STRIPAK directs PP2A activity toward MAP4K4 to promote oncogenic transformation of human cells
Figures
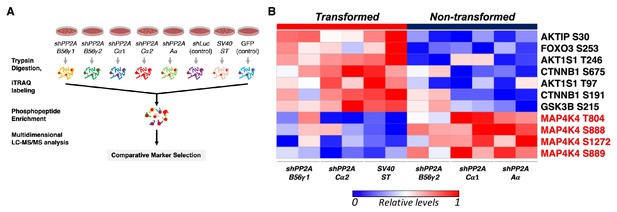
Global phosphoproteomic analysis identifies MAP4K4 dephosphorylation in cells transformed by PP2A perturbation.
(A) Schematic illustrating the global phosphoproteomics experiment. (B) The heatmap depicts phosphopeptides that are either positively or negatively correlated with the transformation phenotype (p<0.05, FDR < 1). Each column represents individual samples that were normalized to shLuc for shPP2A or in the case of ST to GFP control. The sample designations after the normalization and comparative marker selection analysis are shown below the heatmap, with each sample shown in replicates. A selected subset of phosphorylated sites which distinguishes transforming and non-transforming perturbations are shown.

Changes in PP2A levels and AI growth with PP2A knockdown and STRIPAK interactions with ST from HPyV.
(A) PP2A Ca after knockdown using shPP2A Ca1 and shPP2A Ca2 and (B) PP2A B56g after knockdown using shPP2A B56g1 and shPP2A B56g2 as measured by RNAseq (Reads Per Kilobase of transcript, per Million mapped reads). (C) AI colony count following knockdown of the indicated PP2A subunits. AI growth was assessed after PP2A subunits were suppressed using shRNAs specific for the individual subunits. (D) Interactions of polyoma virus STs with STRIPAK and MAP4K4. Co-immunoprecipitation of HA-tagged STs with components of STRIPAK and MAP4K4 confirmed results from proteomic studies. mRNA levels of (student’s t-test: n.s. = not significant, *p<0.01, **p<0.001, ***p<0.0001).
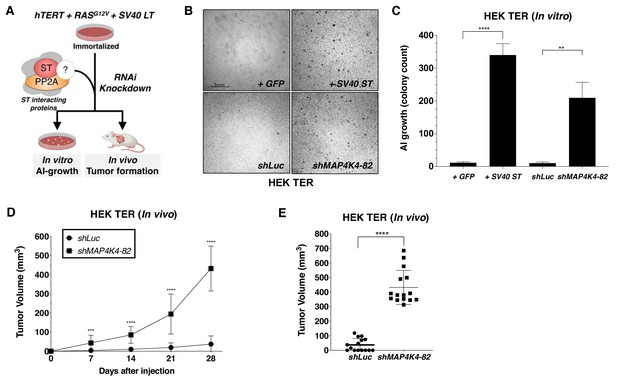
Partial knockdown of MAP4K4 expression promotes oncogenic transformation and tumor formation.
(A) Schematic of experimental design to reveal binding proteins that when depleted can substitute for ST in transformation (B) Representative image of AI growth induced by ST or MAP4K4 partial knockdown (Grid shows 5 mm). (C) Quantification of AI growth following expression of MAP4K4 shRNA-82 (shMAP4K4), SV40 ST or corresponding controls (GFP or shLuc). Graph depicting tumor volume as a function of time (D) or an endpoint at day 28 (E) for subcutaneous xenografts expressing shLuc control or shMAP4K4-82 in HEK TER cells (Student’s t-test, **p<0.001, ***p<0.0001, ****p<0.00001).
-
Figure 2—source data 1
Quantification of soft-agar colony and tumor volume with MAP4K4 knockdown.
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig2-data1-v3.xlsx
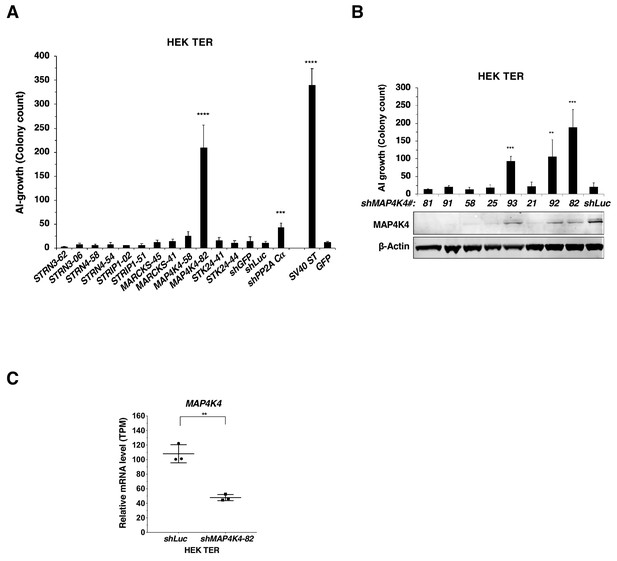
Changes in AI growth with MAP4K4 knockdown.
(A) AI colony count of HEK TER cells expressing the corresponding shRNAs or SV40 ST. Student’s t-test was performed for each shRNA compared to shLuc control. (B) AI colony count of HEK TER cells expressing eight different MAP4K4 shRNAs and MAP4K4 immunoblot showing the corresponding degree of MAP4K4 knockdown. The Student’s t-test was performed against shLuc control. (C) mRNA levels of MAP4K4 after knockdown using shMAP4K4-82 as measured by RNAseq (Transcript Per Million). (student’s t-test: **p<0.001, ***p<0.0001, ****p<0.00001).
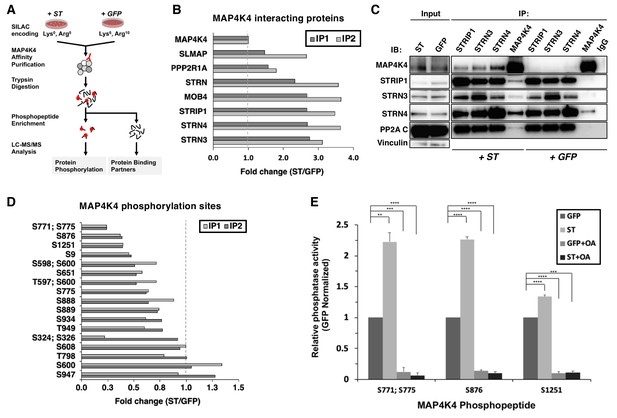
ST promotes MAP4K4 interactions with STRIPAK and MAP4K4 dephosphorylation.
(A) Schematic of targeted proteomic analysis of MAP4K4 phosphorylation and interacting proteins in the presence of ST or GFP control. (B) SILAC experiment in which MAP4K4-associated proteins were assessed in cells expressing ST or a GFP control by SILAC experiments performed as biological replicates (IP1, IP2). The proteins that showed a fold change above one have increased interactions with MAP4K4 in ST expressing cells relative to GFP. All values of the retrieved peptides were normalized to the total number of MAP4K4 peptides, prior to calculating the ratios between ST- versus GFP-expressing cells to account for variations in the amount of MAP4K4 after affinity purification. (C) Immunoblot showing results of a Co-Immunoprecipitation (Co-IP) analysis of components of STRIPAK with MAP4K4 in ST- or GFP-expressing cells. ST induced the association of MAP4K4 with STRIPAK components. (D) Quantification of fold changes in the abundance of MAP4K4 phosphorylation across indicated sites (y-axis) in cells expressing ST relative to the GFP control in two independent experiments (IP1, IP2). The phosphosites with fold changes below one show a decrease of phosphorylation of MAP4K4 in ST expressing cells relative to GFP. All values of the retrieved peptides were normalized to the total number of MAP4K4 peptides, prior to calculating the ratios between ST- versus GFP-expressing cells to account for variations in the amount of MAP4K4 after affinity purification. (E) After immunoprecipitation of STRN4 from ST- versus GFP-expressing cells, in vitro PP2A activity was measured with synthetic MAP4K4 peptides (S771;S775, S876, or S1251) identified in the targeted phosphoproteomic experiments (x-axis). Relative phosphatase activity in ST- relative to GFP-expressing HEK TER cells is shown for each phosphopeptide (y-axis). Okadaic acid (OA) was used to inhibit PP2A activity in parallel conditions (Student’s t-test, **p<0.001, ***p<0.0001, ****p<0.00001).
-
Figure 3—source data 1
Quantification of MAP4K4 interacting proteins and phosphopeptides.
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig3-data1-v3.xlsx

Changes in MAP4K4-STRIPAK interactions with ST expression.
(A) Co-immunoprecipitation analysis in 293T cells of CTAP-tagged SV40 ST wild-type and PP2A binding mutants. Co-immunoprecipitation of STRN3 and Aα is shown. (B) Co-immunoprecipitation analysis of STRN4, STRIP1, PP2A Cα, and MAP4K4 in IMR90 cells in the presence of SV40 ST or GFP. (C) Co-immunoprecipitation analysis of STRN4 and PP2A C subunit. (D) In vitro kinase assay of tandem-affinity purified MAP4K4 (Flag/HA) from HEK TER cells expressing either ST or GFP was performed and MBP was used as a substrate. HA and phospho-MBP were detected by immunoblotting.
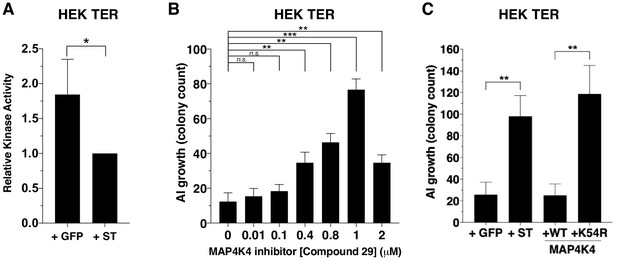
Partial inhibition of MAP4K4 kinase activity elicits transformation.
(A) Quantification of MAP4K4 in vitro kinase activity after MAP4K4 was tandem-affinity purified from cells expressing ST or GFP control. (B) Quantification of AI growth after increasing concentrations of the MAP4K4 inhibitor C29. (C) Quantification of AI growth after expression of MAP4K4 WT, MAP4K4 K54R mutant, ST, or GFP in HEK TER cells (Student’s t-test, *p<0.01, **p<0.001, ***p<0.0001, n.s. = not significant).
-
Figure 4—source data 1
Quantification of AI growth and in vitro MAP4K4 kinase activity.
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig4-data1-v3.xlsx

Changes in AI growth and proliferation with inhibition of MAP4K4 kinase activities.
(A) In vitro kinase assay of tandem-affinity purified MAP4K4 (Flag/HA) from HEK TER cells expressing either ST or GFP was performed and MBP was used as a substrate. HA and phospho-MBP were detected by immunoblotting. (B) In vitro MAP4K4 kinase assay from HEK TER cells using increasing concentrations of the MAP4K4 inhibitor (Compound 29). MBP was used as a substrate and phosphorylation determined by phospho-MBP immunoblotting (bottom). Quantification of relative kinase activity (top). (C) Proliferation of HEK TER cells was measured after exposure to compound 29 at the indicated concentrations. The Student’s t-test was performed based on absorbance values measured between 0 uM and 1 uM at five days. (D) Representative immunoblot showing MAP4K4 in vitro kinase assay using tandem-affinity purified wild-type (WT) or kinase-dead mutant (K54R) MAP4K4. (E) Mean MAP4K4 activity of WT relative to kinase-dead mutant (K54R) assessed using in vitro kinase assay (student’s t-test: n.s. = not significant, **p<0.001, ***p<0.0001).
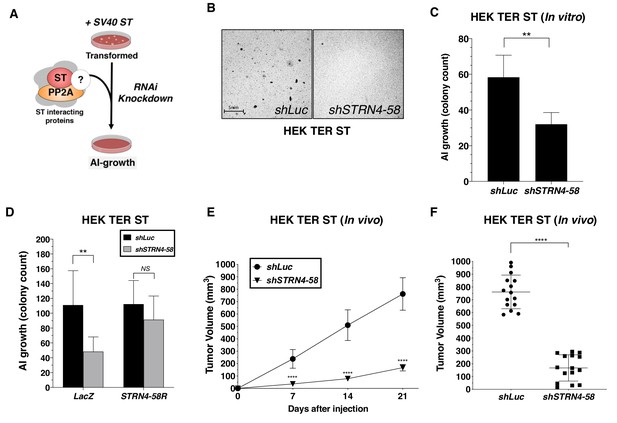
STRN4 is required for ST-mediated transformation and tumor induction.
(A) Schematic of experimental design to reveal binding proteins, that when depleted, inhibit ST-mediated transformation. (B) Representative images of AI colonies observed after knockdown of STRN4 in HEK TER ST cells with shSTRN4-58. (C) Quantification of the number of AI colonies following the introduction of shSTRN4-58 or shLuc control. (D) Quantification of the number of AI colonies after expression of shSTRN4-58 in the presence (STRN4-58R) or absence (LacZ) of an shRNA-resistant STRN4 cDNA. Tumor volume as a function of time (E) or at the endpoint at day 21 (F) for subcutaneous xenografts expressing shLuc control or STRN4 shRNA (shSTRN4-58) in HEK TER ST cells (Student’s t-test, **p<0.001, ****p<0.00001).
-
Figure 5—source data 1
Quantification of soft-agar colony and tumor volume with STRN4 knockdown.
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig5-data1-v3.xlsx

Changes in AI growth with STRN4 knockdown.
(A) AI colony count of HEK TER ST expressing four different STRN4 shRNAs or shLuc control. Immunoblot below depicts corresponding STRN4 expression after the knockdown. (B) Correlation between the relative STRN4 protein levels (quantified and normalized to shLuc) plotted on the x-axis, and AI colony counts on the y-axis. (Pearson Correlation Coefficient and the p-values are shown). (C) Immunoblot of STRN4 in HEK TER ST cells expressing an shSTRN4-58-resistant STRN4 rescue construct (STRN4-58R) or LacZ control together with either shSTRN4-58 or shLuc control. (D) Top. AI colony count of HEK TER ST cells upon CRISPR-Cas9-mediated editing of STRN4 using four different sgRNAs or a Non-Targeting (N.T.) control sgRNA. For comparison, HEK TER cells expressing ST or GFP are included. Bottom. Immunoblot depicting STRN4 protein levels after STRN4 was edited using indicated sgRNAs (student’s t-test: *p<0.01, **p<0.001, ***p<0.0001).
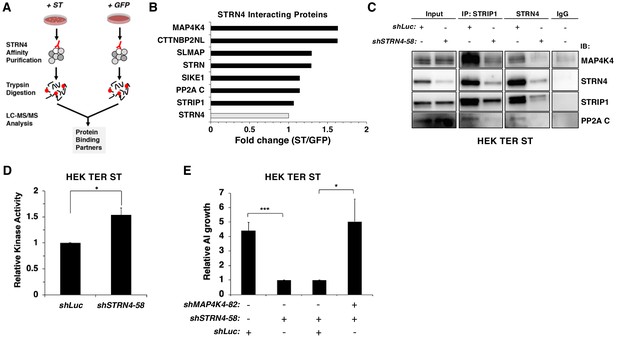
STRN4 is required for STRIPAK to interact with MAP4K4.
(A) Schematic of proteomic analysis of STRN4 interacting proteins in the presence of ST or a GFP control. (B) Fold change in abundance of STRN4 interacting proteins in the presence of ST compared to the GFP control. All values were normalized to STRN4 levels to account for variations in the total amount of STRN4 isolated from cells expression ST or GFP control. Fold-change was calculated to reflect differences in the amount of STRN4 interacting proteins between cells expressing ST relative to GFP. Interactions of a number of proteins in the STRIPAK complex, including PP2A C, MAP4K4, CTTNBP2NL, SIKE1, and SLMAP with STRN4, were increased in ST-expressing cells relative to the GFP control. (C) Immunoblot showing a Co-IP analysis of STRIPAK core components and MAP4K4 after knockdown of STRN4 using shRNA. STRN4 is required for the STRIPAK component STRIP1 to interact with MAP4K4 and PP2A C. (D) Quantification of MAP4K4 in vitro kinase activity after STRN4 knockdown (shSTRN4-58). (E) Quantification of AI growth after expression of STRN4 shRNA with or without co-expression of MAP4K4 shRNA in HEK TER ST cells. Suppression of MAP4K4 expression rescued the transformation defect arising from STRN4 depletion. All experiments were performed in triplicate, and the statistical analyses were performed relative to the controls (Student’s t-test, *p<0.01, ***p<0.0001).
-
Figure 6—source data 1
Qunatification of STRN4 interacting proteins and in vitro MAP4K4 kinase activity and AI growth.
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig6-data1-v3.xlsx

Changes in MAP4K4 kinase activity and STRIPAK interactions with STRN4 knockdown.
(A) Detection of ST from co-immunoprecipitation of STRN4 GFP- and ST-expressing cells through proteomic analysis. The values were normalized to STRN4 counts. ST was not detected (N.D.) in the GFP-expressing cells. (B) Co-immunoprecipitation analysis of endogenous MAP4K4 in HEK TER ST cells with STRN4 or PP2A Cα after cells were depleted for STRN4 or relative to a shLuc control. The ratio was calculated between Co-IP levels of STRN4/MAP4K4 or PP2A Cα/MAP4K4 after blot quantification. (C) Representative immunoblot depicting the results of an in vitro kinase assay of tandem-affinity purified MAP4K4 from HEK TER ST cells expressing shSTRN4-58, shSTRN4-55 or shLuc control. (D) AI colony count of HEK TER ST cells expressing MAP4K4 kinase-dead mutant (K54R) or WT in combination with either shSTRN4-58 or shSTRN4-55 (student’s t-test: n.s. = not significant, *p<0.01).
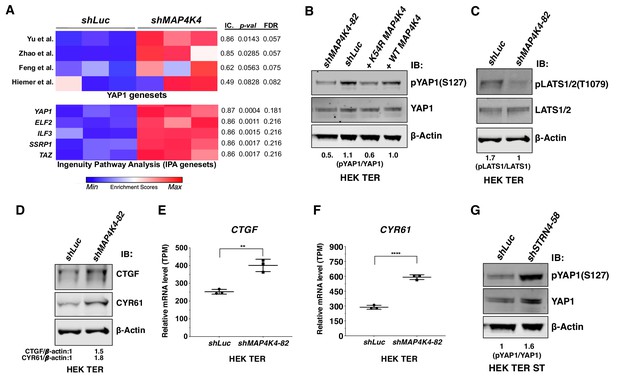
Depletion of MAP4K4 and STRN4 are linked to YAP1 regulation.
(A) Heatmap of Enrichment Scores (ES) from RNA-seq analysis showing that partial suppression of MAP4K4 expression in HEK TER cells upregulates a transcriptional signature closely resembling four published, independently generated YAP1 signatures and two signatures for YAP1/TAZ from Ingenuity Pathway Analysis (IPA) using Information Coefficient (IC) as a similarity metric. ssGSEA was performed and enrichment scores are represented as indicated in the color bar with red indicating relative enrichment and blue depletion. The three columns in the heatmap represent triplicates for each condition. (B) Immunoblot depicting changes in phosphorylation of YAP1 on a key negative regulatory site (S127) following partial MAP4K4 knockdown or expression of MAP4K4 K54R in HEK TER cells. The values below the blot represent quantitation of the YAP1 pSer127 signal relative to the total YAP1 from the immunoblot. (C) Immunoblot showing changes in phospo-LATS1 following partial MAP4K4 knockdown in HEK TER cells. Quantification of the LATS1 Thr1079 signal relative to total LATS1 from the immunoblot is shown below the gel. (D) Immunoblot depicting changes in the YAP1 target genes CTGF and CYR61 following partial MAP4K4 knockdown and the ratios of the levels of CTGF/β-actin, CYR61/β-actin are shown below the blot. β-actin shown was performed in the same blot. Changes in the mRNA levels of YAP1 target genes CTGF (E) and CYR61 (F) upon MAP4K4 suppression. (G) Immunoblot depicting changes in phosphorylation of YAP1 on S127 following STRN4 knockdown in HEK TER ST. The values below the blot depict quantitation of the YAP1 pSer127 signal relative to total YAP1 from the blot (**p<0.001, ****p<0.00001).
-
Figure 7—source data 1
Quantification of CTGF and CYR61 gene expression (TPM).
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig7-data1-v3.xlsx
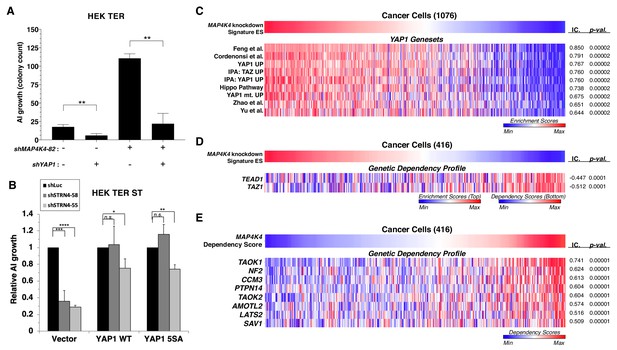
YAP1 is necessary for transformation upon MAP4K4 knockdown and rescues transformation in STRN4 knockdown cells.
(A) Quantification of AI growth obtained following partial MAP4K4 suppression alone or when combined with YAP1 suppression (shYAP1) in HEK TER cells. Transformation induced by partial MAP4K4 suppression depends on YAP1. (B) Quantification of AI growth following STRN4 knockdown with or without co-expression of YAP1 WT or the S5A mutant in HEK TER ST cells. YAP1 rescues the transformation defect of STRN4 suppression by shSTRN4-55 and shSTRN4-58 (immunoblots are shown in Figure 8—figure supplement 1B). (C) Heatmap of ES depicting YAP1 genesets from the literature significantly associated with the MAP4K4 knockdown signature ES using Information Coefficient (IC) as a similarity metric. ssGSEA was performed using these genesets across the CCLE dataset, and enrichment scores are represented as indicated in the color bar, with red indicating relative enrichment and blue depletion (FDR < 0.0001). (D) Heatmap depicting top dependency genes (bottom heatmap) in the Project Achilles dependency profiles that associated with MAP4K4 knockdown signature ES (top heatmap). The top heatmap represents ES from ssGSEA, while the bottom heatmap represent relative dependency (blue indicating strong dependency). Cell lines with low MAP4K4 transcriptional activity (in blue on top) were the most dependent on TEAD1 and TAZ1 (FDR < 0.0001). (E) Heatmap depicting co-dependency analysis of MAP4K4 using IC across the Project Achilles data. The genes most significantly associated with MAP4K4 dependency were enriched for the Hippo/YAP1 pathway, as well as components of the STRIPAK complex (All associations FDR < 0.0001 except TAOK2, SAV1: FDR = 0.002). (Student’s t-test, *p<0.01, **p<0.001, ****p<0.00001, n.s. = not significant).
-
Figure 8—source data 1
Quantification of AI growth with changes in YAP1 and MAP4K4.
- https://cdn.elifesciences.org/articles/53003/elife-53003-fig8-data1-v3.xlsx

Changes in YAP1 and MAP4K4 protein levels and a proposed model.
(A) Immunoblot depicting changes in MAP4K4 and YAP1 protein levels upon either depleting YAP1 alone, or in combination with MAP4K4. (B) Immunoblot depicting the overexpression of YAP1 WT or YAP1 S5A mutant, as well as suppression of STRN4. (C) Proposed model of ST-mediated transformation.
Additional files
-
Supplementary file 1
Key Resources Table.
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp1-v3.docx
-
Supplementary file 2
Normalized iTRAQ phosphoproteomic profiles of changes in phosphopetides upon suppression of PP2A Cα, Aα, B56γ or SV40ST expression.
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp2-v3.xlsx
-
Supplementary file 3
Results of the SILAC experiment representing MAP4K4 interacting proteins.
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp3-v3.xlsx
-
Supplementary file 4
Results of the SILAC experiment representing targeted MAP4K4 phospho-profiling.
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp4-v3.xlsx
-
Supplementary file 5
Results of MudPIT experiment showing STRN4 interacting proteins.
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp5-v3.xlsx
-
Supplementary file 6
RNAseq (TPM) profiles of MAP4K4 knockdown (shMAP4K4-82).
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp6-v3.xlsx
-
Supplementary file 7
Genesets used in the study.
- https://cdn.elifesciences.org/articles/53003/elife-53003-supp7-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53003/elife-53003-transrepform-v3.pdf





