Distinct roles for S. cerevisiae H2A copies in recombination and repeat stability, with a role for H2A.1 threonine 126
Figures
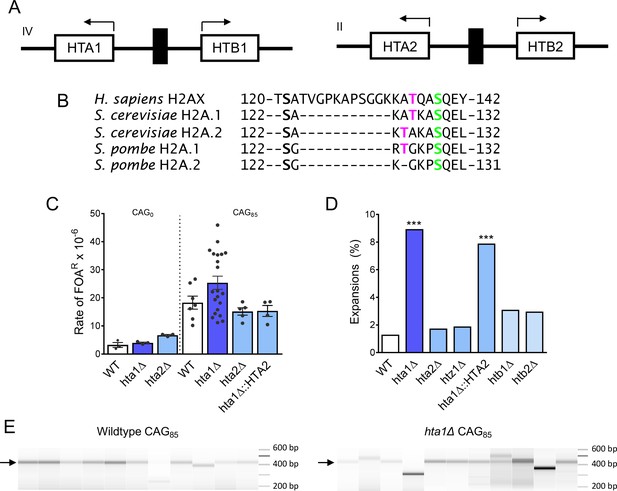
H2A.1 is required to prevent (CAG)85 repeat expansions.
(A) HTA is present in two copies in S. cerevisiae, paired with HTB. The gene pairs are divergently transcribed. The HTA1-HTB1 locus is on chromosome IV, the HTA2-HTB2 locus is on chromosome II. (B) H2A/H2AX protein sequences were aligned by Clustal Omega, only the C-terminal tail residues (past the histone fold) are shown. The threonine residue at position 125/126 is present in human H2AX, conserved in S. cerevisiae and S. pombe H2A.1. (C) Fragility of YAC CF1 (either contained (CAG)0 (no tract) or a (CAG)85 repeat tract) was assessed as previously described (reviewed in Polleys and Freudenreich, 2018) (Figure 1—figure supplement 1A). Rates were evaluated for significant differences by the Student’s t-test; no differences were statistically significant; error bars represent SEM (Supplementary file 1). (D, E) CAG repeat expansion (D) and contraction (E) frequencies were measured by a PCR-based stability assay, using primers P1 and P2, described in Figure 1—figure supplement 1A. Products were run in high-resolution Metaphor agarose or a fragment analyzer (as shown in F) and repeat length changes were evaluated. Raw instability data can be found in Supplementary file 2. Statistical deviation from wild-type was calculated by Fisher’s Exact Test: ***p<0.001 F) Visualization of CAG repeat lengths using an Advanced Analytical fragment analyzer. Arrow indicates length of (CAG)85 PCR product.
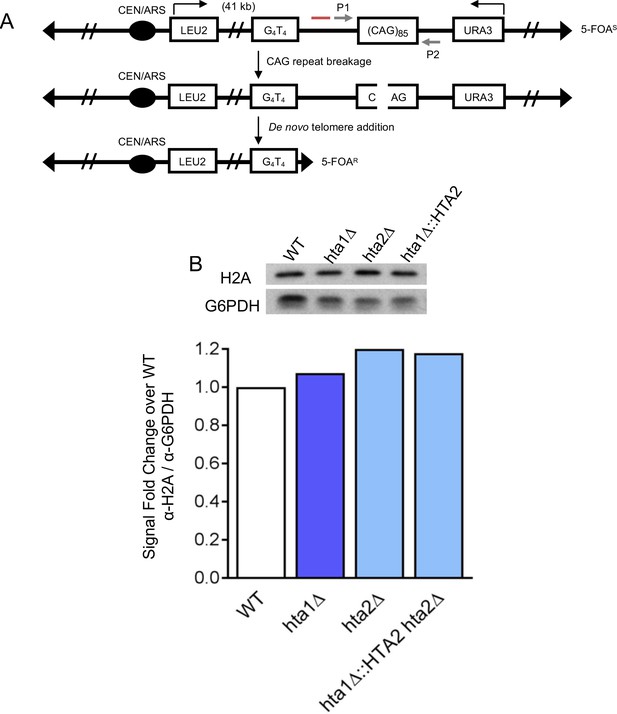
YAC system to evaluate CAG repeat fragility and instability.
(A) YAC CF1 contains an expanded CAG/CTG repeat originally cloned from a myotonic dystrophy patient. Here we have used 85 repeats with the CAG strand on the lagging strand template – referred to as (CAG)85. Breakage at the CAG repeat can result in resection to the backup telomere seed sequence (G4T4)13, resulting in de novo telomere addition and loss of the URA3 gene. This renders the cells resistant to 5-FOA and this rate can be used as a measure of repeat fragility (Polleys and Freudenreich, 2018). Primers (P1 and P2; gray arrows) flank the repeat to screen changes in repeat length by a PCR-based instability assay. Red line denotes location of Southern probe (described in Figure 2A). (B) Total H2A levels in the H2A variant deletion mutants and replacement of H2A.1 with H2A.2. G6PDH was used as a loading control. The y-axis label, fold change in signal from WT, is the fold change in relative quantification values of the mutants from WT, where WT is 1.
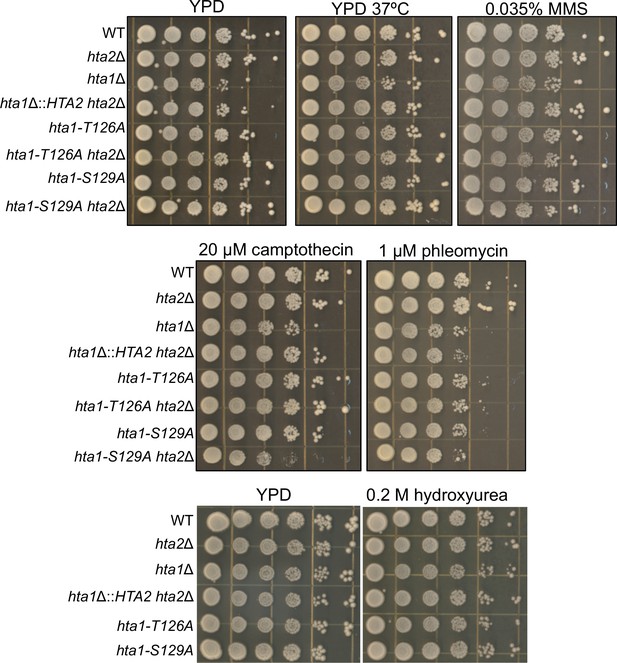
Drug sensitivity panels in H2A mutant strains.
Ten-fold serial dilutions of the listed genotypes were pinned onto either YPD without drug, YPD at 37°C, or YPD with various DNA damaging agents: YPD+0.035% MMS, YPD+20 μM camptothecin, YPD+1 μM phleomycin, and YPD + 0.2 M HU. Plates were incubated for 3 days and photographed. The experiment was repeated 3 times; a representative experiment is shown.
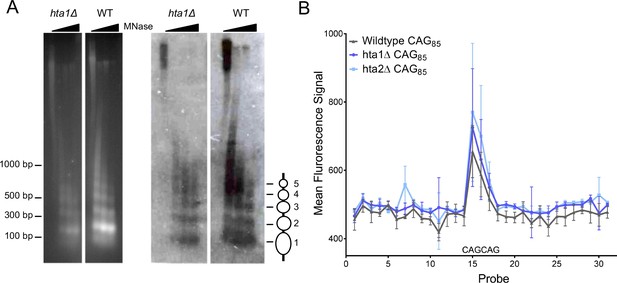
Nucleosome positioning at a (CAG)85 repeat is not altered in the absence of H2A.1 or H2A.2.
A) Indirect end-labeling of nucleosomal DNA upstream of the CAG repeat. MNase (0, 0.25, 2.5, and 7.5 units) digested DNA was run in 1.5% agarose with ethidium bromide (left) and Southern blotted (right) using a probe ~100 bp proximal to the CAG repeat (red line Figure 1—figure supplement 1A). Ovals represent nucleosome positions. The experiment was repeated six times; a representative blot is shown. (B) Illumina array mapping of nucleosome protection at the CAG repeat. Mononucleosomal DNA from strains containing the (CAG)85 repeats was hybridized to a custom array of 30-mer probes spanning 425 bp upstream of the repeat to 436 bp downstream of the repeat in YAC CF1. Probes 14–16 contain CAG repeats; probe 15 is composed purely of CAG repeats (probe sequences in Supplementary file 3). Error bars represent standard deviation of 2–3 independent experiments.
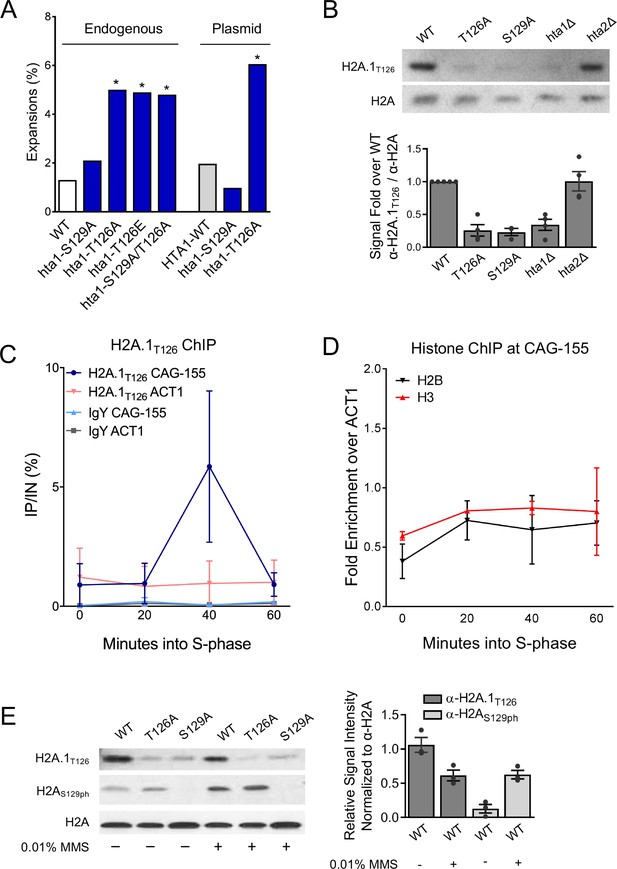
H2A.1-T126 is required to prevent (CAG)85 repeat expansions and is phosphorylated in vivo.
A) CAG repeat length changes were evaluated in strains with H2A.1 C-terminal tail point mutations. The source of H2A.1 is noted above the bars: either the point mutation was in the genomic copy of hta1 (HTA2 still present) or expressed from a plasmid (both endogenous copies of HTA-HTB are deleted). Statistical deviation from the wild-type was calculated by Fisher’s Exact Test, *p<0.05. (B) A Western blot with protein prepared from the indicated strains was probed with antibody raised against the H2A.1T126ph C-terminal tail peptide (H2A.1T126) or anti-H2A. H2A mutations were in the genomic copy of indicated H2A variant; the other variant was still present. Quantification of this blot and other experiments using the same strains (each indicated by a point) is shown at right. The average and SEM are plotted. (C) Incorporation of H2A.1 at a (CAG)155 repeat was measured by ChIP using the H2A.1T126 antibody; IgY was used as a negative control. Cultures were α-factor synchronized and released into fresh medium; time point taken as indicated after release, in S-phase. Enrichment was measured using a primers set that amplified a region spanning 0.4 and 0.6 kb upstream of the CAG repeat or at an ACT1 internal control locus. The average IP/IN and standard error for two independent experiments is plotted. (D) ChIP for histones H2B and H3 at the CAG repeat was performed as in (C). IP/IN values were normalized to the ACT1 control signal. Individual IP/IN and ACT1 normalized values can be found in Supplementary file 4. (E) Western blots using antibodies against H2A.1T126 (top panel), H2AS129ph (middle panel), and total H2A (bottom panel). Total protein was prepared from H2A wild-type and point mutant strains (mutations in the plasmid copy of indicated H2A variant, hta2Δ background) after 2 hr exposure to either no drug (-) or 0.01% MMS (+). Quantification of the H2A.1T126 and H2AS129ph antibody signals normalized to total H2A levels in WT strains are graphed to the right; dots indicate independent experimental replicates and the average and SEM is shown. Quantification of all bands is shown in and listed in Supplementary file 9.
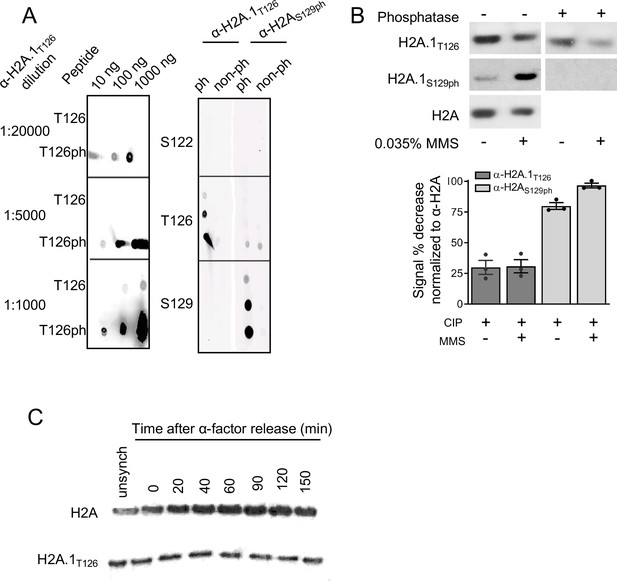
Analyses of H2A.1T126 antibody specificity and H2A.1 levels over the cell cycle.
(A) The H2A.1T126 antibody was raised against the phosphopeptide CZKKSAKA9pT]KASQEL. The antibody specificity was tested against H2A C-terminal tail peptides by peptide dot blot. Whether the peptide was phosphorylated on either S122 or T126 or S129 is indicated to the left of each panel. In the right hand panel, both the anti-T126ph and anti-S129ph were diluted 1:10000. (B) Phophatase treatment of histone extracts from wild-type cells to test the specificity of the H2A.1T126 antibody. Extracts from cells treated with MMS to induce H2A S129 phosphorylation were probed with anti-S129ph as a positive control. For the blot shown, urea-extracted whole cell lyastes were separated in a 15% PAGE gel, blotted onto PVDF, treated with CIP for 1 hr at 37°C, and probed with the indicated antibody. Quantification normalized to total H2A levels is expressed as a percent decrease from no-CIP treatment. On blot phosphatase samples were normalized to non-phosphatase treated H2A as the H2A antibody (Abcam cat #13923) recognizes phosphorylated Serine-2. Quantification is an average of both pre-treatment (in tube) and on-blot CIP treatment values (individual experimental values are indicated by points and are listed in Supplementary file 9). (C) Cell cycle regulation of H2A.1T126 and total H2A levels were evaluated by Western blot of WT α-factor arrested and released cells. Total H2A protein levels increase throughout the cell cycle while H2A.1T126 protein levels remain stable.
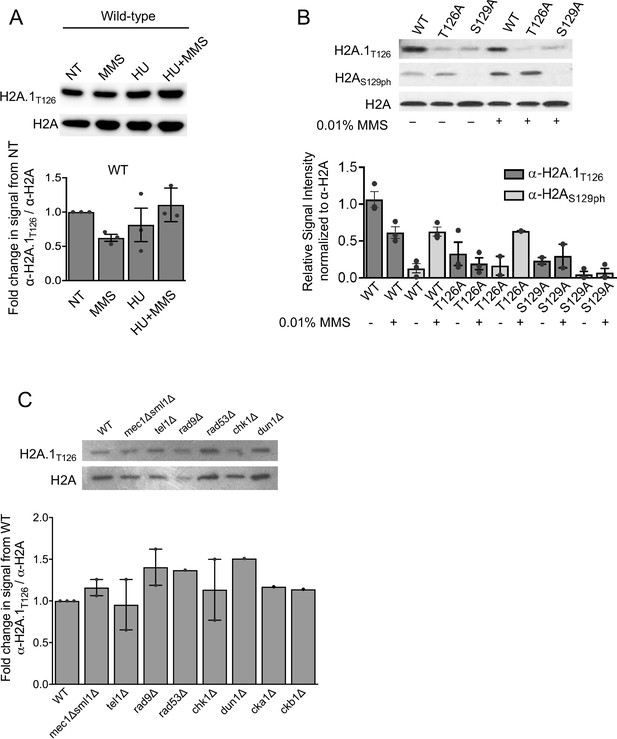
Analyses of H2A.1T126 levels after exposure to DNA damaging agents and deletion of kinases involved in the DNA damage response.
(A) H2A.1T126 and total H2A signal in response to 1 hr exposure to 0.03% MMS, 0.2M HU, or 0.2M HU + 0.03% MMS. The chart shows quantification of the H2A.1T126 antibody signal normalized to total H2A levels and compared to no treatment (NT) for this experiment and independent replicates. Average of individual experiments (marked by a dot) and SEM is shown. (B) H2A.1T126 (top panel), H2AS129h (middle panel), and total H2A (bottom panel) Western blots after 2 hr exposure to 0.01% MMS. Quantification of the H2A.1T126 and H2AS129ph antibody signals normalized to total H2A levels for this experiment and independent replicates is shown below the blot. Average of individual experiments (marked by a dot) and SEM is shown. (C) Quantification of antibody signal for α-H2A.1T126 in kinase mutants. The H2A.1T126 signal intensity was normalized to H2A signal; fold change from WT is plotted. Signal intensity and/or fold change values for these experiments can be found in Supplementary file 9.
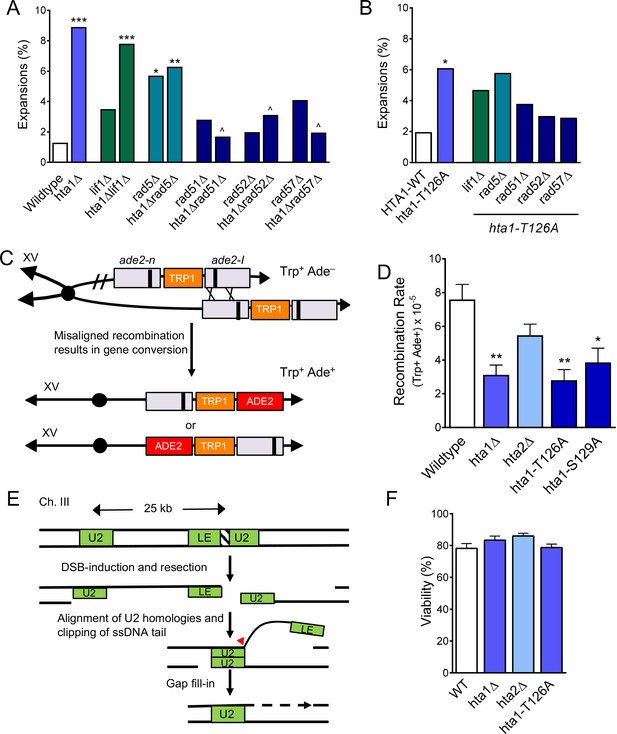
H2A.1 promotes HR fidelity to prevent (CAG)85 expansions and is required for efficient SCR.
(A) Repair proteins were deleted in hta1Δ mutant. Changes in CAG repeat length were assessed as in Figure 1C. Statistical deviations were calculated by Fisher’s Exact Test: *p<0.05 to WT, **p<0.01 to WT, ***p<0.001 to WT; ^p<0.05 suppression from hta1Δ. (B) Repair proteins were deleted in the strain expressing hta1-T126A from the plasmid; no copy of H2A.2 is present in these cells. Changes in CAG repeat length were assessed as in (A). Statistical deviation from the plasmid HTA1 wild-type were calculated by Fisher’s Exact Test: *p<0.05; statistical deviation from the plasmid hta1-T126A was also tested, but none were significantly suppressed. (C) Misaligned recombination during SCR can be measured by gene conversion from Trp+ Ade– to Trp+ Ade+ (House et al., 2014b; Mozlin et al., 2008). If recombination occurs on either side of the ade2-n and ade2-I mutations (indicated by X’s) between aligned mutant alleles, then gene conversion will create a WT ADE2 gene on one of the chromatids. The ade2-I gene conversion resulting from the indicated cross is shown directly below the arrow; an alternate alignment and gene conversion of the ade2-n allele is also possible (bottom chromosome). (D) Rates of spontaneous unequal SCR. For these experiments, the hta1-T126A and hta1-S129A mutations were integrated at the genomic locus, replacing the wild-type copy of HTA1; HTA2 remains intact in these strains. SCR rates for individual experiments are in Supplementary file 5, including for the HTA1 point mutants with HTA2 deleted (presence or absence of HTA2 did not change the results). Average rate and SEM is shown. Statistical differences were calculated using a Student’s t-test: *p<0.05 to WT, **p<0.01 to WT. (E) SSA at a DSB can be measured by viability in the presence of a galactose-induced HO DSB. Upon galactose treatment a DSB at the inserted HO cut site in the LEU2 gene results in resection to the U2 region of homology 25 kB away. Repair occurs via single strand annealing between the U2 segments followed by cleavage of the non-homologous tail (arrowhead) and gap filling (dotted lines), eliminating the HO target site, and cells survive in the presence of galactose (Vaze et al., 2002). (F) Percent viability as measured by the number of colonies on YP-Gal divided by the number of colonies on YPD. Statistical deviation from wild-type was tested by a Student’s t-test; none were significantly different from wild-type (Supplementary file 6).
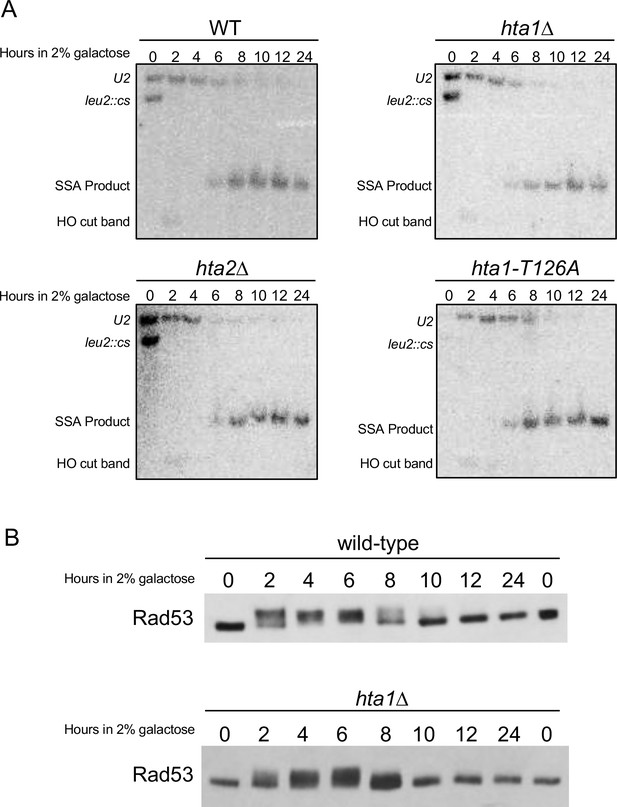
Single strand annealing and checkpoint response.
(A) Using the assay system described in Figure 4E, wild-type, hta1Δ, hta2Δ, and hta1-T126A strains were subjected to DSB induction at the leu2 cut site (leu2::cs) by growth in galactose for the indicated times (hours in 2% galactose). Genomic DNA was purified, restriction digested, separated by gel electrophoresis and Southern blotted as previously described (Vaze et al., 2002). The timing of HO cutting (HO cut band) and repair product formation via SSA (SSA product) is not different between the indicated genotypes. (B) Using the SSA assay described in Figure 4E, Rad53 phosphorylation profiles were determined by Western blot in the WT and hta1Δ strains. The hta1Δ strain shows no significant difference from WT in the kinetics of Rad53 phosphorylation and dephosphorylation after HO cutting and repair.
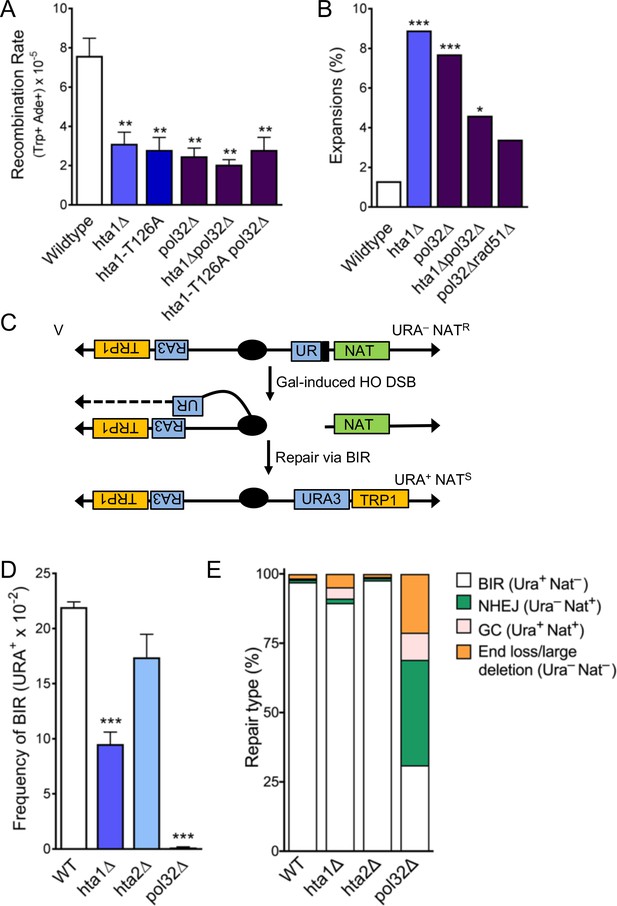
H2A.1 and Pol32 work in the same pathway during SCR, and H2A.1 is required for efficient BIR.
(A) Rates of spontaneous unequal SCR, assessed as in Figure 4D. Statistical deviation from wild-type was calculated using a Student’s t-test, **p<0.01. (B) Changes in CAG repeat length were assessed as in Figure 1C. Statistical deviation from wild-type was calculated by Fisher’s Exact Test: *p<0.05, ***p<0.001. (C) Assay to measure BIR at an HO-induced DSB. The first 400 bp of the URA3 gene (UR; right arm) is upstream of an HO cut site (stripes). Homology driven repair of the HO induced DSB can occur with the remaining 404 bp of the URA3 gene (A3; left arm) and if completed via BIR renders the cell URA+ and NATS (Anand et al., 2014). (D) Frequencies of BIR during repair of an HO-induced DSB; SEM of 4–9 replicates is shown (Supplementary file 7). Statistical deviation from wildtype was calculated using a Student’s t-test, ***p<0.001. (E) The type of repair induced by the HO DSB was evaluated by individually scoring all colonies for growth on YC-URA and YEPD+Nat. Colonies that were URA+ NATS are were repaired via BIR. Other types of possible repair events include gene conversion (URA+ NATR), non-homologous end-joining (URA- NATR), and de novo telomere addition (URA- NATS). Statistical analysis of BIR frequencies and repair types can be found in Supplementary file 7.
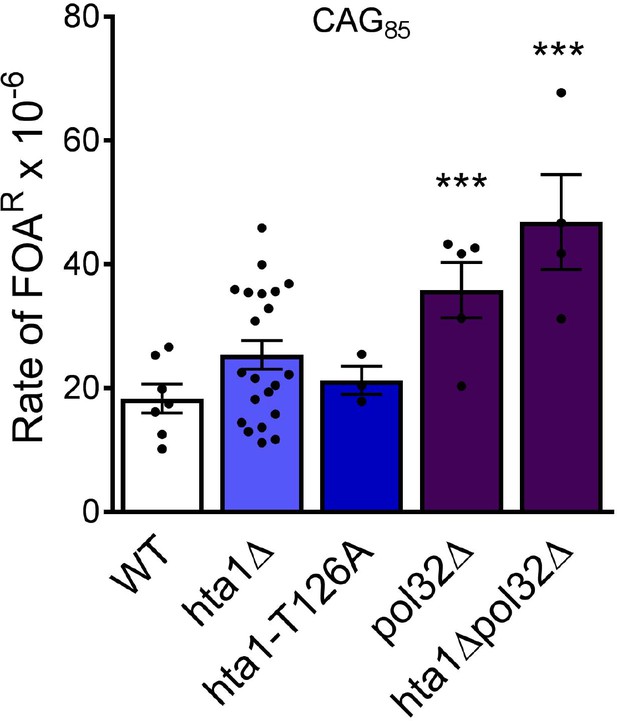
CAG fragility rates in the absence of Pol32.
(CAG)85 repeat fragility was measured in hta1Δ, hta1-T126A and pol32Δ, and hta1Δ pol32Δ strains. Significant deviation from wild-type was measured by Student’s t-test: ***p<0.001 to WT. Values in Supplementary file 1.
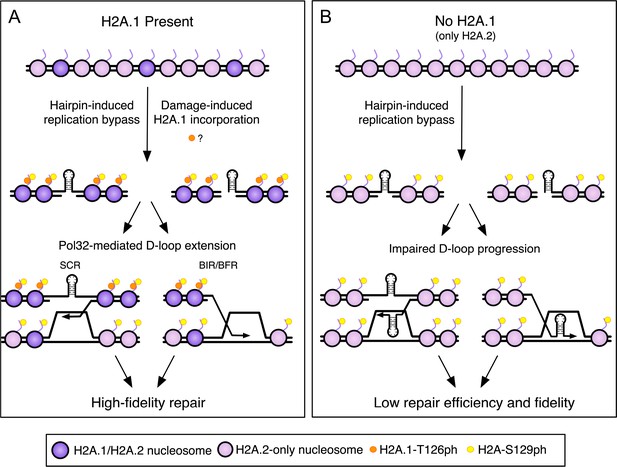
A model for high-fidelity HR-mediated repair promoted by H2A.1 and T126 phosphorylation.
The initiating lesion at the CAG repeat could be a nick, gap, or a one-ended break (e.g. created at a replication fork). (A) Incorporation of H2A.1 at sites of damage (with or without T126ph; with T126ph is shown) promotes repair fidelity. The presence of H2A.1 will promote efficient D-loop extension after invasion into the sister chromatid, leading to repair with fidelity and maintenance of CAG repeat number. Alternatively, the resolution of recombination intermediates may require H2A.1 to prevent multiple reinvasion events that could lead to repeat expansions. Though T126ph is shown as present during BIR/BFR for continuity with the previous diagram, our data suggest that the T126 identity has minimal effect on this step in the context of BIR (BFR was not tested). See discussion for details. (B) Without H2A.1 only H2A.2 with T125 is present and D-loop extension is impeded, allowing hairpins or misalignment during Rad51-dependent invasion, leading to less efficient repair and CAG repeat instability. Purple nucleosomes contain H2A.1, pink nucleosomes are H2A.2-containing only. Orange dots represent H2A.1-T126ph, which could either be a pre-existing modification or induced locally at the damage site. Yellow dots represent H2A-S129ph.
Additional files
-
Supplementary file 1
(CAG)0 and (CAG)85 fragility assay data.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp1-v2.xlsx
-
Supplementary file 2
(CAG)85 repeat instability assay data.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp2-v2.xlsx
-
Supplementary file 3
Illumina Array probe sequences.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp3-v2.xlsx
-
Supplementary file 4
ChIP data.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp4-v2.xlsx
-
Supplementary file 5
Sister chromatid recombination assay data.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp5-v2.xlsx
-
Supplementary file 6
Single strand annealing assay data.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp6-v2.xlsx
-
Supplementary file 7
Break-induced replication assay data.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp7-v2.xlsx
-
Supplementary file 8
HTA2 gene levels by qPCR.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp8-v2.xlsx
-
Supplementary file 9
Western blot quantifications.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp9-v2.xlsx
-
Supplementary file 10
Pol32 mRNA levels.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp10-v2.xlsx
-
Supplementary file 11
Yeast strains used in this study.
- https://cdn.elifesciences.org/articles/53362/elife-53362-supp11-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53362/elife-53362-transrepform-v2.docx





