, RRID:IMSR_JAX:029214 | |
| Strain, strain background (C57BL/6J, Female and male) | Piezo1fl/fl mice | Jackson laboratory | Cat# JAX:029213, RRID:MSR_JAX:029213 | |
| Strain, strain background (C57BL/6J, Female and male) | Piezo2fl/fl mice | Jackson laboratory | Cat# JAX:027720, RRID:IMSR_JAX:027720 | |
| Strain, strain background (C57BL/6J, Female and male) | Piezo2-GFP-IRES-Cre mice | Jackson laboratory | Cat# JAX:027719, RRID:IMSR_JAX:027719 | |
| Strain, strain background (C57BL/6J, Female and male) | Ai9 mice | Jackson laboratory | Cat# JAX:007909, RRID:IMSR_JAX:007909 | |
| Strain, strain background (C57BL/6J, Female and male) | Prrx1-Cre mice | Jackson laboratory | Cat# JAX:005584, RRID:IMSR_JAX:005584 | |
| Strain, strain background (C57BL/6J, Female and male) | Sp7-GFP::Cre mice | Jackson laboratory | Cat# JAX:006361, RRID:IMSR_JAX:006361 | |
| Strain, strain background (C57BL/6J, Female and male) | PC::GCaMP5G-tdTomato mice | Jackson laboratory | Cat# JAX:024477, RRID:IMSR_JAX:024477 | |
| Cell line (Homo sapiens) | HEK 293T | ATCC | Cat# ACS-4500, RRID:CVCL_4V93 | |
| Transfected construct (M. musculus) | mPiezo1 siRNA | Sigma-Aldrich | Cat# SASI_ Hs01_00208584 | |
| Transfected construct (M. musculus) | siRNA Universal Negative Control | Sigma-Aldrich | Cat# SIC001 | |
| Biological sample (M. musculus) | Primary BMSCs | Freshly isolated from M. musculus | ||
| Antibody | Anti-RFP (Rabbit polyclonal) | Rockland | Cat# 600-401-379, RRID:AB_2209751 | 1:200 (IHC), 1:1000 (WB) |
| Antibody | Anti-Sp7 (Rabbit polyclonal) | Abcam | Cat# ab22552, RRID:AB_2194492 | 1:600 (IHC), 1:2000 (WB) |
| Antibody | Anti-SO9 (Rabbit polyclonal) | Millipore | Cat# AB5535, RRID:AB_2239761 | 1:500 (IHC) |
| Antibody | Anti-OPN (Goat polyclonal) | R and D systems | Cat# AF808, RRID:AB_2194992 | 1:200 (IHC) |
| Antibody | Anti-Ctsk (Rabbit polyclonal) | Abclonal | Cat# A1782, RRID:AB_2763824 | 1:100 (IHC) |
| Antibody | Anti-Trap (Rabbit polyclonal) | Abclonal | Cat# A2528, RRID:AB_2764419 | 1:100 (IHC) |
| Antibody | Anti-BrdU (mouse monoclonal) | Biolengd | Cat# 317902, RRID:AB_604040 | 1:100 (IHC) |
| Antibody | Anti-Col10a1 (Rabbit polyclonal) | Abclonal | Cat# A6889, RRID:AB_2767448 | 1:100 (IHC) |
| Antibody | Anti-YAP/TAZ (Rabbit monoclonal) | CST | Cat# 8418, RRID:AB_10950494 | 1:300 (IF), 1:200 (IHC), 1:1000 (WB) |
| Antibody | Anti-β-catenin (mouse monoclonal) | BD Biosciences | Cat# 610154, RRID:AB_397555 | 1:400 (IHC), 1:2000 (WB) |
| Antibody | Anti-β-catenin (Rabbit polyclonal) | Abclonal | Cat# A0316, RRID:AB_2757122 | 1:1000 (WB) |
| Antibody | Anti-NFAT2 (Rabbit polyclonal) | Abclonal | Cat# A1539, RRID:AB_2762296 | 1:200 (IF), 1:100 (IHC), 1:1000 (WB) |
| Antibody | Anti-Phospho-YAP (Ser127) (Rabbit polyclonal) | CST | Cat# 4911, RRID:AB_2218913 | 1:1000 (WB) |
| Antibody | Anti-Phospho-β-catenin (Thr41/Ser45) (Rabbit polyclonal) | CST | Cat# 9565, RRID:AB_331731 | 1:1000 (WB) |
| Antibody | Non-phospho (Active) β-Catenin (Ser33/37/Thr41) (D13A1) Rabbit mAb | CST | Cat# 8814S, RRID:AB_11127203 | 1:1000 (WB) |
| Antibody | β-Actin (8H10D10) Mouse mAb | CST | Cat# 3700, RRID:AB_2242334 | 1:50,000 (WB) |
| Antibody | Anti-LATS1 (Goat polyclonal) | Santa Cruz | Cat# sc-9388, RRID:AB_2133367 | 1:1000 (WB) |
| Antibody | Anti-Phospho-LATS1 (Thr1079) (Rabbit monoclonal) | CST | Cat# 8654, RRID:AB_10971635 | 1:1000 (WB) |
| Antibody | Anti-FAK (mouse monoclonal) | Santa Cruz | Cat# sc-271126, RRID:AB_10614323 | 1:500 (WB) |
| Antibody | Anti-Phospho-FAK (pY397) (mouse monoclonal) | BD Biosciences | Cat# 611806, RRID:AB_399286 | 1:1000 (WB) |
| Antibody | Anti-Mst1 (Rabbit monoclonal) | CST | Cat# 14946, RRID:AB_2798654 | 1:1000 (WB) |
| Antibody | Anti-Phospho-Mst1 (Thr183) (Rabbit monoclonal) | CST | Cat# 49332, RRID:AB_2799355 | 1:1000 (WB) |
| Antibody | Anti-RUNX2 (Rabbit polyclonal) | ABclonal | Cat# A2851, RRID:AB_2764676 | 1:1000 (WB) |
| Antibody | Anti-CTGF (Rabbit polyclonal) | ABclonal | Cat# A11067, RRID:AB_2758390 | 1:1000 (WB) |
| Antibody | Anti-Calcineurin (Rabbit polyclonal) | Abclonal | Cat# A1063, RRID:AB_2758155 | 1:1000 (WB), 1:200 (IP) |
| Antibody | Anti-Flag (M2) (mouse monoclonal) | Sigma | Cat# F1804, RRID:AB_262044 | 1:2000 (WB), 1:1000 (IP) |
| Antibody | Anti-HA (3F10) (Rat monoclonal) | Roche | Cat# 11867423001, RRID:AB_390918 | 1:1000 (WB) |
| Antibody | Anti-c-Myc (9E10) (Mouse monoclonal) | Santa Cruz | Cat# sc-40, RRID:AB_627268 | 1:1000 (WB) |
| Antibody | Anti-GAPDH (Rabbit monoclonal) | CST | Cat# 5174, RRID:AB_10622025 | 1:3000 (WB) |
| Antibody | Anti-DIG-AP conjugate (Sheep) | Roche | Cat# 11093274910, RRID:AB_514497 | 1:500 (ISH) |
| Antibody | Alexa Fluor 488 donkey anti-mouse (polyclonal) | Life Technologies | Cat# A-21202, RRID:AB_141607 | 1:500 (IHC) |
| Antibody | Alexa Fluor 488 donkey anti-rabbit (polyclonal) | Life Technologies | Cat# A-21206, RRID:AB_2535792 | 1:500 (IHC) |
| Antibody | Alexa Fluor 488 donkey anti-goat (polyclonal) | Life Technologies | Cat# A-11055, RRID:AB_2534102 | 1:500 (IHC) |
| Antibody | Alexa Fluor 568 donkey anti-mouse (polyclonal) | Life Technologies | Cat# A10037, RRID:AB_2534013 | 1:500 (IHC) |
| Antibody | Alexa Fluor 568 donkey anti-rabbit (polyclonal) | Life Technologies | Cat# A10042, RRID:AB_2534017 | 1:500 (IHC) |
| Antibody | Alexa Fluor 568 donkey anti-goat (polyclonal) | Life Technologies | Cat# A-11057, RRID:AB_142581 | 1:500 (IHC) |
| Antibody | ECL Donkey anti-rabbit | GE Healthcare Life Science | Cat# NA9340-1ml, RRID:AB_772191 | 1:5000 (WB) |
| Antibody | ECL Sheep anti-mouse | GE Healthcare Life Science | Cat# NA9310-1ml, RRID:AB_772193 | 1:5000 (WB) |
| Antibody | Bovine anti-goat IgG HRP | Santa Cruz | Cat# sc-2350, RRID:AB_634811 | 1:5000 (WB) |
| Antibody | Donkey Anti-Rat IgG Antibody | Sigma | Cat# AP189P, RRID:AB_11214462 | 1:5000 (WB) |
| Recombinant DNA reagent | pcDNA3.0-YAP-HA (plasmid) | This paper | ||
| Recombinant DNA reagent | pcDNA3.0-NFATc1-Flag (plasmid) | This paper | ||
| Recombinant DNA reagent | pcDNA3.0-β-catenin-Myc (plasmid) | This paper | ||
| Recombinant DNA reagent | pGL3-NFAT luciferase (plasmid) | Addgene | RRID:Addgene_17870 | |
| Recombinant DNA reagent | 8XGTIIC-luciferase (plasmid) | Addgene | RRID:Addgene_34615 | |
| Recombinant DNA reagent | Super 8X TOPFlash (plasmid) | Addgene | RRID:Addgene_12456 | |
| Recombinant DNA reagent | pTK-Renilla (plasmid) | Promega | Cat# E2241 | |
| Recombinant DNA reagent | pcDNA3.0-Calcineurin A-S (plasmid) | This paper | ||
| Sequence-based reagent | Primers listed in supplemental table | |||
| Commercial assay or kit | Gel Extraction Kit | Omega | Cat# D2500 | |
| Commercial assay or kit | RNAscope 2.5 HD Reagent Kit | ACD | Cat# 322350 | |
| Commercial assay or kit | RNAscope Probe Mm-Piezo1 | ACD | Cat# 500511 | |
| Commercial assay or kit | RNAscope Probe Mm-Ctgf | ACD | Cat# 314541 | |
| Commercial assay or kit | 1-Step NBT/BCIP Substrate Solution | Life Technologies | Cat# 34042 | |
| Commercial assay or kit | Ion 550 Chip Kit | Life Technologies | Cat# A34541 | |
| Commercial assay or kit | Ion AmpliSeq Transcriptome Mouse Gene Expression Panel, Chef-Ready Kit | Life Technologies | Cat# A36412 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay | Promega | Cat# E1910 | |
| Commercial assay or kit | Lipofectamine 3000 | Life Technologies | Cat# L3000001 | |
| Commercial assay or kit | RAT/Mouse P1NP ELISA kit | Immunodiagnostic Systems Inc | Cat# AC33F1 | |
| Commercial assay or kit | Alexa Fluor 594 Tyramide SuperBoostTM Kit | Life Technologies | Cat# B40944 | |
| Commercial assay or kit | Masson’s Trichrome | Abcam | Cat# ab150686 | |
| Commercial assay or kit | TRAP staining kit | Sigma-Aldrich | Cat# 387A | |
| Commercial assay or kit | TUNEL kit | Life Technologies | Cat# C10617 | |
| Commercial assay or kit | FAST SYBR Green Master Mix | Life Technologies | Cat# 4385612 | |
| Commercial assay or kit | High-Capacity cDNA Reverse Transcription Kit | Life Technologies | Cat# 4374966 | |
| Chemical compound, drug | DIG RNA labeling mix | Roche | Cat# 11277073910 | |
| Chemical compound, drug | Protector RNase inhibitor | Roche | Cat# 03335399001 | |
| Chemical compound, drug | T7 RNA polymerase | Roche | Cat# 10881767001 | |
| Chemical compound, drug | T3 RNA polymerase | Roche | Cat# 11031163001 | |
| Chemical compound, drug | LiCl | Sigma-Aldrich | Cat# L4408 | |
| Chemical compound, drug | Phusion High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0530 | |
| Chemical compound, drug | Proteinase K | Sigma-Aldrich | Cat# P2308 | |
| Chemical compound, drug | Glutaraldehyde solution | Sigma-Aldrich | Cat# G5882 | |
| Chemical compound, drug | Yeast tRNA | Sigma-Aldrich | Cat# R8759 | |
| Chemical compound, drug | Blocking reagent | Roche | Cat# 11096176001 | |
| Chemical compound, drug | BM-purple | Roche | Cat# 11442074001 | |
| Chemical compound, drug | Alizarin Red S | Sigma-Aldrich | Cat# A5533 | |
| Chemical compound, drug | Alcian Blue | Sigma-Aldrich | Cat# A9186 | |
| Chemical compound, drug | RNAzol RT | Sigma-Aldrich | Cat# R4533 | |
| Chemical compound, drug | Calcein | Sigma-Aldrich | Cat# C0875 | |
| Chemical compound, drug | Sliver nitrate | Sigma-Aldrich | Cat# S8157 | |
| Chemical compound, drug | Sodium thiosulfate | Sigma-Aldrich | Cat# 72049 | |
| Chemical compound, drug | BGjb medium | Life Technologies | Cat# 12591038 | |
| Chemical compound, drug | Yoda1 | TOCRIS | Cat# 5586 | |
| Chemical compound, drug | Cyclosporin A | LC Laboratories | Cat# LC-C-6000 | |
| Chemical compound, drug | Gadolinium (III) chloride | Sigma-Aldrich | Cat# 439770 | |
| Chemical compound, drug | β-glycerophosphate | Sigma-Aldrich | Cat# G9422 | |
| Chemical compound, drug | L-ascorbic acid | Sigma-Aldrich | Cat# A5960 | |
| Chemical compound, drug | Sulfo-SANPAH | Life Technologies | Cat# 22589 | |
| Chemical compound, drug | Collagen I, Rat tail | Corning | Cat# 354236 | |
| Chemical compound, drug | BIO | Sigma-Aldrich | Cat# B1686 | |
| Chemical compound, drug | 40% (w/v) acrylamide stock solution | Sigma-Aldrich | Cat# A4058 | |
| Chemical compound, drug | 2% (w/v) bis-acrylamide stock solution | Sigma-Aldrich | Cat# M1533 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 | |
| Software, algorithm | DAVID | DAVID | RRID:SCR_001881 | |
| Software, algorithm | Photoshop | Adobe | RRID:SCR_014199 | |
| Software, algorithm | Transcriptome Analysis Console | Life Technologies | RRID:SCR_016519 |
Additional files
-
Source data 1
Original numbers collected for quantification.
- https://cdn.elifesciences.org/articles/52779/elife-52779-data1-v2.xlsx
-
Supplementary file 1
Quantified results of μCT scanning of the tibia bones from the wild-types control and Sp7-Cre-driven Piezo1 and Piezo1/2 mutant mice (Source data 1).
- https://cdn.elifesciences.org/articles/52779/elife-52779-supp1-v2.docx
-
Supplementary file 2
The sequences of oligo primers used in RT-PCR.
- https://cdn.elifesciences.org/articles/52779/elife-52779-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52779/elife-52779-transrepform-v2.docx





