A big-data approach to understanding metabolic rate and response to obesity in laboratory mice
Figures
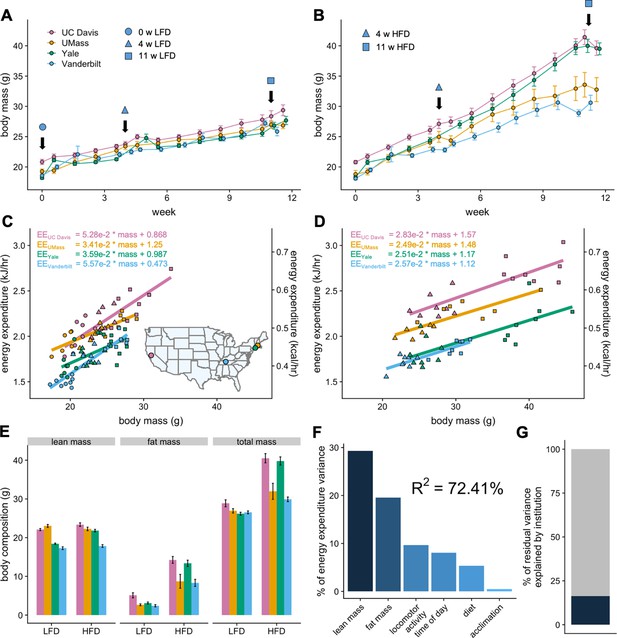
MMPC experiment: institutional variability in response to low-fat or high-fat diet.
Body weights of WT C57BL/6J mice maintained on LFD (A) or HFD (B). Indirect calorimetry was performed at 0 (ο), 4 (Δ), or 11 (□) weeks. EE rates vs total body mass are plotted by site for mice maintained on LFD (C) or HFD (D), with the slope and intercept in kJ/hr indicated for each site. Inset, geographical locations of MMPC sites. Body composition data from mice at 11 weeks on diet (E). The percent variation in EE explained by each of the listed factors (F). The otherwise unattributed institutional variation (16%) (G). n = 6–8 males per group. Error bars represent SEM.
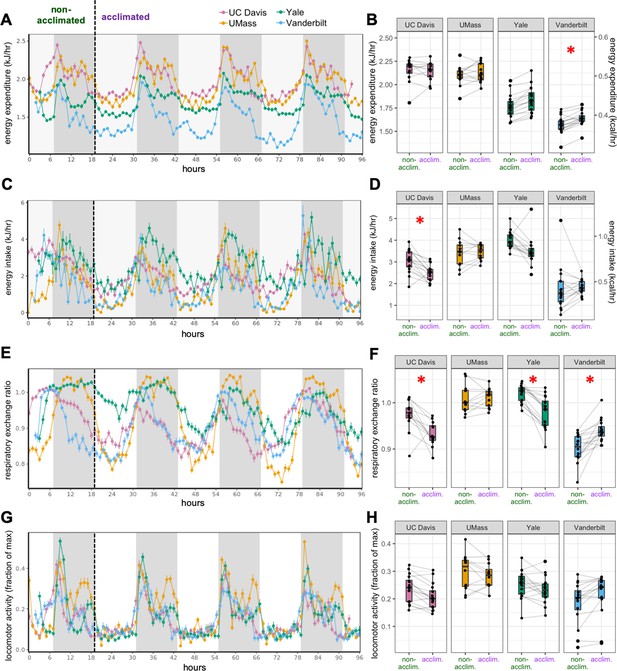
MMPC experiment: effect of acclimation.
Plots of metabolic variables vs time are shown for mice on LFD (0 weeks). EE vs time (A). Mean values for the first dark photoperiod vs the mean of the subsequent three dark photoperiods at each site of the MMPC experiment (B). Energy intake (C, D); RER (E, F); Locomotor activity (G, H). Values are hourly means. *, p<0.05. n = 6–8 males per group. Error bars represent SEM. Shaded regions represent the dark or active phases from 18:00 to 6:00.
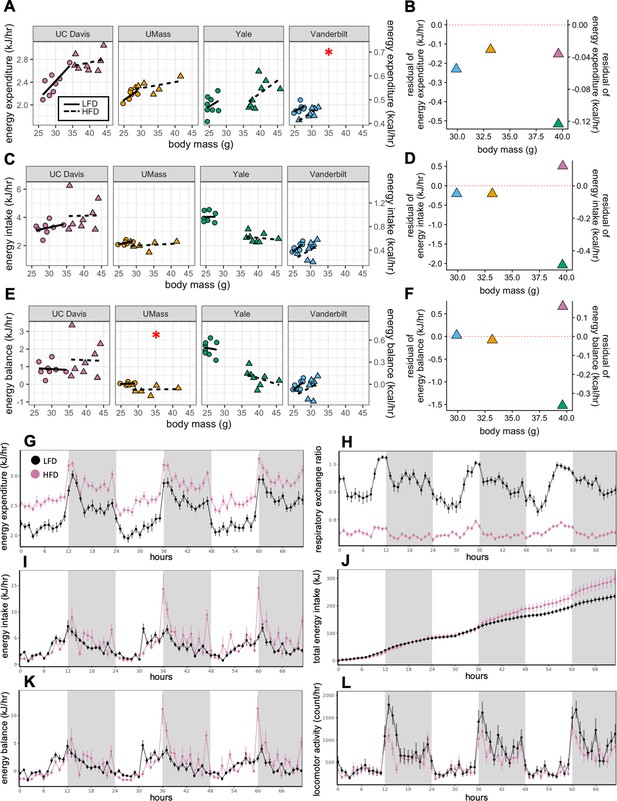
MMPC experiment: institutional variability in metabolic rate.
Regression plots for each of the MMPC sites vs mass (left). A regression model fitted to LFD mice for site, mass, and locomotor activity was established; the deviation from the predicted values is shown as residuals (right). EE (A–B). Energy intake (C–D). Energy balance (E–F). Time-dependent plots for the UC Davis site after 11 weeks on HFD including EE (G), RER (H), hourly energy intake (I), cumulative energy intake (J), energy balance (K), and locomotor activity (L). Values are hourly means (G–L). *, p<0.05. n = 6–8 males per group. Error bars represent SEM. Shaded regions represent the dark photoperiod from 18:00 to 6:00.
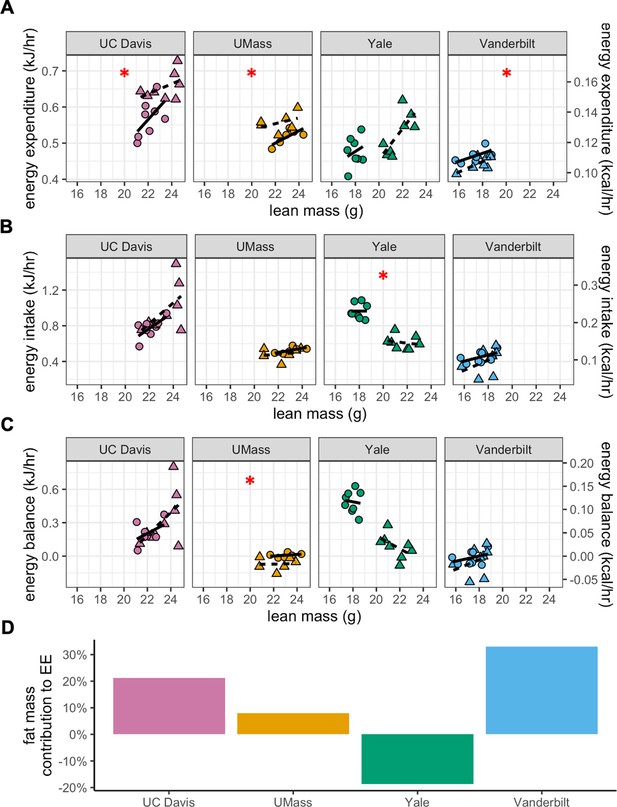
MMPC experiment: energy balance using lean mass as a covariate.
Data as in Figure 3, using the lean mass as the covariate in place of total mass. Regression plots for each of the MMPC sites vs mass. EE (A). Energy intake (B). Energy balance (C). Relative contribution of fat mass to EE for each site from a linear regression model (D). n = 6–8 males per group.
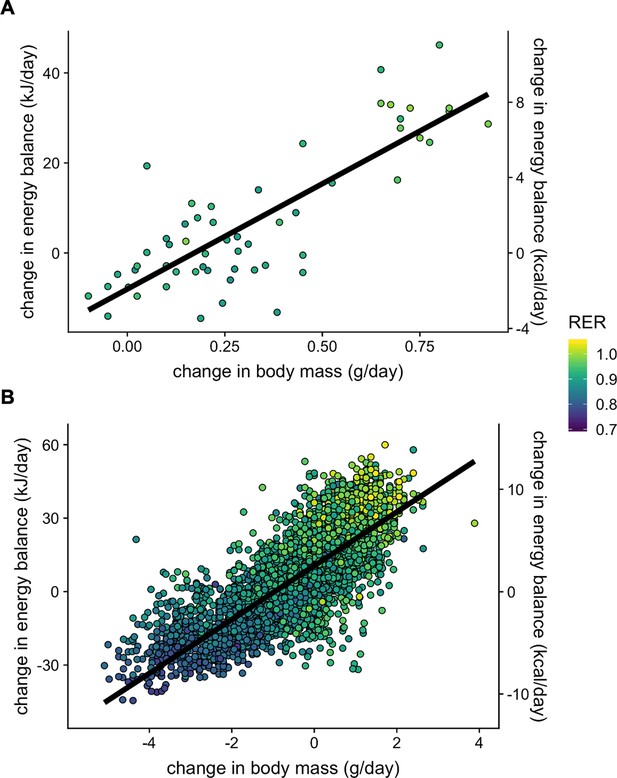
Body weight, energy balance and RER.
Plots of change in energy balance (Energy intake minus EE) vs change in body weight (mass at start of experiment minus mass at end of experiment). MMPC (n = 56 males) (A). IMPC (n = 3738 males and 2140 females) (B).
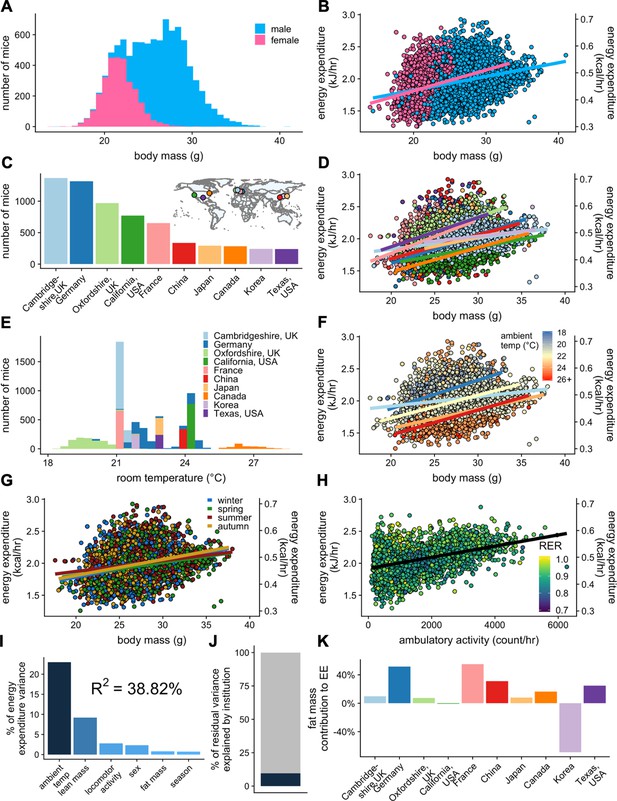
IMPC experiment: institutional variability of metabolic rate.
Distribution of body weights for male and female WT C57BL/6N mice from the IMPC database version 10.0 (A). Relationship of EE vs total body mass for female and male WT mice (B). The numbers of male WT mice examined at each of the 10 IMPC sites (C). Inset: geographical locations of IMPC sites. Relationship of EE vs total body mass for male WT mice at each of the 10 IMPC sites (D). Reported ambient temperatures for the experiments performed at each site (E). Relationship of EE vs total body mass in WT males for temperature (F) and season (G). Relationship of EE vs ambulatory activity in WT mice (n = 3886 males) (H). Unaccounted institutional contribution here is approximately 10% (J). The percent variation in EE explained by each of the listed factors (I). Percent contribution of fat mass to EE by site in WT males (K). n = 6469 males and 2889 females unless otherwise specified.
-
Figure 3—source data 1
Pairwise comparisons of slopes in regression plots of EE vs total mass.
- https://cdn.elifesciences.org/articles/53560/elife-53560-fig3-data1-v3.xlsx
IMPC experiment: body weight and metabolic rate.
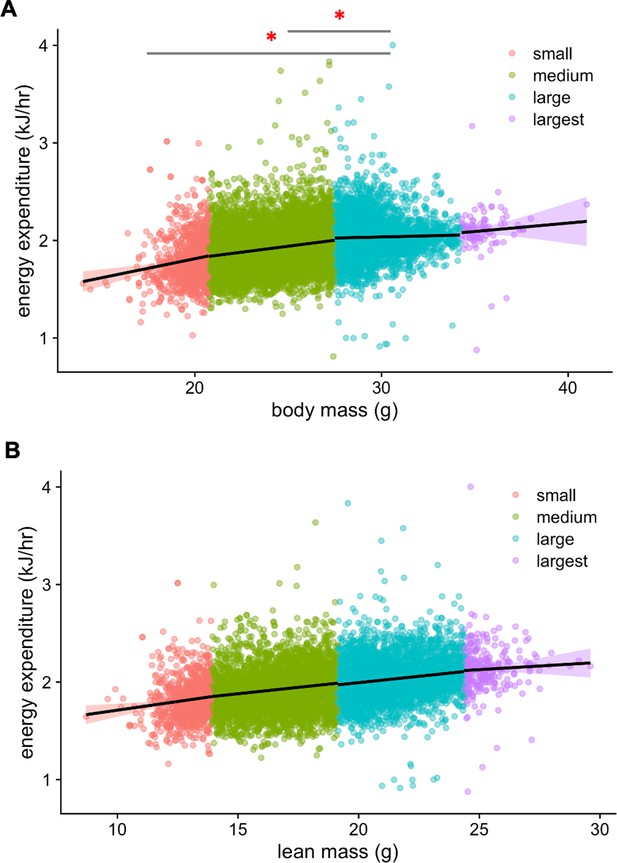
Plot of EE vs total mass for 4 body mass groups (small, 14.00–20.75 g; medium, 20.75–27.50 g; large, 27.50–34.25 g; largest, 34.25–41.00 g) (A). Relationship between the group with large masses is significant from small mass and medium mass groups. No other group relationships are significant. EE vs lean mass (small, 8.70–13.92 g; medium, 13.92–19.14 g; large, 19.14–24.36 g; largest, 24.36–29.60 g). No group relationships are significant. (B). *, p<0.05. n = 6469 males and 2889 females.
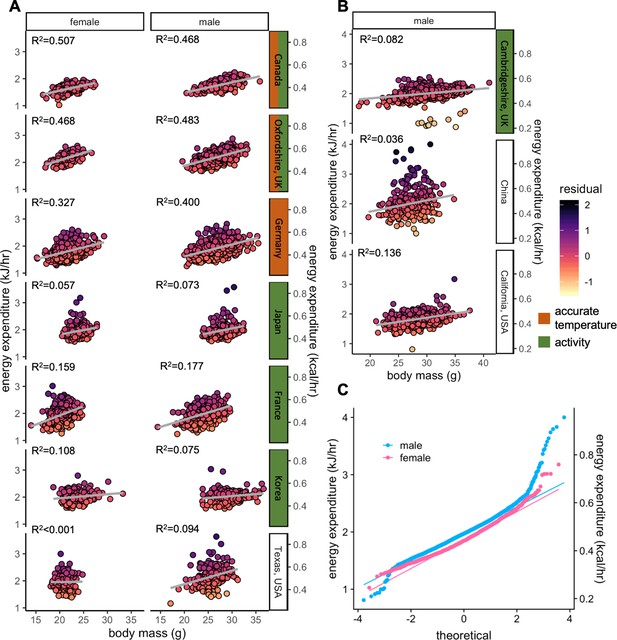
Variation in energy expenditure for WT female and male mice.
Plots of the EE vs total mass showing the quality of the fit (R2 values) and shaded by residual values—the deviation from predicted values for each of the 9358 mice (6469 males and 2889 females). For the seven sites studying both sexes, the R2 values were similar between female and male mice (+/- 9%). Sites with accurate temperature covariate tended to have larger R2 values (A). R2 at the 3 sites reporting only data from male mice (B). A modified quantile-quantile plot of male and female mice from the 7 sites studying both sexes (C).
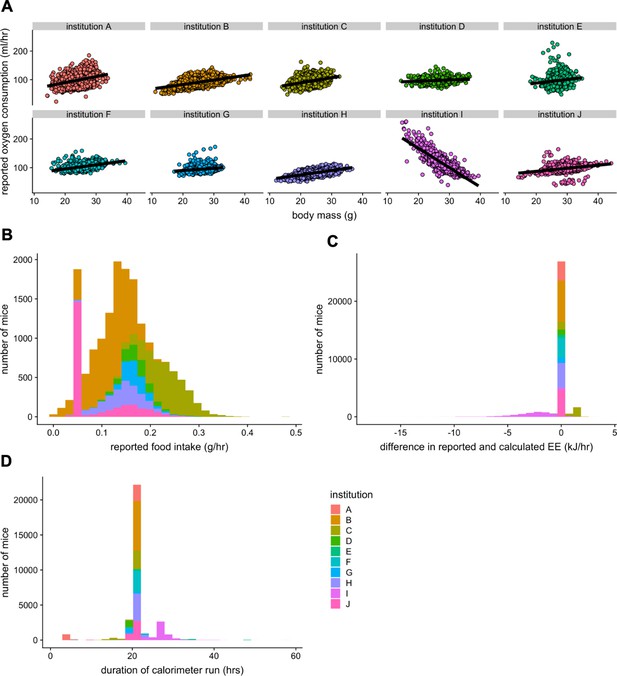
Irregularities in the IMPC dataset.
A plot of VO2 vs total body mass for each of the 10 sites in the IMPC (A). Only one site demonstrates a negative mass effect due to a faulty calculation. Distribution of food intake among the reporting sites. The 0.05 g food intake was erroneously overrepresented (n = 10,021 males and 6818 females) (B). Two sites reported EEs calculated by a different equation from the rest of the dataset (C). Durations of calorimetry run data included in IMPC database (D). These four issues were corrected prior to analysis. n = 25,089 males and 10,725 females unless otherwise specified.
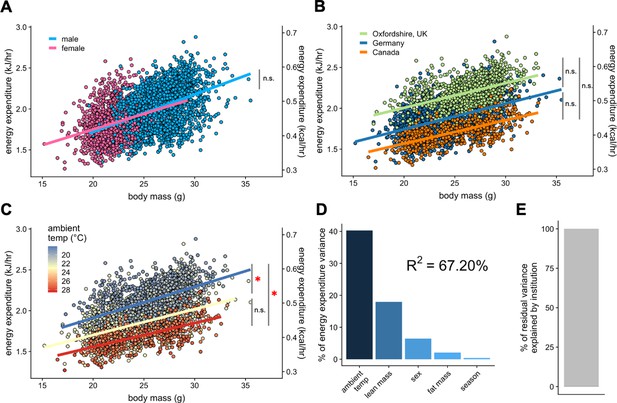
IMPC experiment: Comparison of three sites by sex and temperature.
Regression plots of EE vs total body mass for male and female WT mice (A). Institutional effects for male and female mice on regression plots of EE vs total mass (B). Temperature effects for male and female mice on regression plots of EE vs total mass by ambient temperature (blue, cold regression line <22°C; yellow, mid 22–26°C; red, hot >26°C) (C). The percent variation in EE explained by each of the listed factors (D). Unaccounted institutional variation is less than 1% (E). *, p<0.05. n = 1996 males and 1,153 females.
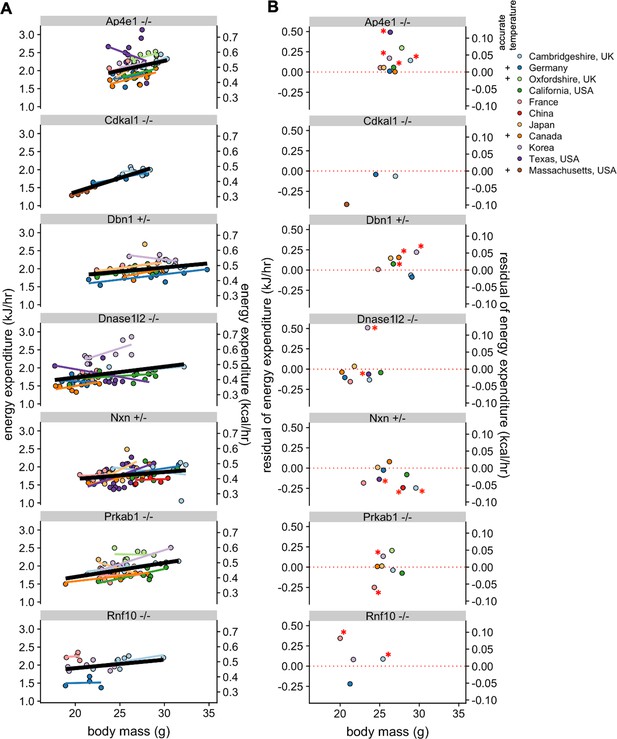
Reproducibility of KO phenotypes.
Plots of EE vs mass for strains that were studied at in least four sites and also for Cdkal1, studied at two IMPC sites and in Massachusetts, USA, with overall best fit regression line shown in bold. (A). Mean residual values from a regression model showing the difference from the controls at the specific site (B). n = 4–21 males per group. *, p<0.05.
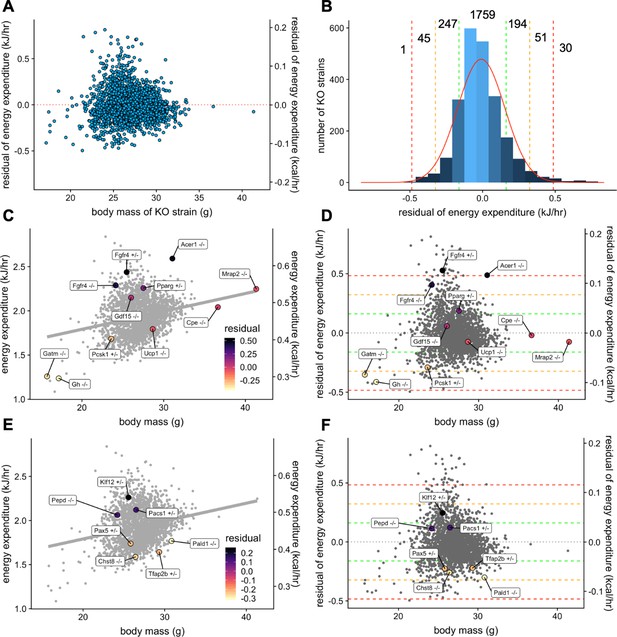
Genetic contribution to EE.
The residual change in EE compared to site-appropriate controls for males of 2329 strains (A). Distribution of residual changes in EE for all KO strains with the numbers of KO strains reaching 1SD (green), 2SD (orange), or 3SD (red). Regression plots of EE vs mass (C) or residual EE vs mass for mice from mouse strains with known metabolic phenotypes (D). Lines showing ranges of standard deviations in EE are overlayed. Similar plots for IMPC strains with genes identified by obesity related GWAS and having phenotypes of more than 1 standard deviation (E, F). n = 23,309 males.
-
Figure 6—source data 1
IMPC dataset for 2329 knockout strains with residual and standard deviation values for EE.
- https://cdn.elifesciences.org/articles/53560/elife-53560-fig6-data1-v3.xlsx
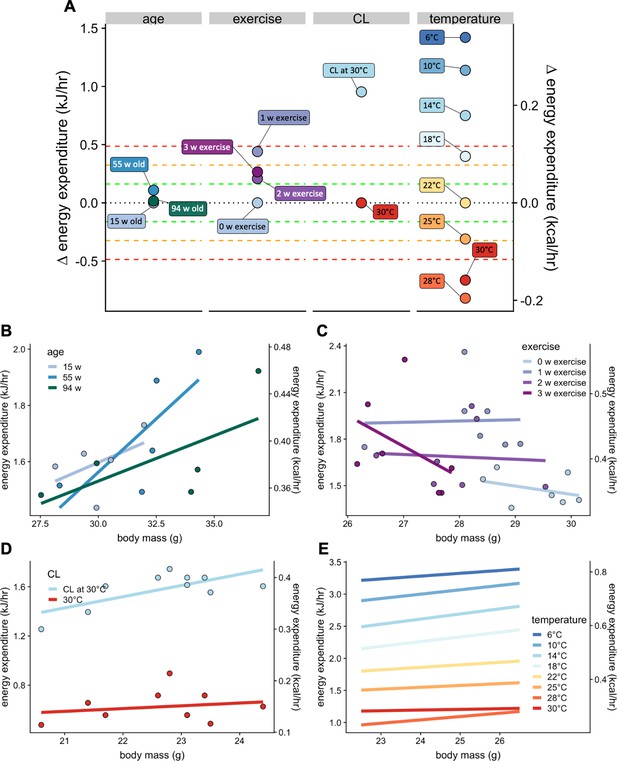
Select physiologic effects on EE.
Changes in EE for physiological challenges including age, voluntary wheel running exercise, β3 adrenergic agonism, warm and cold temperature compared to relative genetic changes as seen in Figure 6B (A). Regression plots for each intervention. Weeks of age (n = 5 per group) (B), weeks of exercise (n = 7) (C), a single dose of β3 adrenergic agonist CL316,243 in mice housed at 30°C (n = 10) (D), and temperature (n = 9) (E).
Tables
Essential information to report on indirect calorimetry studies.
| Location |
| Body mass |
| Body composition |
| Age |
| Accurate ambient temperature |
| Locomotor activity |
| Sex |
| Diet name and composition |
| Strain/Genotype |
| Intervention details |
| Calorimeter model/parameters |
| Body composition method |
IMPC sites and abbreviations.
| Name in this study | Location | IMPC abbreviation |
|---|---|---|
| California, USA | University of California, Davis | UC Davis |
| Cambridgeshire, UK | Wellcome Trust Sanger Institute | WTSI |
| Canada | Toronto Centre for Phenogenomics, The Centre for Phenogenomics | TCP |
| China | Model Animal Research Center of Nanjing University | MARC |
| France | PHENOMIN-Institut Clinique de la Souris | ICS |
| Germany | Helmholtz-Zentrum Muenchen | HMGU |
| Japan | RIKEN BioResource Research Center | RBRC |
| Korea | Korea Mouse Phenotype Consortium | KMPC |
| Oxfordshire, UK | Medical Research Council, Harwell | MRC Harwell |
| Texas, USA | Baylor College of Medicine | BCM |