Specific depletion of the motor protein KIF5B leads to deficits in dendritic transport, synaptic plasticity and memory
Figures
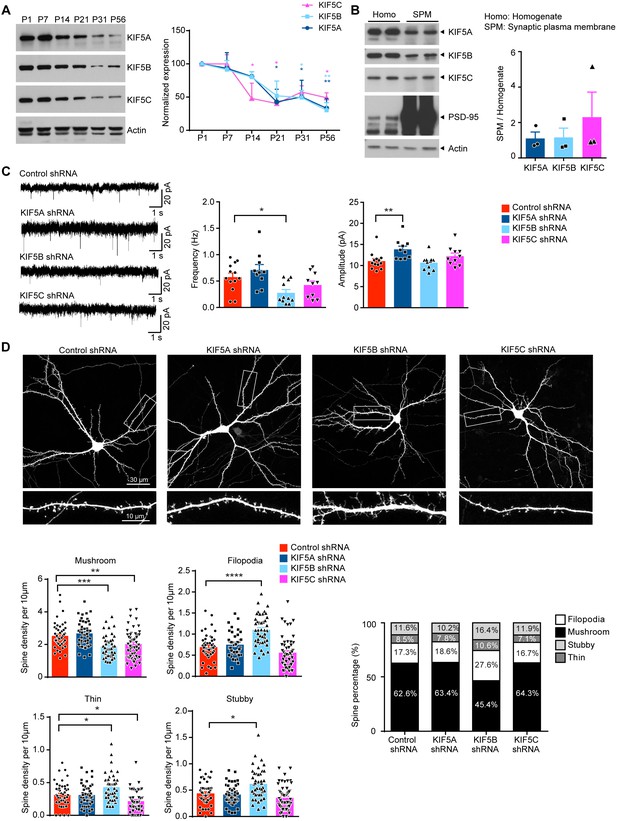
Different roles of KIF5s on dendritic spine morphogenesis and synaptic function.
(A) Western blot showing the developmental expression of different Kinesin I motors in the mouse hippocampus. The expression of all three KIF5s significantly decreased at later postnatal stages as compared to postnatal day 1 (P1); three independent experiments, mean + SEM; *p<0.05; **p<0.01; Two way ANOVA with Dunnett's multiple comparisons test. (B) Presence of all three KIF5s in the synaptic plasma membrane (SPM). Postsynaptic density-95 (PSD-95) served as the positive control for the SPM fraction. (C) Whole-cell patch recording was performed on hippocampal neuron upon individually knocking down KIF5A, KIF5B or KIF5C with short hairpin RNAs (shRNAs). Representative traces of miniature excitatory postsynaptic currents (mEPSCs) were shown. KIF5B knockdown led to significant decrease in mEPSC frequency compared to control. KIF5A knock-down significantly increased mEPSC amplitude compared to control (n = 10–13 neurons for each condition from five independent experiments; mean + SEM; *p<0.05; **p<0.01; Kruskal-Wallis test). (D) Representative images of dissociated rat hippocampal neurons co-transfected with GFP and the shRNA targeting individual KIF5 or a control shRNA. Neurons were transfected at days in vitro (DIV) 13, and fixed and stained by GFP antibody at DIV 16. Knockdown of KIF5B significantly reduced the density of mushroom spines and led to a significant increase in the number of filopodia, thin and stubby spines. Knockdown of KIF5C significantly decreased the number of mushroom and thin spines (42–55 neurons of each group from three independent experiments were quantified; mean + SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; Kruskal-Wallis test).
-
Figure 1—source data 1
Plotted values for Figure 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig1-data1-v2.xlsx

Validation of the knockdown efficiency and specificity of different shRNAs targeting KIF5.
(A) For western blot analysis, cortical neurons were transfected with the different shRNAs by nucleofection. At DIV 5, lysate was collected for Western blotting with antibody that specifically recognized each KIF5 homolog (three independent experiments; mean + SEM; multiple t-test with the Holm-Sidak method, alpha: 0.05). (B) For immunofluorescent staining analysis, different shRNAs were transfected into hippocampal neurons at DIV 11 for 6 days and the soma intensity for the corresponding KIF5 homolog was analyzed. 9 or 11 neurons of each group were quantified. Data are presented in mean + SEM; *p<0.05; Student’s t-test.
-
Figure 1—figure supplement 1—source data 1
Plotted values for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig1-figsupp1-data1-v2.xlsx
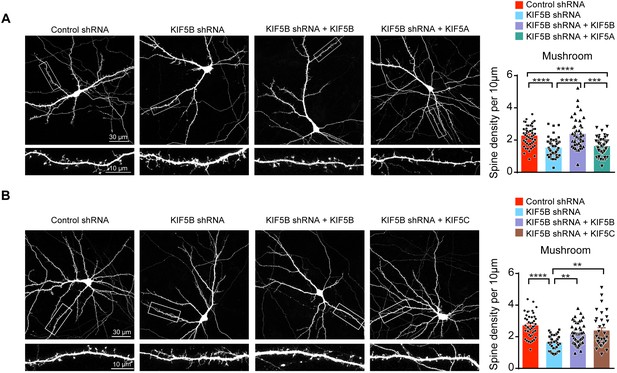
Non-redundant roles of KIF5A and KIF5B on dendritic spine morphogenesis.
(A) Representative images of dissociated rat hippocampal neurons co-transfected with GFP and KIF5B-shRNA with/without KIF5B or KIF5A construct. Co-expression of KIF5B rescued the loss of mushroom spines induced by KIF5B-shRNA, while KIF5A expression failed to reverse the spine phenotypes (42–51 neurons of each group from three independent experiments were quantified; mean + SEM; ***p<0.001, ****p<0.0001; Kruskal-Wallis test). (B) Representative images of dissociated rat hippocampal neurons co-transfected with GFP and KIF5B-shRNA with/without KIF5B or KIF5C construct. Co-expression of KIF5C rescued the loss of mushroom spines induced by KIF5B-shRNA (34–46 neurons of each group from four independent experiments were quantified; mean + SEM; **p<0.01, ****p<0.0001; Kruskal-Wallis test).
-
Figure 2—source data 1
Plotted values for Figure 2.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig2-data1-v2.xlsx
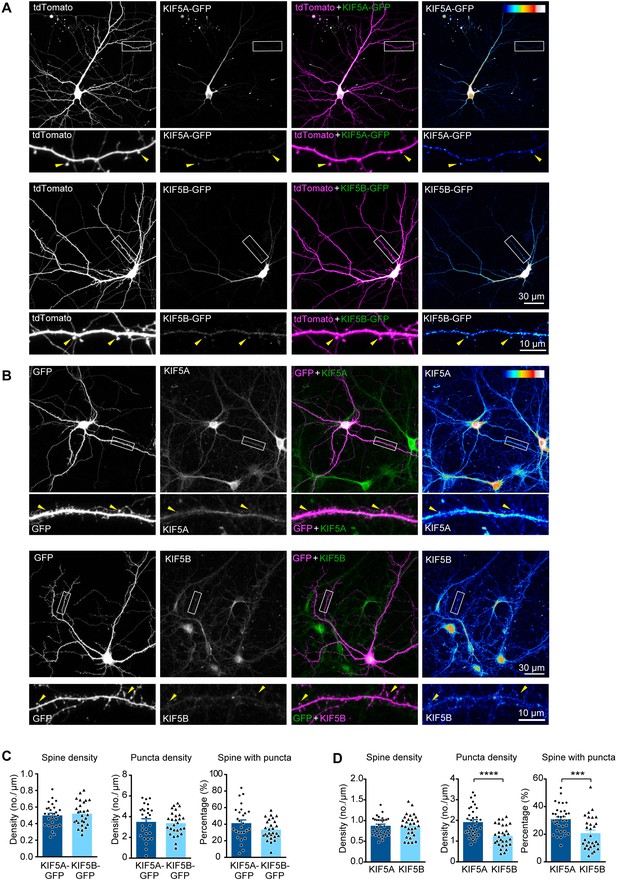
Subcellular localization of KIF5A and KIF5B in hippocampal neurons.
(A) Dissociated hippocampal neurons were transfected with tdTomato and equal amounts of either KIF5A-GFP or KIF5B-GFP. Greyscale images for individual channels, 2-channel merged images (blue for tdTomato and yellow for KIF5-GFP), as well as ‘royal’ (ImageJ) colormap display of KIF5-GFP channel were shown for representative cells and dendrites. Both KIF5A and KIF5B puncta were present in the heads of dendritic spines (arrowheads). (B) Dissociated hippocampal neurons labeled with GFP were immunostained with specific antibodies against KIF5A or KIF5B. Images were displayed in a similar manner, except that GFP was displayed in blue and KIF5 signals were displayed in yellow in merge-channel images. KIF5A and KIF5B puncta were detected in the heads of dendritic spines (arrowheads). (C) Quantification for (A). The densities of spine and the percentage of spines with detected puncta were quantified by manual counting. 27 neurons were quantified for each group. Data are presented in mean + SEM, Student’s t-test. (D) Quatification for (B).The densities of spine and the percentage of spines with detected puncta were quantified by manual counting. 33 neurons were analyzed for KIF5A and 29 neurons were analyzed for KIF5B. Data are presented in mean + SEM; ***p<0.001, ****p<0.0001, Mann-Whitney test.
-
Figure 2—figure supplement 1—source data 1
Plotted values for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig2-figsupp1-data1-v2.xlsx
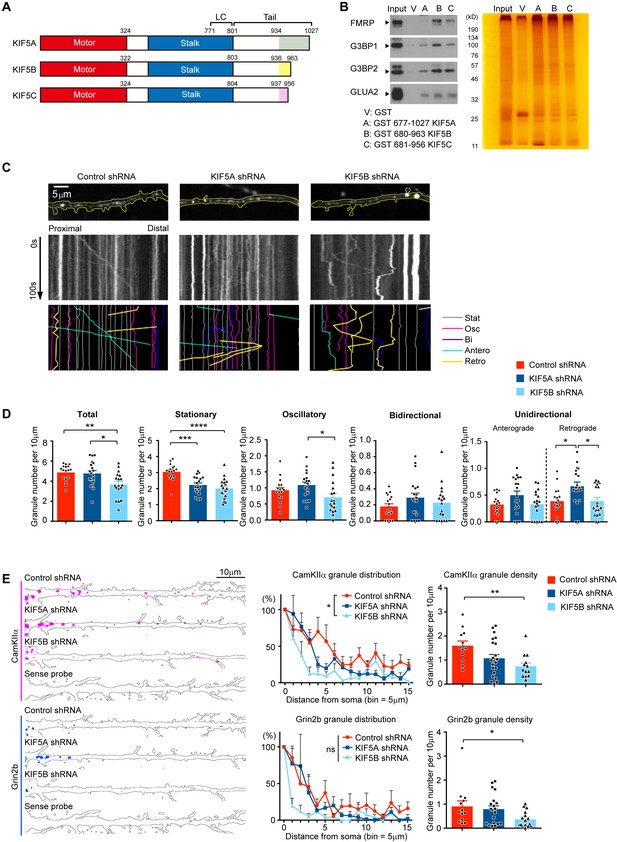
KIF5A and KIF5B differentially regulate the dendritic transport of FMRP and mRNA cargoes.
(A) Schematic diagram of the motor, stalk and tail domains of KIF5A, KIF5B and KIF5C, with the corresponding amino-acid positions indicated. The diverse carboxyl-termini within the tail domains were colored. (B) Pull-down of proteins from brain homogenate by GST-tagged KIF5s. FMRP was preferentially pulled-down from the brain lysate by KIF5B and KIF5C, but not KIF5A. (C) Representative dendrites and kymographs of GFP-FMRP granules from each group of neurons transfected with control shRNA, KIF5A-shRNA or KIF5B-shRNA. Different types of movements were traced manually and displayed in different colors (Stat: stationary; Osc: oscillatory; Bi: bidirectional; Antero: anterograde; Retro: retrograde.). Live imaging was conducted at one frame per second for 100 s. (D) Quantification of the densities of granules showing each category of movement. 17–19 neurons were quantified for each experimental condition. Results were pooled from three independent experiments; mean + SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA, Tukey's multiple comparisons test for total, stationary, oscillatory and anterograde granules; Kruskal-Wallis, Dunn's multiple comparisons test for bidirectional and retrograde granules. (E) Left: representative FISH images of CaMKIIα and Grin2b mRNA puncta along dendrites. Hybridization with the sense probe served as negative control for the in situ hybridization. Middle: distribution of puncta number along dendrites (one bin = 5 μm) from cell body. 12–18 neurons were quantified for Grin2b mRNA and 11–20 neurons were quantified for CaMKIIα mRNA; mean + SEM; *p<0.05, two-way ANOVA, Tukey's multiple comparisons test. Right: quantification of granule density. 12–22 neurons were quantified for CaMKIIα mRNA analysis; mean + SEM; **p<0.01, one-way ANOVA, Tukey's multiple comparisons test. 13–20 neurons were quantified for Grin2b mRNA; mean + SEM; *p<0.05, one-way ANOVA, Kruskal-Wallis test. Results were pooled from three independent experiments.
-
Figure 3—source data 1
Plotted values for Figure 3.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig3-data1-v2.xlsx

Quantification of GFP-FMRP granule motility and percentage of movements after knockdown of KIF5A or KIF5B.
(A) Demonstration of GFP-FMRP granule quantification using kymograph tracing. Different types of movements were traced manually and displayed in different colors. (Stat: stationary; Osc: oscillatory; Bi: bidirectional; Antero: anterograde; Retro: retrograde.) Live imaging was conducted at one frame per second for 100 s. (B) The travel distance, net displacement, maximal run length and maximal velocity were analyzed for each unidirectional and bidirectional granule. 305, 462 and 242 granules from neurons transfected with control shRNA, KIF5A-shRNA, and KIF5B-shRNA were quantified, respectively. Data are presented in mean + SEM, Kruskal-Wallis, Dunn's multiple comparisons test. (C) The percentage of each type of granule movement were quantified. 1571, 1467 and 918 granules from neurons transfected with control shRNA, KIF5A-shRNA, and KIF5B-shRNA were quantified, respectively. The percentage of motile granules was increased after knockdown of KIF5A or KIF5B, while the motility of unidirectional and bidirectional granules was not affected.
-
Figure 3—figure supplement 1—source data 1
Plotted values for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig3-figsupp1-data1-v2.xlsx
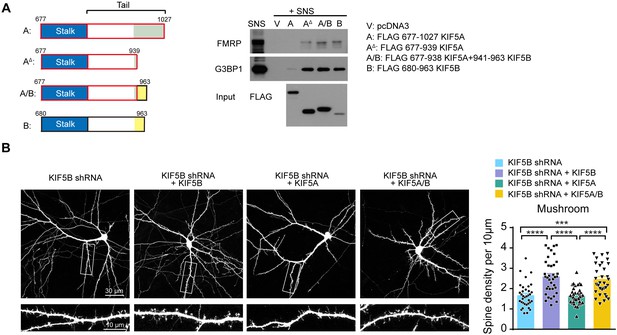
Carboxyl-terminus determines the functional specificity of KIF5A and KIF5B in dendritic spine morphogenesis.
(A) Schematic diagram of the different constructs for pull-down assay (left panel). FMRP and G3BP1 were pulled-down from the SNS by either the truncated KIF5A lacking the 88 amino acids at the carboxyl-terminus, or chimeric KIF5A/B in which the carboxyl-terminus was substituted by that of KIF5B (right panel). (B) Representative images of dissociated rat hippocampal neurons co-transfected with GFP and KIF5B-shRNA with/without KIF5B, KIF5A or chimeric KIF5A/B containing the carboxyl-terminus of KIF5B (1–938 KIF5A+941–963 KIF5B). Co-expression of chimeric KIF5A/B rescued the loss of mushroom spines induced by KIF5B-shRNA (32–37 neurons of each group from three independent experiments were quantified; mean + SEM; ***p<0.001, ****p<0.0001; Kruskal-Wallis test).
-
Figure 4—source data 1
Plotted values for Figure 4.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig4-data1-v2.xlsx
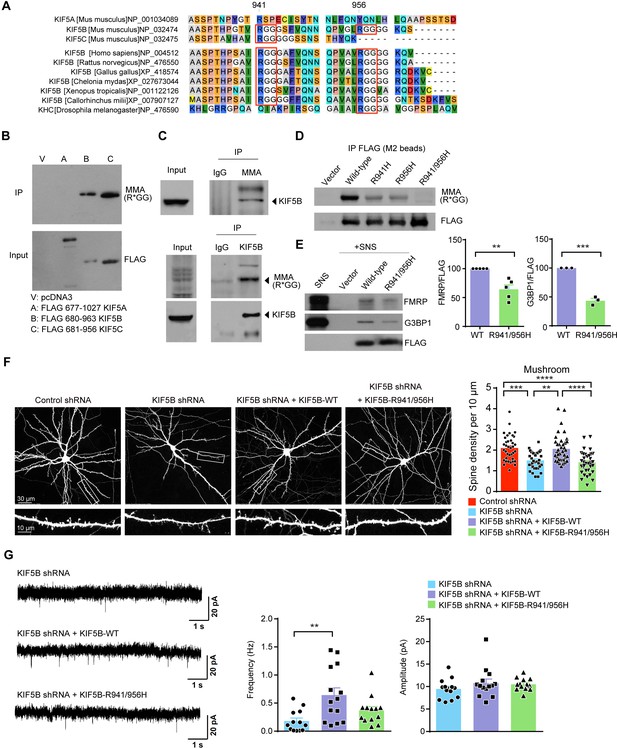
Mono-methylation of arginine near carboxyl-terminus of KIF5B is required for the formation of mushroom spines.
(A) Amino acid alignment of the carboxyl-termini of KIF5A, KIF5B and KIF5C. Two conserved arginine residues (R941 and R956) in KIF5B across different vertebrates were highlighted by red boxes. (B) Carboxyl-terminal portions of different KIF5s were expressed in 293 T cells. After immunoprecipitation with anti-FLAG beads, proteins were immunoblotted by antibody targeting mono-arginine methylation at glycine-rich motifs [MMA (R*GG)]. Transfection with pcDNA3 (vector) served as a negative control. Only KIF5B and KIF5C but not KIF5A were arginine-methylated. (C) Methylation of KIF5B in the synaptoneurosome. Proteins were immunoprecipitated with anti-Mono-Methyl-Arginine antibody (MMA) and immunoblotted with KIF5B antibody (upper panel). Reciprocal immunoprecipitation using KIF5B antibody was performed to verify the arginine methylation pattern (lower panel). (D) Methylation-deficient mutants of KIF5B were constructed by single or double substitution of the two arginine residues to histidine. Vector and FLAG-tagged KIF5B constructs were transfected into 293 T cells. After immunoprecipitation with anti-FLAG beads, proteins were immunoblotted by antibody targeting mono-arginine methylation at glycine-rich motifs [MMA (R*GG)]. Mono-methylation of KIF5B was abolished by substitution of both arginine sites by histidine (R941H/R956H). (E) Reduced amount of FMRP and G3BP1 was pulled down by methylation-deficient (R941/956H) mutant of KIF5B (five independent experiments for FMRP and three independent experiments for G3BP1; mean + SEM; **p<0.01; ***p<0.001; Student’s t-test). (F) Representative images of dissociated rat hippocampal neurons co-transfected with GFP and KIF5B-shRNA together with either RNAi-resistant wild-type or the methylation-deficient (R941/956H) KIF5B construct. Co-expression of wild-type but not the methylation-deficient mutant KIF5B rescued the loss of mushroom spines induced by the KIF5B-shRNA (35–38 neurons of each group from three independent experiments were quantified; mean + SEM; **p<0.01, ***p<0.001, ****p<0.0001; Kruskal-Wallis test). (G) Representative traces of mEPSCs of KIF5B-shRNA co-expressed with RNAi-resistant wild-type or the methylation-deficient (R941/956H) KIF5B construct were shown. Co-expression of wild-type KIF5B, but not methylation-deficient KIF5B, reversed the reduction of mEPSC frequency caused by KIF5B knock-down (13–14 neurons from five independent experiments were quantified for each experimental condition; mean + SEM; **p<0.01; Kruskal-Wallis test).
-
Figure 5—source data 1
Plotted values for Figure 5.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig5-data1-v2.xlsx
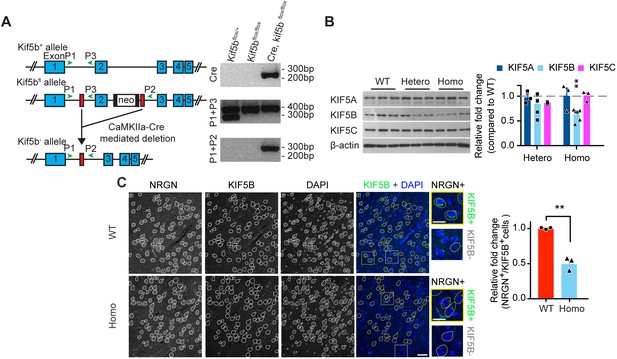
Targeting of Kif5b gene in CaMKII-Cre/Kif5bfl/fl mice and validation of the conditional knockout mice.
(A) Scheme of KIF5B knockout strategy (left panel). The blue rectangles (E1, E2, E3, E4, and E5) annotated exon 1, exon 2, exon 3, exon 4, and exon 5 of the Kif5b gene, respectively. The white rectangle with black bars on each side (each bar representing a flippase recognition target site) represented the 1.7 kb frt-NeoR-frt cassette. The red rectangles represent the LoxP sites (drawing not to scale). The green arrowheads indicated the designated annealing positions of genotyping PCR primers. Genotyping analysis of PCR product from mice DNA with different genotypes of the Kif5b gene (right panel). Cre primers were used to detect Cre recombinase gene. Primers P1 and P3 were used to identify Kif5bfl/+ and Kif5bfl/fl genotypes. Primers P1 and P2 were used to identify Kif5b conditional knockout, Kif5b-/- genotype. (B) Western blot analysis probed with anti-KIF5A, anti-KIF5B, anti-KIF5C and anti-β-actin antibodies. Quantification result in the right panel was presented as the relative fold change compared to wild-type (WT). n = 6 for all groups for the analysis of KIF5B. n = 3 for the analysis of other protein targets. Data were presented in mean + SEM. **p<0.01, One-way ANOVA with post hoc Tukey's HSD. (C) Immunohistochemical staining of sagittal brain sections showed a significant reduction of cells that were both positive for KIF5B and Neurogranin (NRGN) in frontal area on postnatal day (P) 42 ± 1. White traces highlighted NRGN-positive cells. Yellow traces indicated cells that were positive for both NRGN and KIF5B. Yellow and white squares indicate the zoom-in areas of NRGN+/KIF5B+ cells and NRGN+/KIF5B- cells, respectively. Scale bar, 50 μm and 25 μm in magnified inserts. Right panel: quantification of KIF5B knockout in immunohistochemical staining of frontal area. n = 3 for WT. n = 3 for Homo. Data were presented in mean + SEM. **p<0.01,Student’s t-test with Welch's correction.
-
Figure 6—source data 1
Plotted values for Figure 6.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig6-data1-v2.xlsx
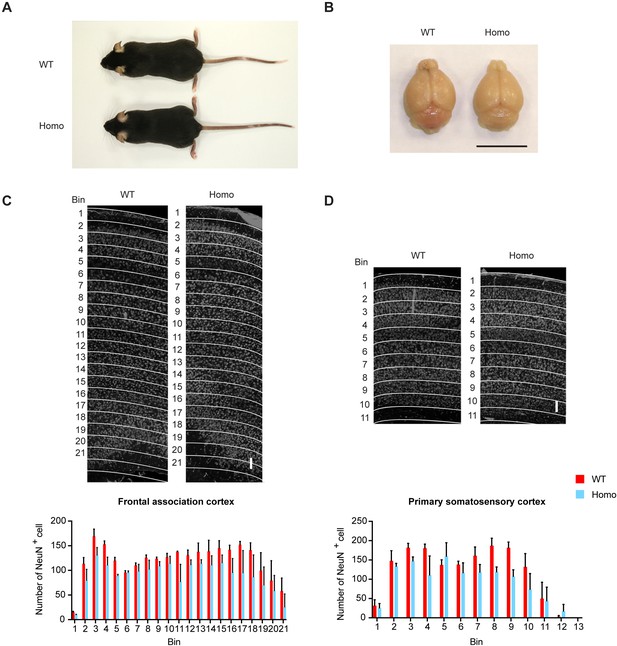
KIF5B conditional knockout mice show normal general appearance and cortical layer architecture.
(A) Representative images of 7-week-old male WT and Homo mice. (B) Representative images of 7 week old WT and Homo mouse brains. Scale bar, 1 cm. (C–D) Immunohistochemical staining of NeuN-positive cells in coronal sections of WT and Homo mouse brains. The cortex was arbitrarily defined into bin layers parallel to the pia surface. Each bin layer had a thickness of 100 μm and the number of NeuN-positive cells within each layer was counted. Images of the frontal association (C) and the primary somatosensory (D) cortices were analyzed and the quantification result was shown in the lower panel. Bin size, 100 μm. Scale bar, 100 μm. n = 3 for WT and Homo. Data are presented in mean + SEM, multiple t-test with false discovery rate approach (FDR, Benjamin, Krieger, yekutieli), Q = 1%.
-
Figure 6—figure supplement 1—source data 1
Plotted values for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig6-figsupp1-data1-v2.xlsx
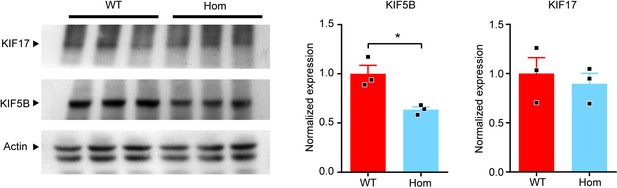
KIF5B conditional knockout mice show no change in KIF17 expression.
Western blot analysis and quantification of KIF17 expression in total brain lysates of homozygous KIF5B conditional knockout mice (Hom) compared with control littermates (WT). n = 3 for WT. n = 3 for Homo. Data are presented in mean + SEM. *p<0.05, Mann-Whitney test.).
-
Figure 6—figure supplement 2—source data 1
Plotted values for Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig6-figsupp2-data1-v2.xlsx
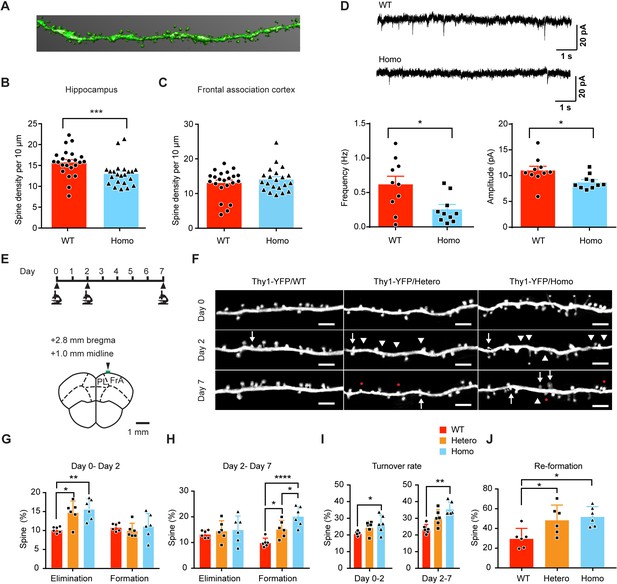
KIF5B conditional knockout mice show increase of dendritic spine instability in the frontal association cortex.
(A) Hippocampal and frontal association cortex slices of Thy1-YFP;CaMKIIα-Cre conditional Kif5bfl/fl knockout mice (Homo) and Thy1-YFP;Kif5b wild-type mice (WT) at postnatal day (P) 44 were fixed. Confocal images of secondary dendrites from apical branches of CA1 hippocampal neurons and prefrontal cortex neurons were captured. 3D reconstruction of individual dendrites was performed for quantification. Representative image of a hippocampal dendrite after 3D reconstruction was shown. (B) The density of dendritic spines in the homozygous KIF5B conditional knockout mice was significantly reduced in CA1 hippocampus as compared to the WT mice (23 dendrites from 2 WT mice and 22 dendrites from two homo mice; mean + SEM; ***p<0.001; Mann-Whitney test). (C) No change in spine density was observed for neurons in the frontal association cortex (22 dendrites from 2 WT mice and 21 dendrites from two homo mice; mean + SEM; not significant; Mann-Whitney test). (D) Representative traces for mEPSCs on CA1 hippocampal neurons from WT and the homozygous KIF5B conditional knockout mice. The frequency and amplitude of mEPSCs from KIF5B conditional knockout neurons showed a significant reduction compared to WT neurons (10 neurons from three mice for each group; mean + SEM; *p<0.05; Student’s t-test for mEPSC amplitude, Mann-Whitney test for mEPSC frequency). (E) Experimental timeline and the diagram of a coronal section of frontal association cortex (FrA) showing the imaging site (green bar). (F) Representative images of dendrites of Thy1-YFP/WT, Thy1-YFP/Hetero and Thy1-YFP/Homo at the imaging time point of Day 0, Day 2, and Day 7. Scale bars, 5 μm. Arrows mark spine formation compared to the previous time point. Arrowheads mark spine elimination compared to the previous time point. Red dots mark re-formation of previously eliminated dendritic spines in close proximity. Asterisks mark filopodia. (G–J) Quantification of spine elimination and formation rates from (G) Day 0 – Day 2, (H) Day 2 – Day 7, (I) total turnover rate and (J) re-formation of eliminated dendritic spines in close proximity on Day 7. n = 6, 947 dendritic spines for WT; n = 6, 906 dendritic spines for Hetero; n = 6, 1078 dendritic spines for Homo. Data were presented in mean + SD. *p<0.05. **p<0.01. ***p<0.001, One-way ANOVA for G-J, except formation in G, Day 2 – Day 7 turnover rate in I used Kruskal-Wallis test.
-
Figure 7—source data 1
Plotted values for Figure 7.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig7-data1-v2.xlsx
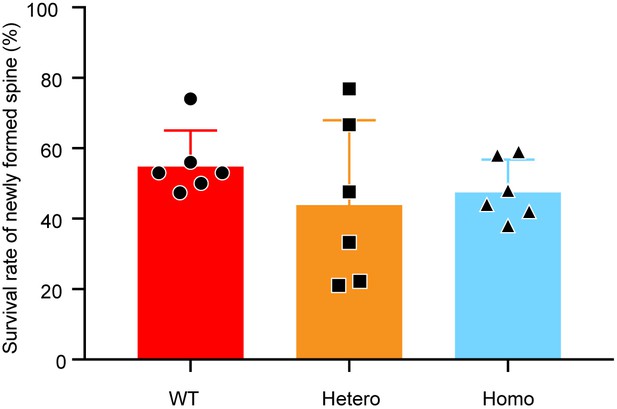
KIF5B conditional knockout mice show no significant difference in survival rate of newly formed dendritic spines.
Survival rate of newly formed dendritic spines. The number of dendritic spines newly formed on Day 2 that persisted to Day 7 was counted as stable new spines. The stability percentage was the number of such spines divided by the total number of dendritic spines that were formed on Day 2. n = 6 for WT, Hetero, and Homo. Data are presented in mean + SEM, Kruskal-Wallis test.
-
Figure 7—figure supplement 1—source data 1
Plotted values for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig7-figsupp1-data1-v2.xlsx
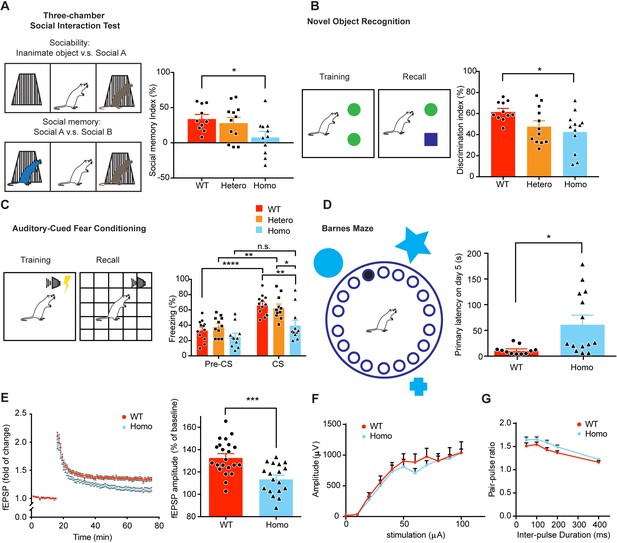
KIF5B conditional knockout mice show deficits in synaptic plasticity, learning and memory.
(A) Schematic diagram shows the set-up of three-chamber social interaction test and the quantification of social memory index in the right panel. n = 10 for WT. n = 11 for Hetero and Homo. Data were presented in mean + SEM. *p<0.05. (B) Schematic diagram shows the set-up of novel object recognition test and the quantification of discrimination index during recall phase in the right panel. n = 11 for WT. n = 12 for Hetero. n = 12 for Homo. Data were presented in mean + SEM. *p<0.05. (C) Schematic diagram shows the set-up of fear conditioning and the quantification of freezing time before and during the recall tone was played in the right panel. n = 12 for WT. n = 11 for Hetero. n = 11 for Homo. Data were presented in mean + SEM. *p<0.05, **p<0.01, ****p<0.0001, n.s.: not significant, Two Way ANOVA with post hoc Tukey’s multiple comparison test. (D) Schematic diagram shows the set-up of Barnes maze. Quantification of primary latency during recall on 5 days after training. n = 11 for WT. n = 13 for Homo. Data were presented in mean + SEM. *p<0.05, Mann-Whitney test. (E) Hippocampal LTP was reduced in KIF5B homozygous conditional knockout mice as shown by the reduced field EPSP amplitude (five mice, 18 slices) compared with wild-type (six mice, 22 slices).***p<0.001, mean + SEM, Student’s t-test. (F) Input/output and (G) pair-pulse ratio curves from hippocampal slices of WT (three mice, 11 slices) and KIF5B homozygous conditional knockout mice (four mice, 21 slices). No significant difference between WT and Homo, two-way ANOVA.
-
Figure 8—source data 1
Plotted values for Figure 8.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig8-data1-v2.xlsx

KIF5B conditional knockout mice showed no significant abnormalities in anxiety-related behavior tests.
(A) Schematic diagram shows the experimental set-up of open field test and the quantification of time spent in center area and the total distance travelled. n = 11 for WT. n = 12 for Hetero and Homo. Data are presented in mean + SEM, One-way ANOVA and Kruskal-Wallis test. (B) Schematic diagram shows the experimental set-up of elevated plus maze and the quantification of the time spent in open arm. n = 11 for WT. n = 12 for Hetero and Homo. Data are presented in mean + SEM, Kruskal-Wallis test. (C) Schematic diagram shows the experimental set-up of marble burying test and the quantification of marble burying. n = 7 for WT. n = 5 for Hetero. n = 9 for Homo. Data are presented in mean + SEM, One-way ANOVA. (D) Schematic diagram shows the experimental set-up of rotarod and the quantification of performance of mice in seven days of rotarod training. Performance is represented in the highest speed (rate per minute, r.p.m.) the mice achieved before they fell from the rotarod. n = 5 for WT. n = 2 for Hetero. n = 7 for Homo. Data are presented in mean + SEM, Two-way ANOVA.
-
Figure 8—figure supplement 1—source data 1
Plotted values for Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig8-figsupp1-data1-v2.xlsx

Sociability, fear acquisition, and primary error of Barnes maze in KIF5B conditional knockout mice.
(A) Quantification of animal behavior in stage 1 of social interaction test. The left panel is the quantification of the total interaction time of mice with either object in stage 1 of the three-chamber social interaction test. n = 10 for WT. n = 11 for Hetero and Homo. Data are presented in mean + SEM, One-way ANOVA. The middle panel is the quantification of interaction time of mice with either the inanimate object or the social object (mouse 1) during stage 1 of three-chamber social interaction test. ****p<0.0001. Two-way ANOVA. The right panel shows the quantification of sociability index, which is the ratio of the difference between the interaction time of the social object and the inanimate object to the total interaction time of stage 1. n = 10 for WT. n = 11 for Hetero and Homo. Data are presented in mean + SEM. **p<0.01. One-way ANOVA with post hoc Tukey's HSD. (B) Quantification of animal behavior in stage 2 of social interaction test. The left panel is the quantification of the total interaction time of mice with either object in stage 2 of the three-chamber social interaction test. n = 10 for WT. n = 11 for Hetero and Homo. Data are presented in mean + SEM, Kruskal-Wallis test. The right panel is the quantification of interaction time of mice with either the novel social object (mouse 2) or the familiar social object (mouse 1) during stage 2 of three-chamber social interaction test. n = 10 for WT. n = 11 for Hetero and Homo. Data are presented in mean + SEM. **p<0.01. ****p<0.0001, Two-way ANOVA with post hoc Tukey's HSD. (C) Quantification of freezing time during acquisition stage of fear conditioning. The percentage of freezing of every 25 s interval are quantified and plotted along the time of fear conditioning. The appearance of conditioned stimulus (tone) and unconditioned stimulus (foot shock) is indicated. n = 12 for WT. n = 11 for Hetero. n = 11 for Homo. Data are presented in mean + SEM, Two-way ANOVA with post hoc Tukey's HSD. #p<0.05, comparison between WT and Hetero; **p<0.01, comparison between WT and Homo. (D) Left panel showing heat map showing the percentage of time the representative mouse of each genotype spent on the maze during acquisition phase. Black circles indicate non-target holes and white circles accompanied with asterisks indicate the target escape holes. WT mouse explored more non-target holes with higher number of primary error in the initial few trials, while Homo mouse stayed in the non-target hole with lower number of primary error during the acquisition phase. Middle panel showing quantification of primary error during acquisition phase and quantification of primary error on day five during recall in the right panel. (n = 11 for WT, n = 13 for Homo) Data are presented in mean + SEM. *p<0.05, Two-way ANOVA with post hoc Tukey's multiple comparison test.
-
Figure 8—figure supplement 2—source data 1
Plotted values for Figure 8—figure supplement 2.
- https://cdn.elifesciences.org/articles/53456/elife-53456-fig8-figsupp2-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus) | C57/6J | The University of Hong Kong Laboratory Animal Unit | ||
| Genetic reagent (M. musculus) | Thy1-YFP-H | Jackson Laboratory. | 003782 |thy1-YFP-H | |
| Genetic reagent (M. musculus) | CaMKIIα-Cre | Jackson Laboratory. | 005359 |T29-1 | |
| Genetic reagent (M. musculus) | Kif5bfl/fl | Jiandong Huang | PMID: 20870970 | |
| Genetic reagent (M. musculus) | CaMKIIα-Cre;Kif5bfl/fl | This paper | generated from breeding of CaMKIIα-Cre and Kif5bfl/fl mice | |
| Transfected construct (M. musculus) | FLAG-677–1027 KIF5A | This paper | aa 677–1027 of mouse KIF5A with a N-terminal FLAG tag was inserted into pcDNA3 | |
| Transfected construct (M. musculus) | FLAG-677–939 KIF5A | This paper | aa 677–939 of mouse KIF5A with a N-terminal FLAG tag was inserted into pcDNA3 | |
| Transfected construct (M. musculus) | FLAG-677–938 KIF5A+941–963 KIF5B | This paper | aa 677–938 of mouse KIF5A and aa 941–963 of mouse KIF5B with a N-terminal FLAG tag was inserted into pcDNA3 | |
| Transfected construct (M. musculus) | FLAG-680–963 KIF5B | This paper | aa 680–963 of mouse KIF5B with a N-terminal FLAG tag was inserted into pcDNA3 | |
| Transfected construct (R. norvegicus) | KIF5A shRNA | This paper | 5’-TGGAAACGCCACAGATATC-3’ | |
| Transfected construct (M. musculus) | KIF5B shRNA | This paper | 5’-GGACAGATGAAGTATAAAT-3’ | |
| Transfected construct (R. norvegicus) | KIF5C shRNA | This paper | 5’-GACCCTGGCAGATGTGAAT-3’ | |
| Transfected construct (R. norvegicus) | control shRNA | Lin et al., 2017 | PMID: 28442576 | 5’-GGCTACCTCCATTTAGTGT-3’ |
| Transfected construct (M. musculus) | pKin1A | Anthony Brown | RRID:Addgene_31607 | |
| Transfected construct (M. musculus) | pcDNA3-KIF5B | Jiandong Huang | PMID: 23293293 | |
| Transfected construct (M. musculus) | pGFP-Kif5c | Michelle Peckham | RRID:Addgene_71853 | |
| Biological sample (R. norvegicus) | Primary hippocampal neuron; primary cortical neuron | Lin et al., 2017 | PMID: 28442576 | Procedures of preparing primary neurons were described in Lin et al. (2017) |
| Biological sample (M. musculus) | Synaptoneurosome | Scheetz et al., 2000 | PMID: 10700251 | Procedures of preparing synaptoneurosome were described by Scheetz et al. (2000) |
| Biological sample (M. musculus) | Synatpic plasma membrane | Bermejo et al., 2014 | PMID: 25226023 | Procedures of preparing synaptic plasma mebrane were described by Bermejo et al. (2014) |
| Antibody | KIF5B | Jiandong Huang | PMID: 20870970 | |
| Antibody | KIF5A | Abcam | RRID:AB_2132218 | |
| Antibody | KIF5C | Abcam | RRID:AB_304999 | |
| Antibody | FMRP | Abcam | RRID:AB_2278530 | |
| Antibody | KIF17 | Sigma | RRID:AB_477148 | |
| Antibody | FLAG | Sigma | RRID:AB_262044 | |
| Antibody | G3BP1 | Bethyl | RRID:AB_1576539 | |
| Antibody | G3BP2 | Bethyl | RRID:AB_1576545 | |
| Antibody | GluA2 | Millipore | RRID:AB_2113875 | |
| Antibody | RNMT | Millipore | RRID:AB_11215450 | |
| Antibody | NRGN | Millipore | Cat#AB5620 | |
| Antibody | NeuN | Millipore | Cat#AB377 | |
| Antibody | PSD-95 | NeuroMab | RRID:AB_2292909 | |
| Antibody | methylated mono-arginine R*GG | Cell Signaling | RRID:AB_10896849 | |
| Antibody | Mouse IgG2a anti-GFP | Invitrogen | RRID:AB_221568 | |
| Antibody | Rabbit anti-RFP | Rockland | RRID:AB_2209751 | |
| Antibody | Alexa 488 anti mouse IgG2a | Invitrogen | RRID:AB_2535771 | |
| Antibody | Alexa 546 anti rabbit IgG | Invitrogen | RRID:AB_2534077 | |
| Antibody | horseradish peroxidase-conjugated goat anti-rabbit IgG | Cell Signaling | RRID:AB_2099233 | |
| Antibody | horseradish peroxidase-conjugated goat anti-mouse IgG | Cell Signaling | RRID:AB_330924 | |
| Recombinant DNA reagent | pFRT-TODestFLAGHAhFMRPiso1 | Thomas Tuschl | RRID:Addgene_48690 | |
| Recombinant DNA reagent | tdTomato | Michael Davidson | RRID:Addgene_54653 | |
| Recombinant DNA reagent | GST-fused KIF5A | This paper | aa 677–1027 of mouse KIF5A was inserted into pGEX-6P-2 | |
| Recombinant DNA reagent | GST-fused KIF5B | This paper | aa 680–963 of mouse KIF5B was inserted into pGEX-6P-2 | |
| Recombinant DNA reagent | GST-fused KIF5C | This paper | aa 681–956 of mouse KIF5C was inserted into pGEX-6P-2 | |
| Recombinant DNA reagent | pEGFP-N1-KIF5A | This paper | Constructed by inserting PCR-amplified mouse KIF5A coding sequences into the pEGFP-N1 plasmid using KpnI and BamHI | |
| Recombinant DNA reagent | pEGFP-N1-KIF5B | This paper | Constructed by inserting PCR-amplified mouse KIF5B coding sequences into the pEGFP-N1 plasmid using KpnI and BamHI | |
| Sequenced-based reagent | Grin2b transcript probe (NM_012574.1, type 1) | ThermoFisher | Cat#VC1-16464 | |
| Sequenced-based reagent | Type one sense probe | ThermoFisher | Cat#VC1-20903 | |
| Sequenced-based reagent | CaMKIIa transcript probe (NM_012920.1, type 6) | ThermoFisher | Cat#VC6-11639 | |
| Sequenced-based reagent | Type six sense probe | ThermoFisher | Cat#VC6-16372 | |
| Chemical compound, drug | FLAG beads | Sigma | RRID:AB_10063035 | |
| Chemical compound, drug | glutathione sepharose four fast flow beads | GE Healthcare | Cat#17-5132-01 | |
| Chemical compound, drug | Protein A-Sepharose beads | GE Healthcare | Cat#17-5280-01 | |
| Commercial assay or kit | Lipofectamine LTX with Plus Reagent | ThermoFisher Scientific | Cat#15338100 | |
| Commercial assay or kit | SilverQuest Silver Staining Kit | Life technologies | Cat#LC6070 | |
| Commercial assay or kit | ViewRNA ISH Cell Assay Kit | ThermoFisher | Cat#QVC0001 | |
| Commercial assay or kit | Neon transfection system | ThermoFisher Scientific | Model MPK5000 | |
| Software, algorithm | Volocity | Quorum Technologies | RRID:SCR_002668 | |
| Software, algorithm | Zen digital imaging software | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | Actimetrics FreezeFrame software | Coulbourn Instruments | RRID:SCR_014429 | Version 2.2 |
| Software, algorithm | ANY-maze software | ANY-maze | RRID:SCR_014289 | |
| Software, algorithm | MetaMorph software | Molecular Devices | SCR_002368 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 6 |
| Software, algorithm | FIJI | FIJI (https://imagej.net/Fiji) | RRID:SCR_002285 | |
| Software, algorithm | KymoResliceWide | Eugene Katrukha (https://github.com/ekatrukha/KymoResliceWide) | ||
| Software, algorithm | Straighten | Eva Kocsis (https://imagej.nih.gov/ij/plugins/straighten.html) | PMID: 1817611 | |
| Software, algorithm | Mini Analysis Program | Synaptosoft | RRID:SCR_002184 | |
| Software, algorithm | CLC Main Workbench | Qiagen (https://www.qiagenbioinformatics.com/products/clc-main-workbench/) | RRID:SCR_000354 |
Additional files
-
Supplementary file 1
Table for statistical tests.
- https://cdn.elifesciences.org/articles/53456/elife-53456-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53456/elife-53456-transrepform-v2.pdf





