Mutations associated with human neural tube defects display disrupted planar cell polarity in Drosophila
Figures
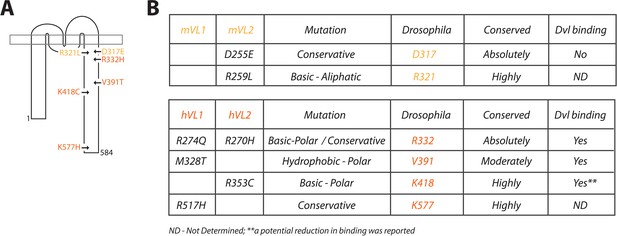
Description of Vang mutations.
(A) Schematic showing the location of investigated mutations along the C-terminal tail of Vang. Mutations highlighted in yellow were originally identified in the mouse, mutations highlighted in orange were found in human NTD patients (see supplement for references). (B) Table displaying information concerning the selected mutations from mouse (top) and human patients (below). Details include: original mammalian mutation, nature of the mutational change (e.g. D to E is listed as conservative as both are acidic residues), equivalent residue in Drosophila Vang, whether the mutated residue is conserved among species, and whether the respective mutated Vangl protein was able to retain binding to the effector protein Dishevelled (Dvl).
-
Figure 1—source data 1
Summary of NTD-associated mammalian mutations found to date within the C-terminal tail of Vangl/VANGL genes.
- https://cdn.elifesciences.org/articles/53532/elife-53532-fig1-data1-v2.docx
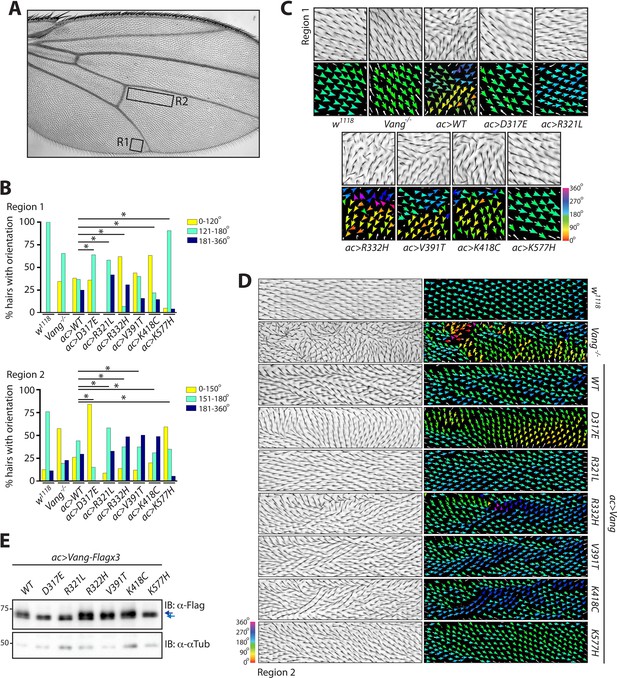
Behavior of over-expressed Drosophila Vang carrying C-terminal mutations.
(A) Overview of an adult wing highlighting the analyzed regions of hair reorientation. (B) Quantification of the percentage of actin hairs oriented within a specific angle range upon actin-Gal4 driven overexpression (ac>) of the different Vang mutant proteins and wild-type Vang. Also included for comparison is Vang-/- and w1118 (a genetically wild-type wing that displays no phenotype). Graph is shown for region 1 (upper) and region 2 (lower). Angles from three independent wings were combined, and data binned to allow analysis using a Chi-squared test. * indicates p<0.001 (C) Example images of hair reorientation in region 1 in the indicated genotypes. Top panel shows adult wing, bottom panel shows the corresponding angles of hair orientation visualized through color (see 360 degree color scale, bottom right). Angles were determined using the Fuji plugin FijiWingsPolarity (Dobens et al., 2018). (D) Example images of hair reorientation in region 2. Left panel shows adult wing, and right panel the corresponding angles of hair orientation. (E) Western blot of wing disc lysates from the indicated genotypes. While similar levels of expression are observed, a difference in Vang mobility is notable for D317E, R321L and K577H, as indicated by blue arrows.
-
Figure 2—source data 1
Raw data from the quantification of hair reorientation summarized in Figure 2.
- https://cdn.elifesciences.org/articles/53532/elife-53532-fig2-data1-v2.xlsx
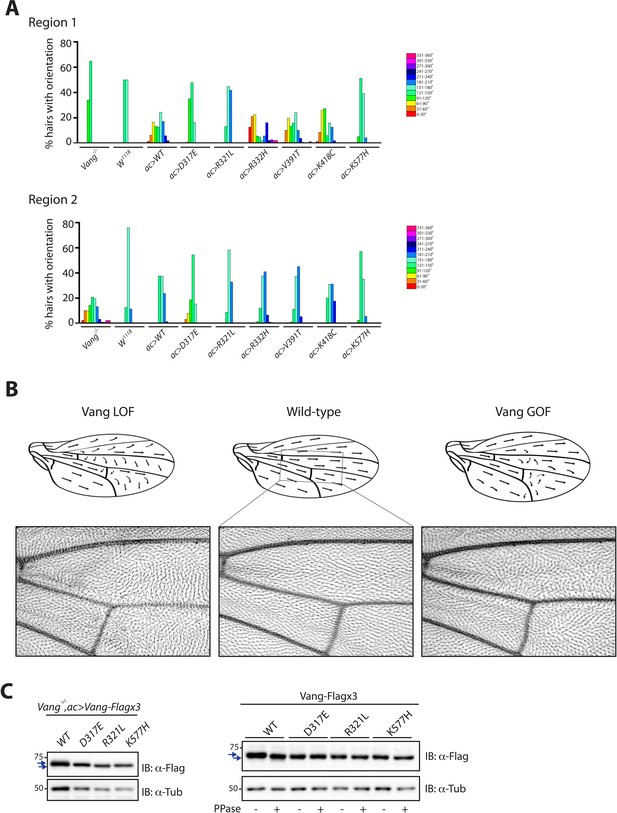
Expression of Vang transgenes mimics features of Vang gain- and loss-of-function phenotypes.
(A) Graphs showing the percentage of hairs in each 30 degree segment within region 1 (above) and region 2 (below) after overexpression of wild-type Vang, Vang carrying the different mutations, or in control wings (color scheme as in Figure 2). (B) Schematic representation of changes to hair orientation in indicated conditions. Representative image of a ROI (region of interest) within an adult wing is shown next to the schematic. (C) Western blot showing changes to Vang mobility after introduction of the indicated mutations and after phosphatase treatment where indicated, arrows highlight differences. Samples were extracted from wing discs of the indicated genotypes (left) or after S2 cell transfection (right).
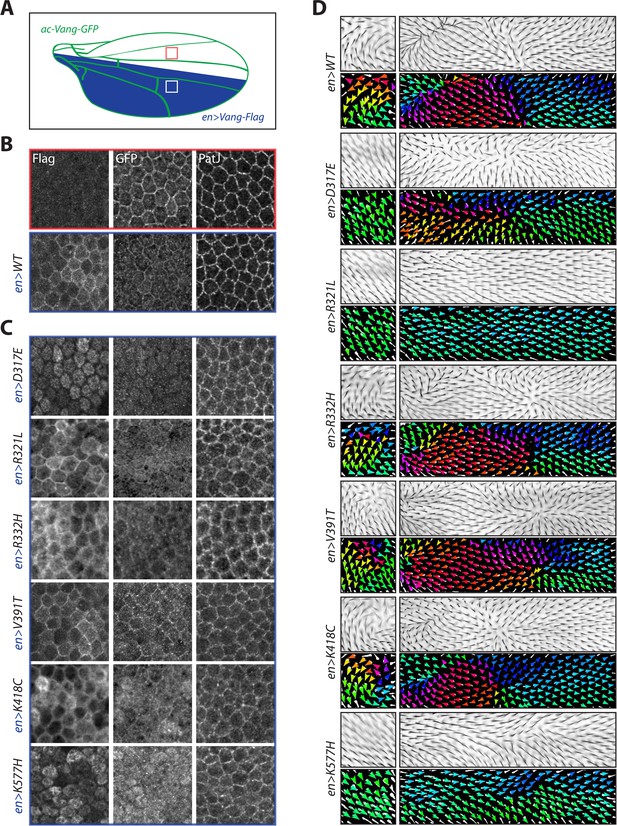
Overexpression of Vang transgenes affects localization of wild-type Vang.
(A) Schematic showing regions of expression of different transgenes along the Drosophila wing blade. A direct actin-promoter driven (ac-Vang-GFP) construct is expressed at homeostatic levels throughout the wing blade, engrailed-Gal4 drives expression of wild-type Vang-Flag or each of the associated mutant proteins in the posterior compartment of the wing. (B) Representative immunofluorescence images of regions from the anterior and posterior regions of a pupal wing overexpressing wild-type Vang-Flag in the posterior region. Note recruitment of Vang-Flag to the membrane in the posterior region (blue), and concomitant reduction in membrane localization of Vang-GFP in the posterior (blue) vs. anterior (red) compartment. The junctional marker PatJ shows consistent membrane labeling in both regions. (C) Representative immunofluorescence images of regions from the posterior compartment of pupal wings overexpressing the indicated Vang-Flag constructs. Note differences in localization of Vang-Flag mutants as compared to wild-type Vang (B), as well as altered wt-Vang-GFP localization. The junctional marker PatJ was unaffected in each case. (D) Regions of adult wing showing hair reorientation phenotypes in the indicated genotypes. Note the similarity in pattern between overexpression of WT, R332H, V391T and K418C mutants (swirls, cf. GOF in Figure 2—figure supplement 1), as compared to R321L (wild-type orientation), and D317E and K577H (downwards reorientation, cf. LOF in Figure 2—figure supplement 1) in posterior region of the wing. Color range is same as in Figure 2.
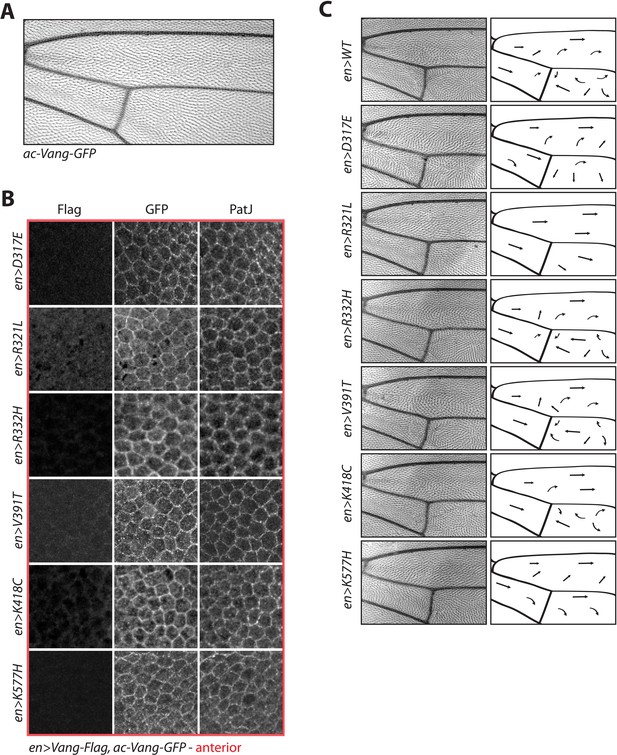
Expression of ac-Vang-GFP within the Drosophila wing blade does not interfere with polarity.
(A) Image of adult wing expressing a direct driven ac-Vang-GFP transgene. Note that no hair reorientation defects are observed confirming that expression is within the homeostatic range. (B) Representative immunofluorescence images of regions from the anterior compartment of wings, the region of the wing in which engrailed-Gal4 driven constructs are not expressed. Note similarity in staining between Vang-GFP and PatJ in each condition. (C) Schematic of adult wing showing hair reorientation phenotypes in the indicated genotypes. Arrows show general flow of hair reorientation, note the similarity in pattern between overexpression of WT, R332H, V391T and K418C mutants (swirls, cf. GOF in Figure 2—figure supplement 1), as compared to R321L (wild-type orientation), and D317E and K577H (downwards reorientation, cf. LOF in Figure 2—figure supplement 1) in posterior region of the wing.
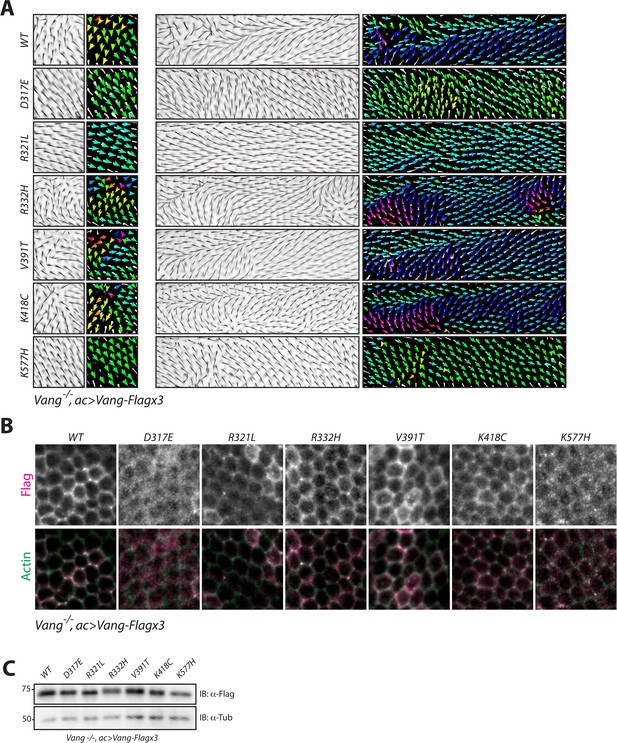
NTD mutations affect membrane localization of Vang.
(A) Regions 1 and 2 of the adult wing (see Figure 2A) showing hair reorientation phenotypes in the indicated genotype. Note similarity in pattern between WT, R332H, V391T and K418C mutants (swirls, cf. GOF in Figure 2—figure supplement 1), as compared to R321L (wild-type orientation), and D317E and K577H (downwards reorientation, cf. LOF in Figure 2—figure supplement 1). Color range is same as in Figure 2. (B) Representative immunofluorescence images of pupal wings upon expression of Vang-Flag or indicated mutant Vang proteins using actin-Gal4 driver. Note changes to Vang localization as shown through Flag staining (red), and the degree of overlap with actin (stained with Phalloidin, green) which marks the membrane (C) Western blot of wing discs from the indicated genotypes. Note that all transgenes were expressed at similar levels. Differences in mobility are also observed as in Figure 2 and Figure 2—figure supplement 1.
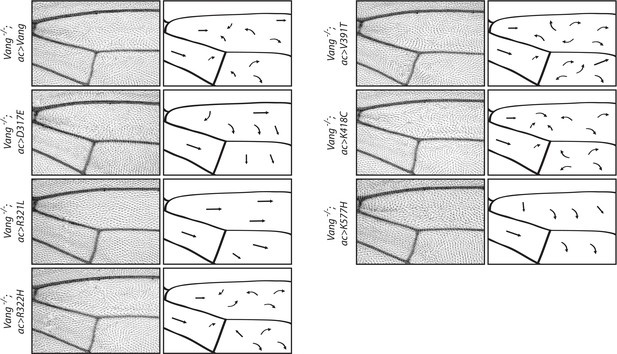
Schematic representation of wings shows flow of hair reorientation in different mutant conditions.
Regions 1 and 2 of the adult wing (see Figure 2A) showing hair reorientation phenotypes in the indicated genotype. Arrows show general flow of hair reorientation; note similarity in pattern between WT, R332H, V391T and K418C mutants (swirls, cf. GOF in Figure 2—figure supplement 1), as compared to R321L (wild-type orientation), and D317E and K577H (downwards reorientation, cf. LOF in Figure 2—figure supplement 1).
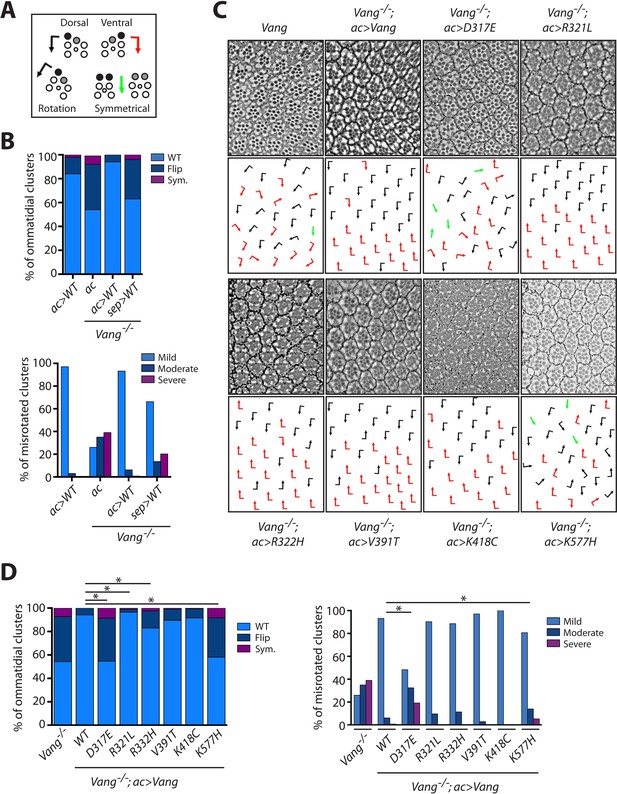
Vang-/- rescue experiments with individual mutations reveal the nature of how they affect Vang function.
(A) Schematic representation of the chiral forms of ommatidial clusters within the Drosophila eye. Dorsal chirality is represented by black arrows, ventral chirality by red arrows, and symmetrical (achiral) clusters by green arrows. The varying degree to which each cluster rotated during development is denoted by the angle of each arrow. (B) Quantification of the percentage of different chiral forms of clusters in the indicated genotypes (top). These include wild-type (WT), a flipped chiral form (Flip), or a symmetrical cluster (Sym.). Quantification of the percentage of misrotated clusters in the associated genotypes (bottom). Misrotation was split into three categories, mild is wild-type +/- 10 degrees, moderate is +/- 10–30 degrees from wild-type, and severe is +/- > 30 degrees misrotation from wild-type. Note that rescue with actin-Gal4 results in a mild overexpression phenotype due to the expression strength with this driver, (also Figure 5—figure supplement 1), while sep-Gal4 shows a weaker rescue due to its weaker and restricted (not all R-cells) expression. (C) Tangential eye sections of the region flanking the dorso-ventral midline (equator) in the indicated genotypes after rescue with actin-Gal4 driver, anterior is left and dorsal is up. (D) Quantification of percentage of different chiral forms of clusters in the indicated genotypes (left). Quantification of percentage of misrotated clusters in the indicated genotypes (right). Data were analyzed using a Chi-squared test: * indicates p<0.001.
-
Figure 5—source data 1
Raw data from the quantification of eye chirality and rotation defects summarized in Figure 5.
- https://cdn.elifesciences.org/articles/53532/elife-53532-fig5-data1-v2.xlsx
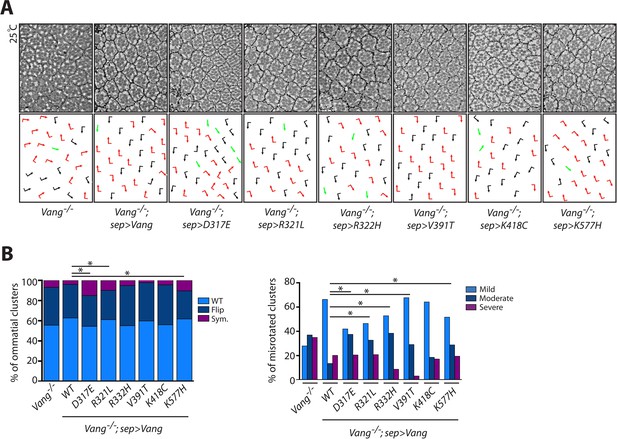
Phenotypes observed upon partial rescue with sep-Gal4 are consistent with results obtained using actin-Gal4.
(A) Tangential eye sections of region flanking the equator in the indicated genotypes upon rescue with sep-Gal4, anterior is left and dorsal is up. (B) Quantification of the percentage of different chiral forms of clusters in the indicated genotypes (left). Quantification of percentages of misrotated clusters in the indicated genotypes (right). Data were analyzed using a Chi-squared test: * indicates p<0.001.
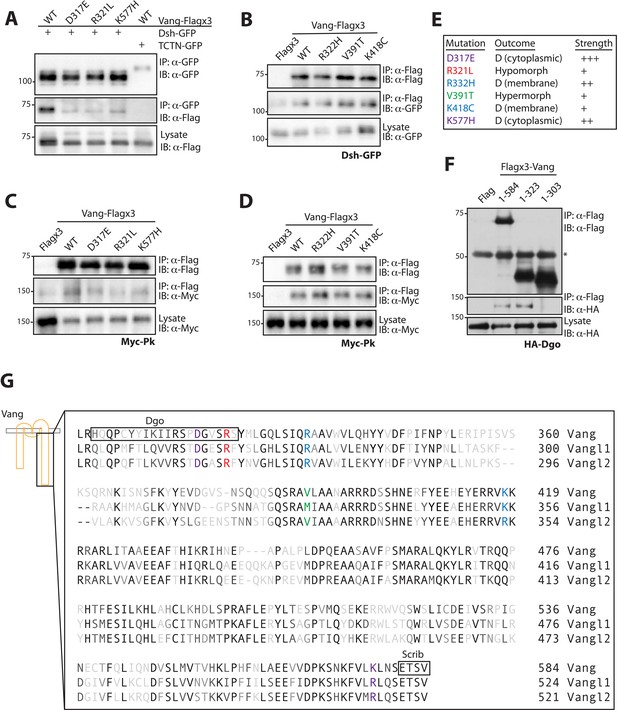
Mutations associated with LOF phenotypes display reduced effector binding.
(A) Western blot showing binding between Dsh-GFP and Vang-Flag wild-type or the indicated mutants. Note that Vang-Flag wild-type binds to Dsh-GFP but not the negative control Tctn-GFP. The D317E, R321L and K557H mutants show reduced binding as compared to wild-type Vang. (B) Western blot showing binding between Vang-Flag wild-type or the indicated mutants and Dsh-GFP. The Vang constructs are able to bind to Dsh to the same degree, while no binding is observed with Flag alone. (C) Western blot showing binding between Vang-Flag wild-type or the indicated mutants and Myc-Pk. The D317E, R321L and K577H mutants show a markedly weaker interaction with Myc-Pk as compared to wild-type Vang. (D) Western blot showing binding between Vang-Flag wild-type or the indicated mutants and Myc-Pk. The Vang constructs are able to bind Pk to the same degree. (E) Summary of the phenotypic outcome of investigated mutations, D, (cytoplasmic) and (membrane) distinguish between the two types of dominant mutations observed, which is based on their cellular localization. Colors are assigned based on different outcomes, also shown is the relative phenotypic strength of each mutation (see Figures 3 and 4). (F) Western blot showing the regions of Vang required for interaction with Dgo. Note that full-length Vang (1-584) and a construct containing residues 1–323 of Vang are able to interact with HA-Dgo. While a construct containing residues 1–303 does not interact, suggesting residues 304–323 in Vang are essential for its interaction with Dgo. (G) Schematic showing the C-terminal sequence of Vang, and human Vangl1 and Vangl2. Mutated residues are highlighted in colors associated with their phenotypic outcome as in E. Also highlighted are regions of Vang essential for its interaction with Dgo and Scribble, note their proximity to specific residues.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Vang | Pubmed ID: 35922 | Dmel_CG807, CG8075, Dmel\CG8075, Stbm, Strabismus, Van Gogh | Chromosome 2R, NT_033778.4 (9103238…9106796) |
| Genetic reagent (Drosophila melanogaster) | w[1118] | FlyBase ID: FBal0018186 | Reference control | |
| Genetic reagent (Drosophila melanogaster) | Vang[6] | BloomingtonDrosophilaStock Center | 6918 | Null allele |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang- Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]-attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang-D317E-Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]-attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang-R321L-Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]- attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang-R332H-Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]-attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang- V391T-Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]-attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang-K418C-Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]-attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | UAS-Vang-K577H-Flagx3 | This Paper | Insertion into BDSC stock 9752 - PBAC{yellow[+]-attP-3B}VK00037 Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-D317E-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-R321L-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-R332H-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-V391T-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-K418C-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | Vang[6],UAS-Vang-K577H-Flagx3 | This Paper | Recombined stock Can be obtained from Mlodzik Laboratory, ISMMS | |
| Genetic reagent (Drosophila melanogaster) | actin-Gal4 | BloomingtonDrosophilaStock Center | 3954 | |
| Genetic reagent (Drosophila melanogaster) | ac-Vang-GFP | Gift from David Strutt, University of Sheffield, UK | ||
| Genetic reagent (Drosophila melanogaster) | en-Gal4 | BloomingtonDrosophilaStock Center | 1973 | |
| Genetic reagent (Drosophila melanogaster) | sep-Gal4 | (Fanto et al., 2000) | ||
| Cell line (Drosophila melanogaster) | S2 | Thermo Fisher Scientific | 69007 | Stock tested for contamination, characterized by isozyme and karyotype analysis |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang-Flagx3 | This Paper (Bischof et al., 2007) | pUASTattB vector | Cloned using NotI-XbaI Can be obtained from Mlodzik Laboratory, ISMMS |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang-D317E-Flagx3 | This Paper | Made using SDM Can be obtained from Mlodzik Laboratory, ISMMS | |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang- R321L-Flagx3 | This Paper | Made using SDM Can be obtained from Mlodzik Laboratory, ISMMS | |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang- R332H-Flagx3 | This Paper | Made using SDM Can be obtained from Mlodzik Laboratory, ISMMS | |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang-V391T-Flagx3 | This Paper | Made using SDM Can be obtained from Mlodzik Laboratory, ISMMS | |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang-K418C-Flagx3 | This Paper | Made using SDM Can be obtained from Mlodzik Laboratory, ISMMS | |
| Transfected construct (Drosophila melanogaster) | pUAS-Vang-K577H-Flagx3 | This Paper | Made using SDM Can be obtained from Mlodzik Laboratory, ISMMS | |
| Transfected construct (Drosophila melanogaster) | pAc5.1-Gal4 | Gift from Andreas Jenny, AECOM, USA | ||
| Transfected construct (Drosophila melanogaster) | pAc5.1-Dsh-GFP | (Simons et al., 2009) | ||
| Transfected construct (Drosophila melanogaster) | pAc5.1-Myc-Pk | Gift from Andreas Jenny, AECOM, USA | ||
| Transfected construct (Drosophila melanogaster) | pAc5.1-HA-Dgo | Gift from Andreas Jenny, AECOM, USA | ||
| Transfected construct (Drosophila melanogaster) | pTub-Tctn-GFP | This Paper | pCaSpeRTubGFP vector with pUAST MCS | Cloned using BglII-XhoI Can be obtained from Mlodzik Laboratory, ISMMS |
| Transfected construct (Drosophila melanogaster) | pAc5.1-Flagx3 | Gift from Andreas Jenny, AECOM, USA | ||
| Transfected construct (Drosophila melanogaster) | pAc5.1-Flag-Vang | This Paper | pAc5.1-Flag vector | Cloned using NotI-XbaI Can be obtained from Mlodzik Laboratory, ISMMS |
| Transfected construct (Drosophila melanogaster) | pAc5.1-Flag- Vang 1–323 | This Paper | pAc5.1-Flag vector | Cloned using NotI-XbaI Can be obtained from Mlodzik Laboratory, ISMMS |
| Transfected construct (Drosophila melanogaster) | pAc5.1-Flag-Vang-1–303 | This Paper | pAc5.1-Flag vector | Cloned using NotI-XbaI Can be obtained from Mlodzik Laboratory, ISMMS |
| Antibody | Flag | Sigma Aldrich | M2 | 1:5000 IB/I:50 IF |
| Antibody | Gamma- Tubulin | Sigma Aldrich | GTU-88 | 1:1000 |
| Antibody | GFP | Roche | 7.1 and 13.1 | 1:1000 |
| Antibody | GFP | Invitrogen | A11122 | 1:100 |
| Antibody | PatJ | Gift from Jun Wu, ISMMS, USA | 1:500 | |
| Antibody | Myc | Santa Cruz Biotechnology | 9E10 | 1:1000 |
| Antibody | HA | Roche | 3F10 | 1:1000 |
| Sequence- based reagent | D317E | GATCATTCGCTCCCCGGAAGGCGTTTCGCGCTCCTAC | PCR primer | |
| Sequence-based reagent | R321L | GACGGCGTTTCGCTCTCCTACATGTTG | PCR primer | |
| Sequence- based reagent | R332H | GTCAGCTGAGCATCCAACATGCGGCTGTGTGGGTGCTAC | PCR primer | |
| Sequence- based reagent | V391T | CCAGAGTCGAGCAACTCTAGCAGCCAACG | PCR primer | |
| Sequence-based reagent | K418C | GTACGAACGTCGTGTGTGTAAACGGCGTGCCCGTC | PCR primer | |
| Sequence-based reagent | K577H | AAGCAACAAATTTGTTCTTCACTTGAACTCCGAAACATCC | PCR primer | |
| Sequence-based reagent | TCTN-f | GGAAGATCTATGAAGGAAGTG | PCR primer | |
| Sequence-based reagent | TCTN-r | CCGCTCGAGGCAAAGTTG | PCR primer | |
| Sequence-based reagent | Vang-f | TATGCGGCCGCTCATGGAAAACGAATCCGTC | PCR primer | |
| Sequence-based reagent | Vang-584-r | ATATCTAGATTATACGGATGTTTCGGAGTT | PCR primer | |
| Sequence-based reagent | Vang-323-r | ATATCTAGATTAGTAGGAGCGCGAAACGCC | PCR primer | |
| Sequence-based reagent | Vang-303-r | ATATCTAGATTAGTGTCGCAGCTCTAGTAA | PCR primer | |
| Commercial assay or kit | Effectene | Qiagen | 301427 | |
| Commercial assay or kit | GFP-Trap Agarose | Chromotek | gta | |
| Software, algorithm | FijiWingsPolarity | (Dobens et al., 2018) | ||
| Other | Lambda Protein Phosphatase | NEB | P0753S |





