Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation
Figures
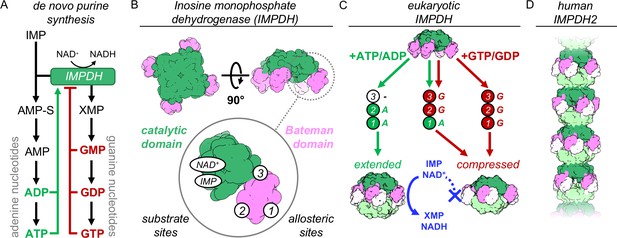
IMPDH structure and function.
(A) De novo purine nucleotide biosynthesis pathways. (B) IMPDH consists of a catalytic domain with two substrate binding sites (green), and a regulatory Bateman domain (pink) with three allosteric binding sites on the Bateman domain numbered 1,2,3. (C) Bound nucleotides promote regulatory domain dimerization, forming reversible IMPDH octamers that may be active or inhibited. Opposing tetramers colored light green and light pink. (D) Human IMPDH2 assembles into filaments composed of canonical octamers.
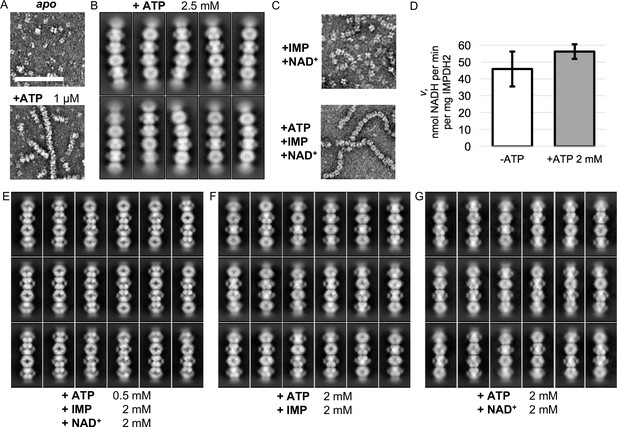
Electron microscopy of uninhibited IMPDH2 filaments.
(A) Negative stain EM of purified human IMPDH2. Treatment with 1 μM ATP induces filament assembly. Scale bar 100 nm. (B) Representative 2D class averages from the 2.5 mM ATP cryo-EM dataset. (C) Negative stain EM of actively catalyzing IMPDH2 (2 mM IMP, 2 mM NAD+), with and without 2 mM ATP. (D) Initial velocity of enzyme (2 mM IMP, 2 mM NAD+), with and without 2 mM ATP. Average of three replicates, error bars + /- 1 s.D. (E–G) Representative 2D class averages from the three uninhibited enzyme cryo-EM datasets, with nucleotide concentrations as indicated at bottom.

A cryo-EM image processing workflow for structure determination of flexible IMPDH2 filaments.
(A-D) Representative cryo-EM micrographs of IMPDH2 treated with 2.5 mM ATP (A), 0.5 mM ATP, 2 mM IMP, and 2 mM NAD+ (B), 2 mM ATP and 2 mM IMP (C), or 2 mM ATP and 2 mM NAD+ (D). Full datasets contained 480, 2169, 2289, and 2178 micrographs, respectively. Scale bars 100 nm. (E) Template-based picking, unmasked refinement, density subtraction, and masked refinement results in a reconstruction of the eight symmetrically arranged catalytic domains that make up the filament assembly interface. (F) Reverting to the un-subtracted particles, expanding the D4 symmetry, and classifying without alignment using a mask including a single filament segment identifies different segment conformations. (G) The best resolved map of each filament segment class was obtained by pooling similar classes, re-extracting and re-centering the refinement from the assembly interface onto to the canonical octamer, collapsing the symmetry expansion by deleting all Euler angle priors and removing overlapping particles, and re-refining from scratch, with additional classification and application of point-group symmetry resulting in further improvements in resolution.

Kinetic data of IMPDH2.
(A) Example spectrophotomer trajectories from IMPDH2 kinetic assays at different protein concentrations (1 mM NAD+, 1 mM IMP). (B) The same curves from A), showing only the range used to calculate reaction rates (3 min to 8 min after reaction start), as well as linear fits. (C) Experimental values for Kcat at either 24 or 37 degrees Celsius, for different enzyme concentrations (three replicates, error bars +/1 1 s.D.).
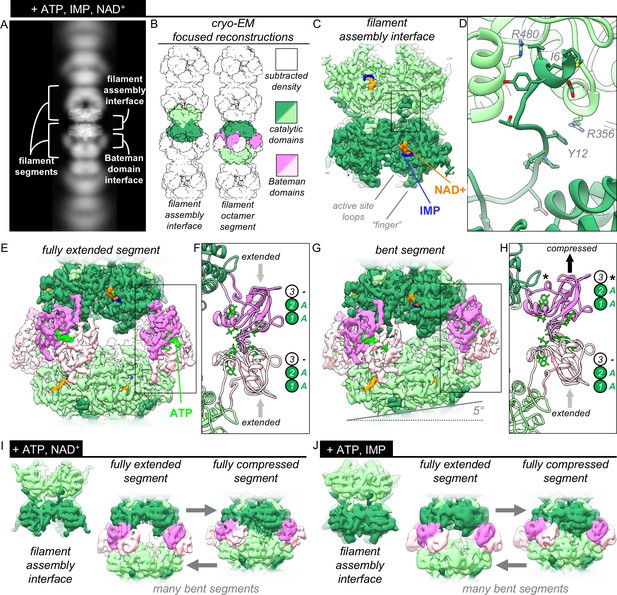
The structures of uninhibited IMPDH2 filaments.
(A) Cryo-EM of IMPDH2 filaments with both substrates (representative 2D class average). (B) We resolved two types of structures from IMPDH filaments: the consensus filament assembly interface, and various conformations of filament segments. (C) Cryo-EM density for the ATP/IMP/NAD+ consensus filament assembly interface, consisting of two tetramers bound back-to-back (dark and light green) (0.5 mM ATP, 2 mM IMP, 2 mM NAD+). (D) The filament assembly interface is mediated by the vertebrate-specific N-terminus, in particular a key bridge between Y12 and R356. (E) Cryo-EM density for the ATP/IMP/NAD+ fully extended filament segment. Opposing catalytic tetramers (dark and light green), are held separate by their symmetrically extended Bateman domains (dark and light pink). ATP (bright green) is resolvable in the Bateman domains. (F) In the fully extended Bateman domains, sites 1 and 2 are occupied by ATP, and site 3 is unformed. (G) Cryo-EM density for the best resolved ATP/IMP/NAD+ bent filament segment, in which the two catalytic tetramers are not parallel. (H) Filament segment bending results from asymmetric compression of Bateman domains. In this reconstruction, one protomer from each of the two tetramers is compressed, and allosteric site 3 is formed, but unoccupied (black asterisk). (I) Summary of the ATP/NAD+ cryo-EM dataset (2 mM ATP, 2 mM NAD+). The filament assembly interface is unchanged, and filament segments varied from fully extended, to bent, to fully compressed. In the absence of IMP, the flexible active site loops are disordered. (J) Summary of the ATP/IMP cryo-EM dataset (2 mM ATP, 3 mM IMP).
-
Figure 3—source data 1
Statistics of cryo-EM data collection, reconstruction and model refinement for the ATP/IMP/NAD+ dataset.
- https://cdn.elifesciences.org/articles/53243/elife-53243-fig3-data1-v2.docx
-
Figure 3—source data 2
Statistics of cryo-EM data collection, reconstruction and model refinement for the ATP/NAD+ dataset.
- https://cdn.elifesciences.org/articles/53243/elife-53243-fig3-data2-v2.docx
-
Figure 3—source data 3
Statistics of cryo-EM data collection, reconstruction and model refinement for the ATP/IMP dataset.
- https://cdn.elifesciences.org/articles/53243/elife-53243-fig3-data3-v2.docx
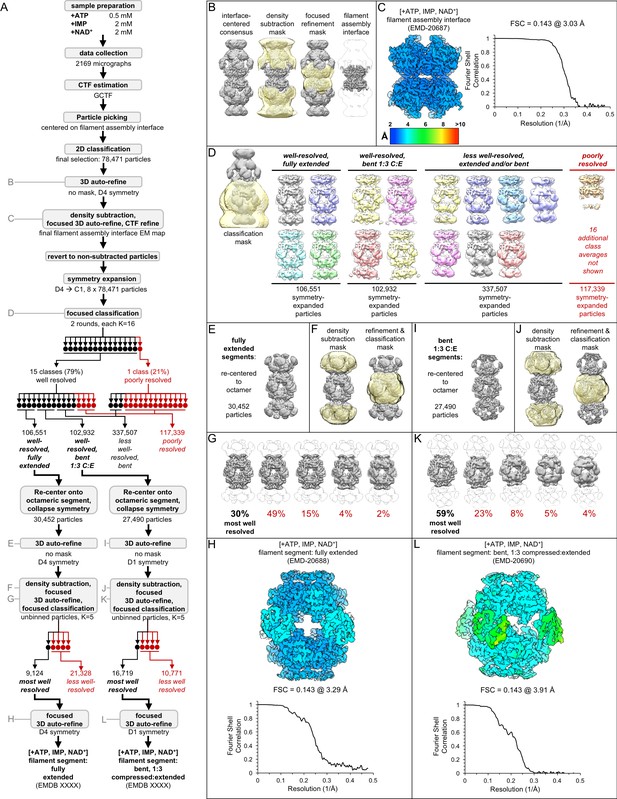
Image processing of the IMPDH2 +ATP, IMP, NAD+ cryo-EM dataset.
Nucleotide concentrations for this dataset: 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. (A) Flow chart summarizing data processing strategy. (B) Density subtraction and focused refinement of the consensus filament assembly interface. (C) Local resolution estimation and FSC curve (via relion postprocessing) for the ATP/IMP/NAD+ consensus filament assembly interface. (D) Final class averages from symmetry expanded classification of filament segments. (E) Unmasked refinement from all fully extended segments, pooled and re-centered. (F) Masks used for continued processing of fully extended segments. (G) Final classification of the best-resolved fully extended filament segment class H) Local resolution estimation and FSC curve for the ATP/IMP/NAD+ fully extended filament segment I-L) Same as E-H, but for the best-resolved ATP/IMP/NAD+ bent filament segment.

Model/Map FSC curves for the IMPDH2 +ATP, IMP, NAD+ cryo-EM dataset.
Nucleotide concentrations for this dataset: 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. For each structure, model/map Fourier shell correlations were calculated between the final map and model (left), as well as between a model refined against half-map 1 and either half-map 1 (FSC-work) or half-map 2 (FSC-test) (right). (A) Final model/map FSC curves for the ATP/IMP/NAD+ consensus filament assembly interface. (B) Final model/map FSC curves for the ATP/IMP/NAD+ fully extended filament segment. (C) Final model/map FSC curves for the ATP/IMP/NAD+ bent filament segment.

The vertebrate-specific N-terminus mediates IMPDH2 assembly of ATP-bound IMPDH2 filaments, in which individual protomers can extend or compress freely.
(A) The conformation of the N-terminus seen in assembled filaments of human IMPDH2 is unique among solved IMPDH structures, including other human structures. Boxed regions correspond to views in panel C. (B) Sequence alignment of human IMPDH1 (human 1) and IMPDH2 (human 2) and other IMPDH homologues. (C) Comparison of N-terminus conformations from cryo-EM of assembled filament (green) and published crystallized IMPDH2 (blue, PDB ID 6I0O). (F) Rotated views of the cryo-EM density for the best resolved ATP/IMP/NAD+ bent structure, colored as in Figure 2. The asymmetric unit is a tetramer, and each of the four chains can be viewed by rotating incrementally by 90 degrees. Gray letters and arrows indicate chain symmetry mates and Bateman domain conformations.
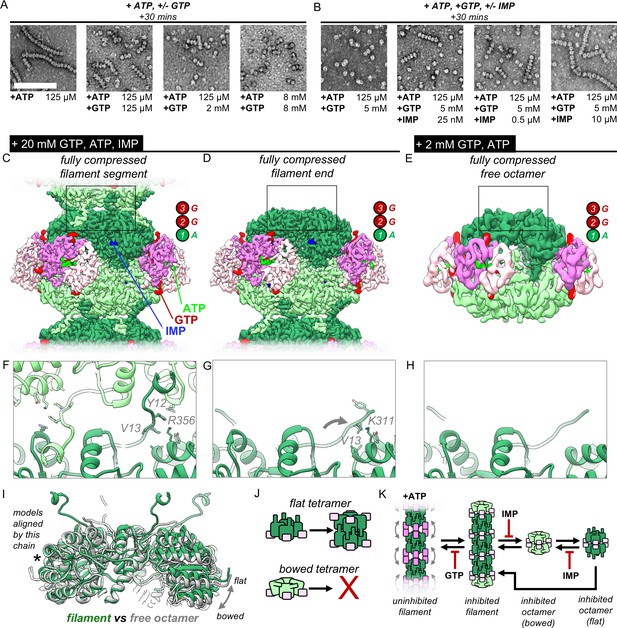
IMP and GTP allosterically modulate filament assembly and disassembly.
(A) Roughly 2 mM GTP inhibits filament assembly of IMPDH by ATP. Negative stain EM, protein concentration 2 uM. Scale bar 100 nm. Final nucleotide concentrations for each EM grid were as indicated below each image. (B) Roughly 10 uM IMP inhibits filament assembly by GTP. Final nucleotide concentrations for each EM grid were as indicated below each image. (C) Composite cryo-EM density of the GTP/ATP/IMP filament assembly interface and fully compressed filament segment maps (20 mM GTP, 0.5 mM ATP, 1 mM IMP). (D) Cryo-EM density of the fully compressed filament end map. (E) Cryo-EM density of the GTP/ATP non-filament fully compressed free octamer map (2 mM GTP, 2 mM ATP). (F–H) Close-up ribbon views of the assembled and unassembled filament interfaces the maps in A-C. (I) Comparison between the tetramer conformations of the ‘flat’ assembled filament interface (green) and the ‘bowed’ unassembled free octamer (gray). (J) Cartoon of the relationship between tetramer bowing and filament assembly. (K) Model of the regulation of filament assembly by GTP and IMP.
-
Figure 4—source data 1
Statistics of cryo-EM data collection, reconstruction and model refinement for the ATP, 2 mM GTP dataset.
- https://cdn.elifesciences.org/articles/53243/elife-53243-fig4-data1-v2.docx
-
Figure 4—source data 2
Statistics of cryo-EM data collection, reconstruction and model refinement for the ATP/IMP, 20 mM GTP dataset.
- https://cdn.elifesciences.org/articles/53243/elife-53243-fig4-data2-v2.docx
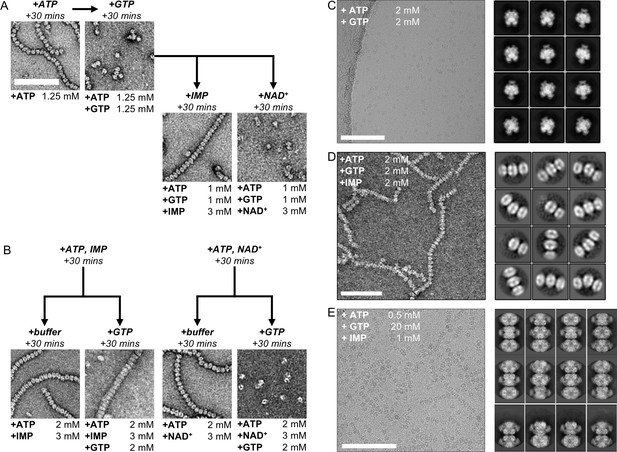
Electron microscopy of human IMPDH2 treated with ATP, GTP, IMP, and NAD+.
(A) IMP, but not NAD+, promotes re-assembly of GTP-disassembled filaments. Reagents added sequentially, with 30 min room-temperature incubation steps between. Final nucleotide concentrations for each EM grid were as indicated below each image. (B) IMP, but not NAD+, protects against disassembly of filaments by GTP. Reagents added sequentially, with 30 min room-temperature incubation steps between. Final nucleotide concentrations for each EM grid were as indicated below each image. (C) Cryo-EM of IMPDH2 treated with 2 mM ATP and 2 mM GTP. Representative micrograph (of 1159) and 2D class averages. (D) Negative stain EM of IMPDH2 treated with 2 mM ATP, 2 mM IMP, and 2 mM GTP. Representative micrograph and 2D class averages. (E) Cryo-EM of IMPDH2 treated with 0.5 mM ATP, 20 mM GTP and 1 mM IMP. Representative micrograph (of 2248), and 2D class averages. All scale bars 100 nm.

Image processing of the IMPDH2 +ATP, IMP, 20 mM GTP cryo-EM dataset.
Nucleotide concentrations for this dataset: 20 mM GTP, 0.5 mM ATP and 1 mM IMP. (A) Flow chart summarizing data processing strategy. (B) Density subtraction and focused refinement of the consensus filament assembly interface. (C) Local resolution estimation and FSC curve (via relion postprocessing) for the ATP/IMP/[20 mM]GTP consensus filament assembly interface. (D) 2D classification to separate filament segments and filament ends. Representative 2D class averages. (E) Unmasked refinement from all fully compressed segments, pooled and recentered. (F) Masks used for continued processing of fully compressed segments. (G) Final classification of the best-resolved fully compressed filament segment class H) Local resolution estimation and FSC curve for the ATP/IMP/[20 mM]GTP fully compressed filament segment I-L) Same as E-H, but for the best-resolved ATP/IMP/[20 mM]GTP filament end.
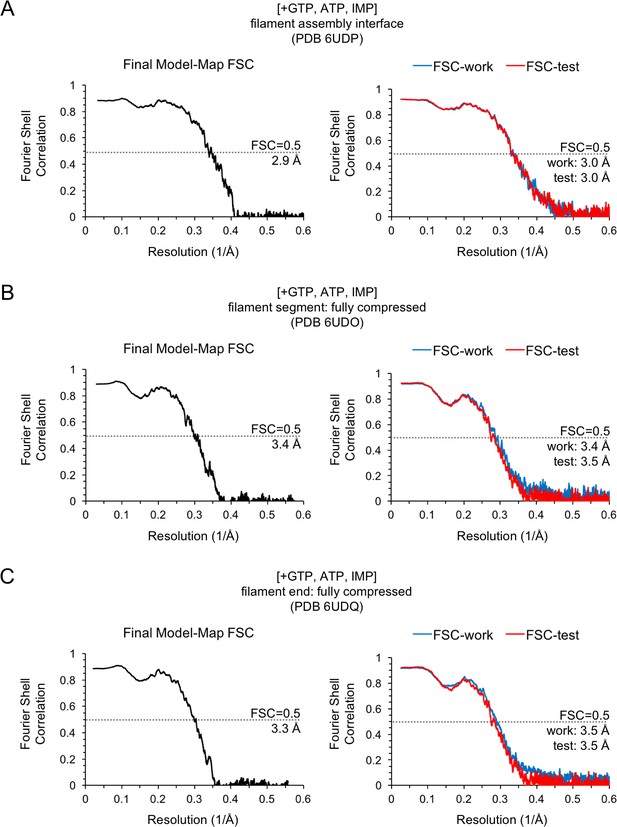
Model/Map FSC curves for the IMPDH2 +ATP, IMP, 20 mM GTP cryo-EM dataset.
Nucleotide concentrations for this dataset: 20 mM GTP, 0.5 mM ATP and 1 mM IMP. For each structure, model/map Fourier shell correlations were calculated between the final map and model (left), as well as between a model refined against half-map 1 and either half-map 1 (FSC-work) or half-map 2 (FSC-test) (right). (A) Final model/map FSC curves for the for the ATP/IMP/[20 mM]GTP consensus filament assembly interface. (B) Final model/map FSC curves for the ATP/IMP/[20 mM]GTP fully compressed filament segment. (C) Final model/map FSC curves for the ATP/IMP/[20 mM]GTP filament end.
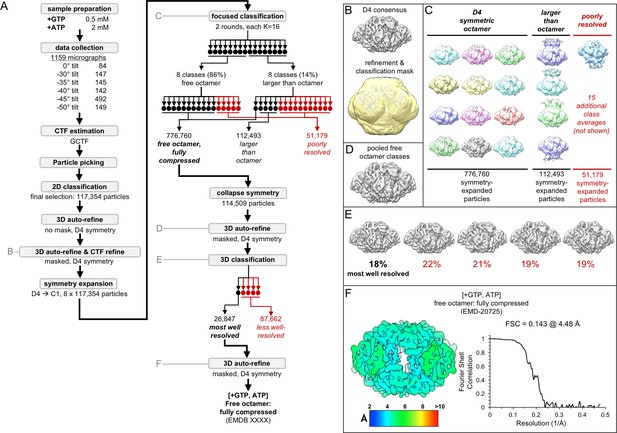
Image processing of the IMPDH2 +ATP, 2 mM GTP cryo-EM dataset.
Nucleotide concentrations for this dataset: 2 mM GTP and 2 mM ATP. (A) Flow chart summarizing data processing strategy. (B) Masked 3D refinement and all particles from 2D classification/refinement consensus filament assembly interface. Mask also used for all further processing. D4 symmetry enforced during refinement. (C) Final class averages from symmetry expanded classification of free octamers. (D) Masked refinement from all fully compressed free octamers. (E) Final classification of the best-resolved fully compressed free octamers H) Local resolution estimation and FSC curve for the ATP/[2 mM]GTP fully compressed free octamer.
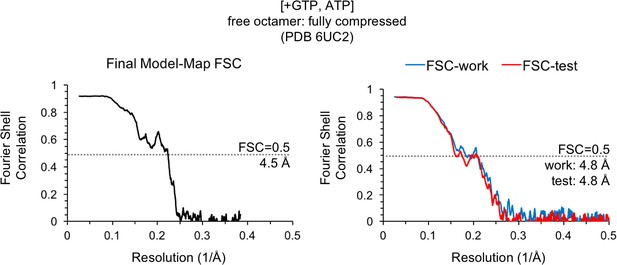
Model/Map FSC curves for the IMPDH2 +ATP, 2 mM GTP cryo-EM dataset.
Nucleotide concentrations for this dataset: 2 mM GTP and 2 mM ATP. Model/map Fourier shell correlations were calculated between the final map and model (left), as well as between a model refined against half-map 1 and either half-map 1 (FSC-work) or half-map 2 (FSC-test) (right).

The assembled IMPDH2 filament interface is not compatible with the ‘bowed’ tetramer conformation seen in the unassembled GTP-bound free octamer.
(A) The protomer from the +GTP crystal structure 6I0O (green), with applied symmetry from the filament assembly interface ‘flat’ tetramer (gray). N-terminus residues that now clash are colored magenta. Inset: closeup of clashing N-terminus. Red lines indicate specific steric clashes. (B) Two identical protomers of the filament assembly interface dimer (green) with applied symmetry from the ‘bowed’ tetramer of the +GTP crystal structure 6I0O (gray), with clashing residues colored magenta. Inset: tetramer bowing separates the key residues Y12 and R356 (distances shown are between the gamma carbons of the two residues, indicated by dotted blue line).
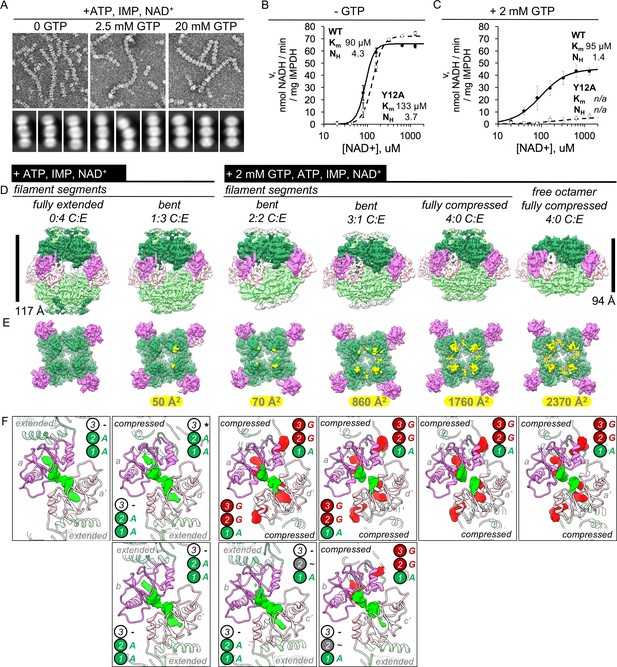
IMPDH2 filaments resist GTP inhibition by promoting bent octamer conformations that separate opposing active sites.
(A) Negatively stained EM of uninhibited (left), partially inhibited (center), and fully inhibited (right) IMPDH2. Representative micrographs and reference free 2D class averages. Prepared with 1 mM IMP, 1 mM NAD+, and either 0, 2.5 mM, or 20 mM GTP. (B–C) NAD+ saturation curves of uninhibited WT IMPDH2 (solid line), and the non-assembly mutant Y12A (dashed line). Reactions performed with 0.5 mM ATP, 1 mM IMP, and varying NAD+. (C) NAD+ saturation curves of WT filaments treated with 0.5 mM ATP, 1 mM IMP, varying NAD+ and 2 mM GTP. (D) Six cryo-EM maps from two datasets (uninhibited ATP/IMP/NAD+ and partially inhibited ATP/IMP/NAD+/[2 mM]GTP) exhibiting a range of Bateman domain conformations. The uninhibited filaments were prepared with 0.5 mM ATP, 2 mM IMP, and 2 mM NAD+. The partially inhibited filaments were prepared with 0.5 mM ATP, 2 mM IMP, and 2 mM NAD+, and 2 mM GTP. (E) A view of a single tetramer from the inside of each octamer. The lighter colored tetramer from panel A is hidden, with the surface area buried between tetramer active sites colored in yellow, with the indicated total buried surface area. (F) Corresponding views of Bateman domain conformations. Protein displayed as ribbon, with the two interacting Bateman domains colored orchid and light pink. Cryo-EM density for non-protein ligand densities is colored green and red, for ATP and GTP, respectively. Symmetry identities labeled with gray letters. In the extended conformation, allosteric site 3 is distorted, and does not bind ligands (black dashes). Allosteric site 3 is formed in compressed protomers, but in the absence of guanine nucleotides it remains unoccupied (black asterisk). In the extended protomers of some bent octamers, is is possbile that allosteric site 2 is only partially occupied (black tilde).
-
Figure 5—source data 1
Statistics of cryo-EM data collection, reconstruction and model refinement for the ATP/IMP/NAD+, 2 mM GTP dataset.
- https://cdn.elifesciences.org/articles/53243/elife-53243-fig5-data1-v2.docx
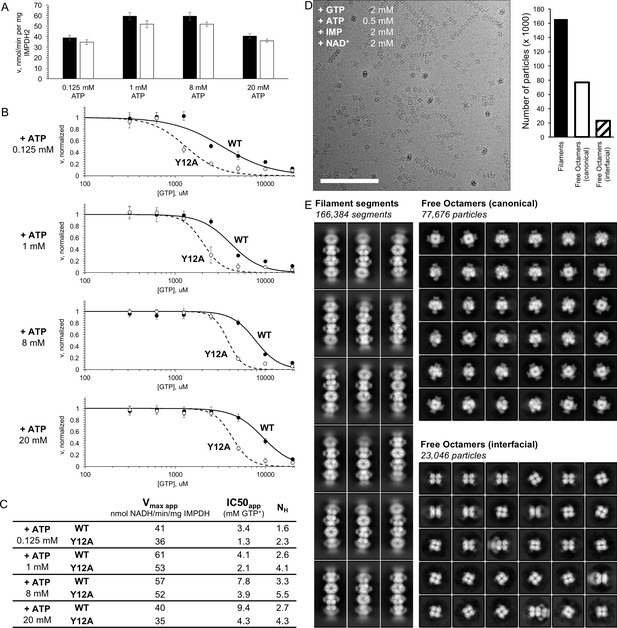
IMPDH2 filaments resist GTP inhibition.
(A) Initial reaction rates for uninhibited WT enzyme and the filament non-assembly mutant Y12A under various ATP concentrations. High concentrations of ATP likely inhibit by competing with co-substrate NAD+. Reactions prepared using 2 mM IMP, 2 mM NAD+, and either 0.125, 1, 8, or 20 mM ATP. (B) Apparent GTP inhibition under saturating substrates for a range of ATP concentrations. Under all conditions, WT was more resistant to GTP inhibition than Y12A. Reactions prepared using 2 mM IMP, 2 mM NAD+, either 0.125, 1, 8, or 20 mM ATP, and varying GTP. (C) Estimated Hill equation parameters for data in panel B. (D) Cryo-EM of partially inhibited IMPDH2 filaments. Enzyme treated with 0.5 mM ATP, 2 mM IMP, 2 mM NAD+, and 2 mM GTP. Representative micrograph, 2944 total. Scale bar 100 nm. (E) Representative 2D class averages of ATP/IMP/NAD+/[2 mM]GTP cryo-EM dataset. Three particle types were observed: filament segments, canonical ‘face-to-face’ free octamers, and interfacial ‘back-to-back’ octamers.

Image processing of the IMPDH2 +ATP, IMP, NAD+, 2 mM GTP cryo-EM dataset, part 1: initial processing, and processing of filament segments.
Nucleotide concentrations for this dataset: 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. (A) Flow chart summarizing data processing strategy. (B) Density subtraction and focused refinement of the consensus filament assembly interface. (C) Local resolution estimation and FSC curve (via relion postprocessing) for the ATP/IMP/NAD+/[2 mM]GTP consensus filament assembly interface. (D) Final class averages from symmetry expanded classification of filament segments. (E) Unmasked refinement from all fully compressed segments, pooled and recentered. (F) Masks used for continued processing of fully compressed segments. (G) Final classification of the best-resolved fully compressed filament segment class H) Local resolution estimation and FSC curve for the ATP/IMP/NAD+/[2 mM]GTP fully compressed filament segment I-K) Same as E-G, but for the best-resolved ATP/IMP/NAD+/[2 mM]GTP bent filament segment. (L–M) Local resolution estimation and FSC curves for the two different ATP/IMP/NAD+/[2 mM]GTP bent filament segments.
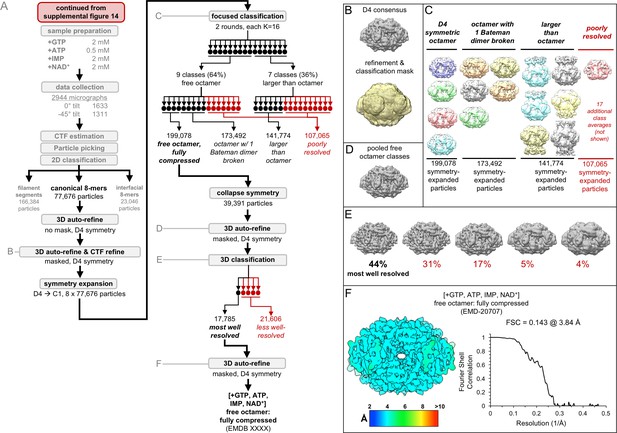
Image processing of the IMPDH2 +ATP, IMP, NAD+, 2 mM GTP cryo-EM dataset, part 2: the free canonical octamers.
Nucleotide concentrations for this dataset: 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. (A) Flow chart summarizing data processing strategy. (B) Masked 3D refinement and all particles from 2D classification/refinement consensus filament assembly interface. Mask also used for all further processing. D4 symmetry enforced during refinement. (C) Final class averages from symmetry expanded classification of free octamers. (D) Masked refinement from all fully compressed free octamers. (E) Final classification of the best-resolved fully compressed free octamers H) Local resolution estimation and FSC curve for the ATP/IMP/NAD+/[2 mM]GTP fully compressed free canonical octamer.
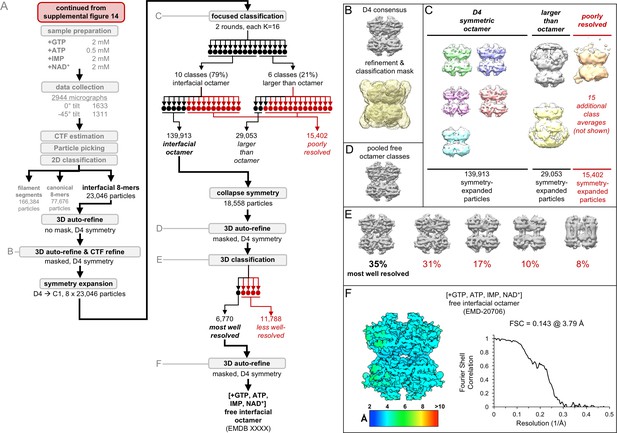
Image processing of the IMPDH2 +ATP, IMP, NAD+, 2 mM GTP cryo-EM dataset, part 3: the free interfacial octamers.
Nucleotide concentrations for this dataset: 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. (A) Flow chart summarizing data processing strategy. (B) Masked 3D refinement and all particles from 2D classification/refinement consensus filament assembly interface. Mask also used for all further processing. D4 symmetry enforced during refinement. (C) Final class averages from symmetry expanded classification of free octamers. (D) Masked refinement from all fully compressed free octamers. (E) Final classification of the best-resolved fully compressed free octamers H) Local resolution estimation and FSC curve for the ATP/IMP/NAD+/[2 mM]GTP free interfacial octamer.

Model/Map FSC curves for the IMPDH2 +ATP, IMP, NAD+, 2 mM GTP cryo-EM dataset (filament structures).
Nucleotide concentrations for this dataset: 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. For each structure, model/map Fourier shell correlations were calculated between the final map and model (left), as well as between a model refined against half-map 1 and either half-map 1 (FSC-work) or half-map 2 (FSC-test) (right). (A) Final model/map FSC curves for the for the ATP/IMP/NAD+/[2 mM]GTP consensus filament assembly interface. (B) Final model/map FSC curves for the ATP/IMP/NAD+/[2 mM]GTP fully compressed filament segment. (C–D) Final model/map FSC curves for the ATP/IMP/NAD+/[2 mM]GTP bent filament segments.
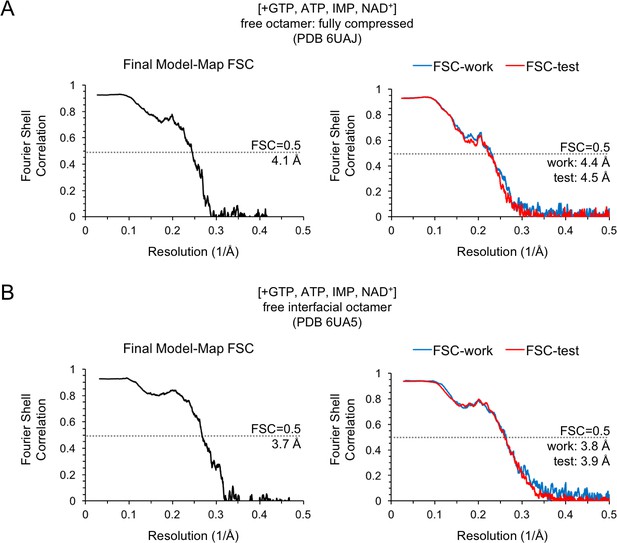
Model/Map FSC curves for the IMPDH2 +ATP, IMP, NAD+, 2 mM GTP cryo-EM dataset (non-filament structures).
Nucleotide concentrations for this dataset: 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. For each structure, model/map Fourier shell correlations were calculated between the final map and model (left), as well as between a model refined against half-map 1 and either half-map 1 (FSC-work) or half-map 2 (FSC-test) (right). (A) Final model/map FSC curves for the for the ATP/IMP/NAD+/[2 mM]GTP fully compressed free canonical octamer. (B) Final model/map FSC curves for the ATP/IMP/NAD+/[2 mM]GTP free interfacial octamer.
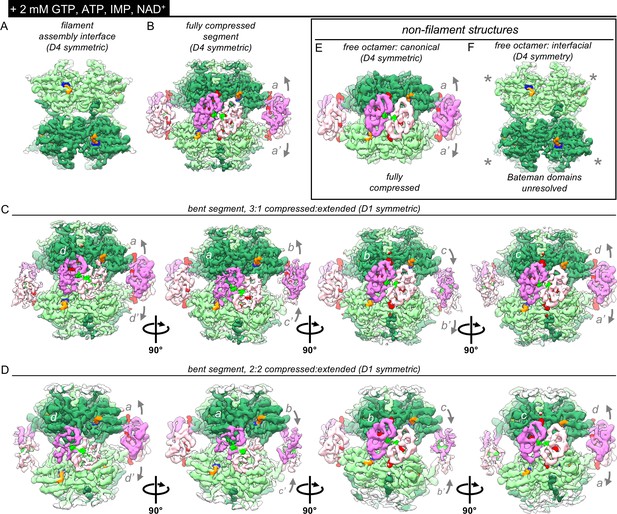
Bateman domains of partially inhibited IMPDH2 filaments are in a mix of compressed and extended states.
Nucleotide concentrations for this dataset: 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+. (A) Cryo-EM density of the consensus filament assembly interface from the ATP/IMP/NAD+/[2 mM]GTP dataset, with density for bound IMP (blue), and NAD+ (orange). (B) Cryo-EM density of the ATP/IMP/NAD+/[2 mM]GTP fully compressed filament segment, with (putative) ATP density in Bateman site 1 (bright green) and GTP in sites 2 and 3 (red). (C–D) Two D1-symmetric bent filament segments, with a mix of extended and bent protomers. The asymmetric unit of each of these is a tetramer. (E–F) Cryo-EM densities of the two different types of D4 symmetric non-filament free octamers resolved from this dataset. The Bateman domains of the free interfacial octamer were unresolved (gray asterisks).
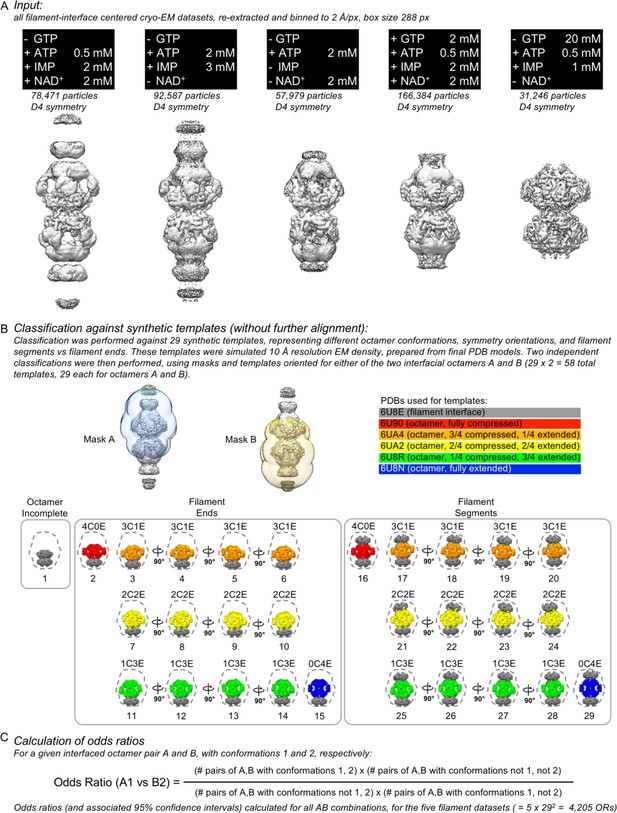
Classification of interfaced octamer pairs against synthetic templetes, and calculation of interfaced octamer odds ratios.
(A) The five interface-centered filament datasets were re-extracted to a common pixel size and box size, large enough to contain both interface octamers (nucleotide concentrations were as shown). (B) Low resolution synthetic templates were prepared by combining chains from six of the final molecular models and simulating the EM density, and the interface datasets were classified against these templates using masks to focus on a single octamer. (C) Formula used for calculation of odds ratios.

Interfaced octamer odds ratios for the 0.5 mM ATP, 2 mM IMP, 2 mM NAD+ cryo-EM dataset.
(A) Class distribution histograms and odds ratios for all combinations of classes of octamer A vs octamer B. Only ORs for which the associated 95% confidence interval does not include 1 are displayed. (B) A simplified summary of odds ratio data, made by pooling all classes depicted in A into the 21 different categories at left, irrespective of orientation, end vs segment, or A/B order. Center, histogram of # of octamer pairs per category. Right, odds ratios (error bars indicate 95% confidence interval). Abbreviations are as follow: 4C0E = fully compressed; 3C1E = 3/4 compressed, ¼ extended; 2C2E = 2/4 compressed, 2/4 extended; 1C3E = 1/4 compressed, 3/4 extended; 0C4E = fully extended.

Interfaced octamer odds ratios for the 2 mM ATP, 3 mM IMP cryo-EM dataset.
(A) Class distribution histograms and odds ratios for all combinations of classes of octamer A vs octamer B. Only ORs for which the associated 95% confidence interval does not include 1 are displayed. (B) A simplified summary of odds ratio data, made by pooling all classes depicted in A into the 21 different categories at left, irrespective of orientation, end vs segment, or A/B order. Center, histogram of # of octamer pairs per category. Right, odds ratios (error bars indicate 95% confidence interval). Abbreviations are as follow: 4C0E = fully compressed; 3C1E = 3/4 compressed, ¼ extended; 2C2E = 2/4 compressed, 2/4 extended; 1C3E = 1/4 compressed, 3/4 extended; 0C4E = fully extended.
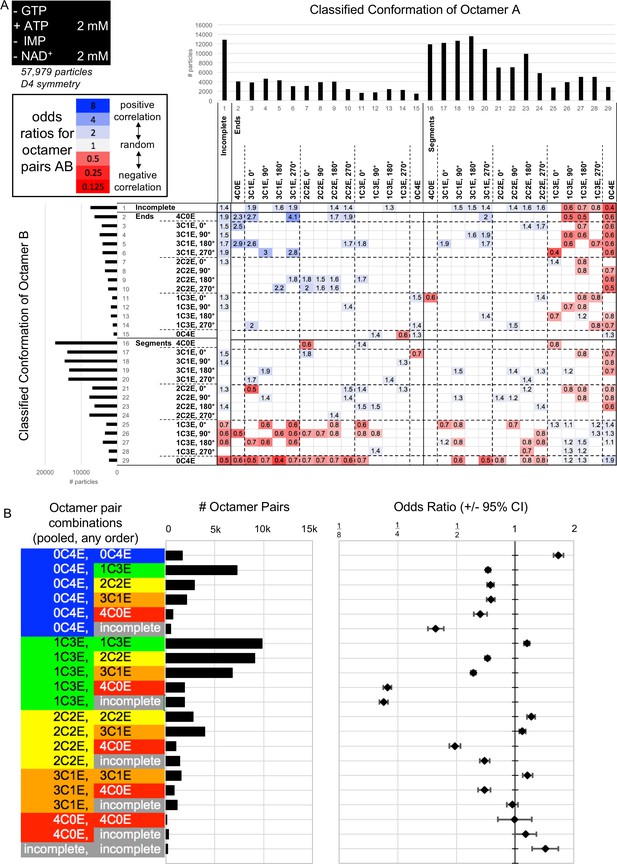
Interfaced octamer odds ratios for the 2 ATP, 2 mM NAD+ cryo-EM dataset.
(A) Class distribution histograms and odds ratios for all combinations of classes of octamer A vs octamer B. Only ORs for which the associated 95% confidence interval does not include 1 are displayed. (B) A simplified summary of odds ratio data, made by pooling all classes depicted in A into the 21 different categories at left, irrespective of orientation, end vs segment, or A/B order. Center, histogram of # of octamer pairs per category. Right, odds ratios (error bars indicate 95% confidence interval). Abbreviations are as follow: 4C0E = fully compressed; 3C1E = 3/4 compressed, ¼ extended; 2C2E = 2/4 compressed, 2/4 extended; 1C3E = 1/4 compressed, 3/4 extended; 0C4E = fully extended.

Interfaced octamer odds ratios for the 2 mM GTP, 0.5 mM ATP, 2 mM IMP, 2 mM NAD+ cryo-EM dataset.
(A) Class distribution histograms and odds ratios for all combinations of classes of octamer A vs octamer B. Only ORs for which the associated 95% confidence interval does not include 1 are displayed. (B) A simplified summary of odds ratio data, made by pooling all classes depicted in A into the 21 different categories at left, irrespective of orientation, end vs segment, or A/B order. Center, histogram of # of octamer pairs per category. Right, odds ratios (error bars indicate 95% confidence interval). Abbreviations are as follow: 4C0E = fully compressed; 3C1E = 3/4 compressed, ¼ extended; 2C2E = 2/4 compressed, 2/4 extended; 1C3E = 1/4 compressed, 3/4 extended; 0C4E = fully extended.
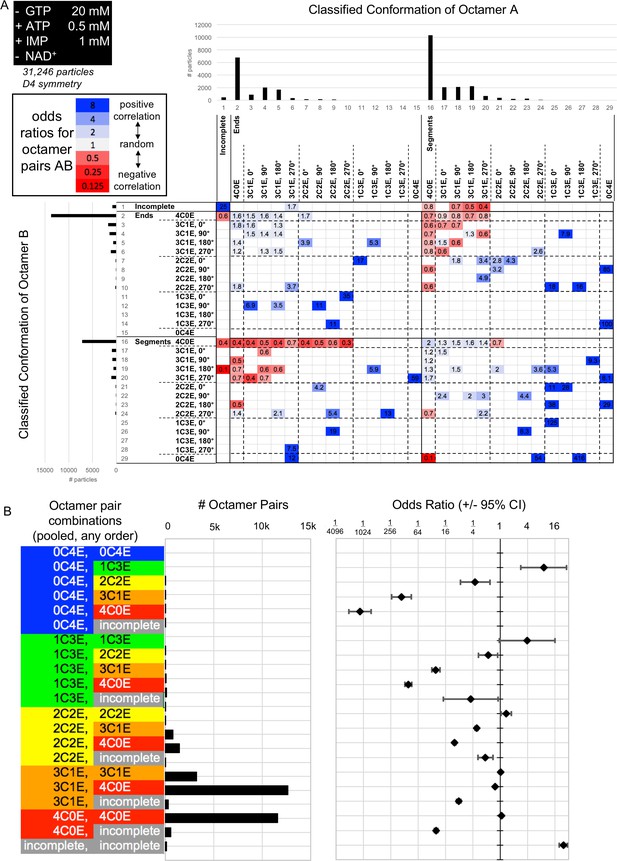
Interfaced octamer odds ratios for the 20 mM GTP, 0.5 mM ATP, 1 mM IMP cryo-EM dataset.
(A) Class distribution histograms and odds ratios for all combinations of classes of octamer A vs octamer B. Only ORs for which the associated 95% confidence interval does not include 1 are displayed. (B) A simplified summary of odds ratio data, made by pooling all classes depicted in A into the 21 different categories at left, irrespective of orientation, end vs segment, or A/B order. Center, histogram of # of octamer pairs per category. Right, odds ratios (error bars indicate 95% confidence interval). Abbreviations are as follow: 4C0E = fully compressed; 3C1E = 3/4 compressed, ¼ extended; 2C2E = 2/4 compressed, 2/4 extended; 1C3E = 1/4 compressed, 3/4 extended; 0C4E = fully extended.
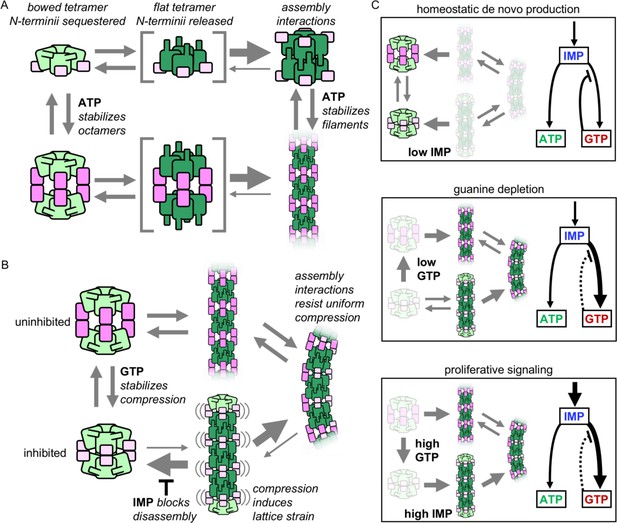
Model of IMPDH2 assembly and filaments’ role in guanine nucleotide regulation.
(A) Filament assemble when octamer interactions are stabilized by ATP binding to the regulatory domain (pink), and the N-terminal residues (blue) are released by flattening of the catalytic tetramer (green). Filament assembly interactions stabilize the flat conformation. (B) GTP binding stabilizes an inhibited, compressed conformation. Filament is less sensitive to GTP-induced compression, maintaining a population of octamers in mixed activity states. Filaments in the fully compressed GTP-bound state are strained, which promotes disassembly that is inhibited by substrate IMP binding. (C) The equilibrium in (B) explains the different cellular conditions in which IMPDH polymerization occurs. (i) Under homeostatic conditions IMPDH2 is dispersed and activity is regulated by GTP binding to octamers, which balances low levels of de novo synthesis between adenine and guanine pathways. ii) When guanine nucleotides are depleted the equilibrium shifts toward filaments. iii) Proliferative signaling can directly shift the equilibrium toward filaments, where higher flux is maintained through the guanine pathway under elevated GTP concentrations due to reduced sensitivity of the filaments to GTP inhibition (dashed line).
Videos
Comparison between the ‘flat’ tetramer of assembled filaments and the ‘bowed’ tetramer of free octamers.
Morph comparison between catalytic tetramers of the GTP/ATP/IMP filament assembly interface and the GTP/ATP free octamer. For visualization purposes, we have depicted the complete N-terminus in both conformations, however in the free octamer model, some residues were unresolved (gray). For both models, opposing tetramers, Bateman domains, and active site loops have been hidden from view.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Eschericia coli) | BL21(DE3) | Thermo Scientific | EC0114 | competent cells |
| Recombinant DNA reagent | pSMT3-hIMPDH2-WT (pJK053) | https://doi.org/10.1091/mbc.E17-04-0263 | expression plasmid | |
| Recombinant DNA reagent | pSMT3-hIMPDH2-Y12A (pJK061) | https://doi.org/10.1091/mbc.E17-04-0263 | expression plasmid | |
| Peptide, recombinant protein | Ubiquitin-like-specific protease 1 (ULP1) | https://doi.org/10.1091/mbc.E17-04-0263 | purified in house | |
| Peptide, recombinant protein | human IMPDH2 | https://doi.org/10.1091/mbc.E17-04-0263 | purified in house | |
| Chemical compound, drug | LB broth mix | LabExpress | 3003 | bacterial growth media |
| Chemical compound, drug | IPTG | GoldBio | I2481C100 | |
| Chemical compound, drug | MgCl2 | Fisher Scientific | BP215-500 | |
| Chemical compound, drug | KPO4 | Fisher Scientific | BP362-500 | |
| Chemical compound, drug | KCl | Fisher Scientific | BP217-3 | |
| Chemical compound, drug | imidazole | Sigma Aldrich | SLBT7469 | |
| Chemical compound, drug | urea | Fisher Scientific | BP169-212 | |
| Chemical compound, drug | DTT | Fisher Scientific | 172–25 | |
| Chemical compound, drug | HEPES | Fisher Scientific | BP310-1 | |
| Chemical compound, drug | ATP | Sigma Aldrich | A2383-10G | |
| Chemical compound, drug | GTP | Sigma Aldrich | G8877-1G | |
| Chemical compound, drug | IMP | Sigma Aldrich | 57510–5G | |
| Chemical compound, drug | NAD+ | Sigma Aldrich | N6522-1G | |
| Chemical compound, drug | uranyl formate | Electron Microscopy Sciences | 22450 | negative stain EM |
| Software, algorithm | GCTF | https://doi.org/10.1016/j.jsb.2015.11.003 | ||
| Software, algorithm | Relion | https://doi.org/10.7554/eLife.42166 | ||
| Software, algorithm | MotionCor2 | https://doi.org/10.1038/nmeth.4193 | ||
| Software, algorithm | SWISS-MODEL | https://doi.org/10.1093/nar/gky427 | ||
| Software, algorithm | UCSF Chimera | https://doi.org/10.1002/jcc.20084 | ||
| Software, algorithm | COOT | https://doi.org/10.1107/S0907444910007493 | ||
| Software, algorithm | PHENIX | https://doi.org/10.1107/97809553602060000865 | ||
| Software, algorithm | LocScale | https://doi.org/10.7554/eLife.27131 | ||
| Software, algorithm | Molprobity | https://doi.org/10.1107/S0907444909042073 | ||
| Software, algorithm | EMRinger | https://doi.org/10.1038/nmeth.3541 | ||
| Other | C-flat 2/2 holey carbon films on Cu 200 mesh grid | Protochips, Inc | CF-2/2–2C | cryo-EM sample preparation |
| Other | HisTrap FF Crude 5 ml | GE Life Sciences | 17528601 | protein purification |
| Other | Superose 6 Increase 10/300 GL | GE Life Sciences | 29-0916-96 | protein purification |
| Other | Amicon Ulta-15 30K MWCO centrifugal filters | Millipore | UFC903008 | protein concentration |





