Ordered patterning of the sensory system is susceptible to stochastic features of gene expression
Figures
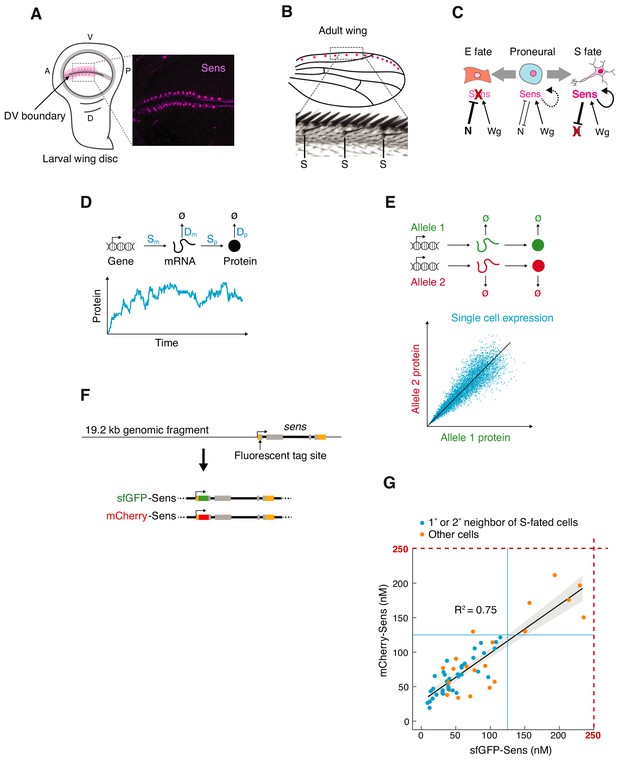
Measuring sens gene expression stochasticity during sensory organ fate selection.
(A) Sens protein is expressed in two stripes of cells bordering the dorsoventral (DV) boundary of the wing disc. The pattern refines into a periodic pattern of S-fated cells in the anterior region, which can be seen as expressing high levels of Sens protein. Anterior (A), left. Ventral (V), top. Right panel is a micrograph of Sens protein immunofluorescence. (B) This generates the highly ordered pattern of sensory bristles along the anterior margin of the adult wing. S denotes chemosensory bristles that had been determined at the stage visualized in (A). (C) Cells are induced by Wg to a proneural state expressing moderate levels of Sens. Notch-mediated lateral inhibition causes cells to switch to either low stable expression (E fate) or high stable expression (S fate) of Sens. Cell-autonomous positive feedback by Sens and non-autonomous feedback by mutual inhibition are key to this process. (D) Gene expression output is inherently variable due to stochastic synthesis and decay of mRNA and protein molecules. Therefore, single cell protein counts fluctuate stochastically around the expected steady state expression level. The magnitude of these fluctuations is determined by the rate constants of individual steps (in blue). (E) Stochasticity can be measured by tagging the two alleles of a gene with distinct fluorescent proteins and measuring fluorescence correlation in individual cells. Each datapoint is red and green fluorescence in one cell. Cells with greater gene expression stochasticity deviate further from the expected average fluorescence (black line). (F) A genomic fragment containing sens was N-terminally tagged with either single sfGFP or mCherry tags. These were used to rescue sens mutant animals by site-specific insertion into genomic location 22A3 (Figure 1—figure supplement 1). (G) Single-cell mCherry and sfGFP protein numbers counted by FCS in sfGFP-sens/mCherry sens wing cells (see also Figure 1—figure supplements 2–3).
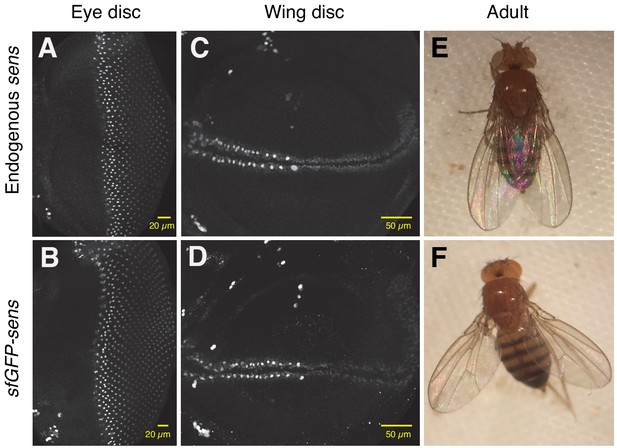
The sens transgene inserted at 22A3 expresses Sens similarly to the endogenous sens gene and it rescues all mutant phenotypes.
(A–D) Larval imaginal discs stained with anti-Sens antibody. Scale bars represent 20 μm for (A, B) and 50 μm for (C, D). Endogenous sens expression in wildtype eye (A) and wing (C) discs. sfGFP-sens expression in eye (B) and wing (D) discs. Almost no endogenous protein is present in discs with sfGFP-sens due to both endogenous alleles being mutated. (E, F) Adults with two copies of the sens transgene rescuing mutant endogenous sens (F) show no phenotypic differences compared to adults with wildtype endogenous sens (E). The animals have the same number and pattern of sensory organs (bristles, eyes, and antennae) as wildtype. Mutant sens animals at a comparable stage cannot be shown because they are embryonic lethal.
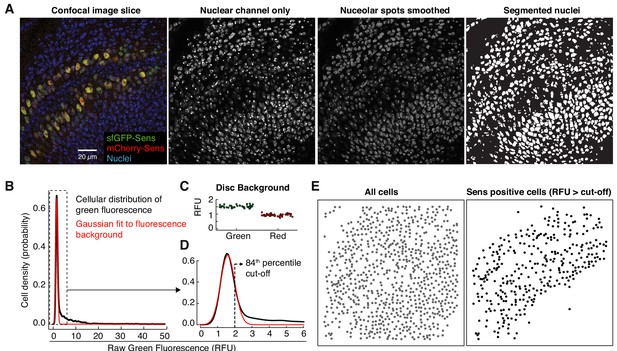
Image analysis of developing wing tissue to quantify Sens protein in single cells.
(A) The nuclear (DAPI) channel from individual confocal image slices was used to computationally identify and segment individual nuclei. Average single cell sfGFP and mCherry fluorescence signals were then estimated from segmented nuclei. (B) Raw sfGFP signal (green fluorescence channel) histogram from the wing disc in (A) is shown as an example. The Gaussian-fitted fluorescence background is shown in red. (C) Mean fluorescence background was calculated for green or red channels in individual images. Each datapoint is one disc. (D) Magnified view of histogram in (B). Dashed line marks the cut-off signal value above which cells were considered Sens-positive. (E) Computationally identified Sens-positive cells displayed the expected expression pattern and were used for further analysis.
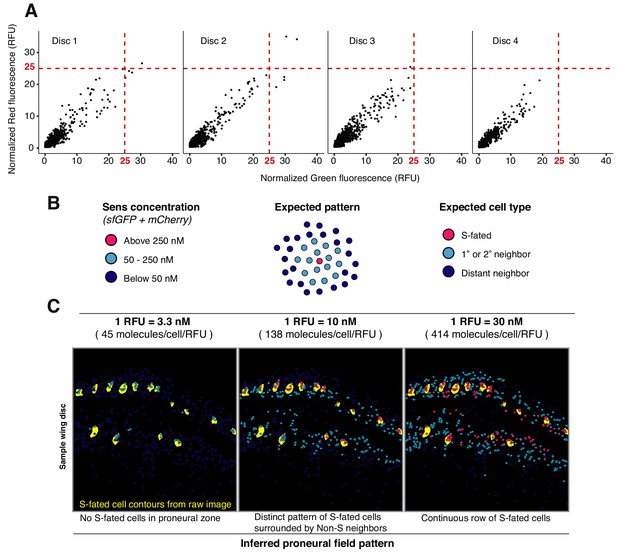
Calibrating a conversion factor between confocal microscopy fluorescence and FCS molecule counting.
(A) Relative fluorescence intensity (RFU) for cells in four fixed wing discs is shown. Red dashed lines mark the estimated maxima for Sens expression at 25 RFU, corresponding to 250 nM maxima measured by FCS in Figure 1G (B) Expected arrangement of S-fated cells (magenta), first or second degree neighbors (cyan), and distant neighbors (dark blue) within Sens-positive stripes adjacent to the DV boundary. Cells were categorized into distinct classes based on total Sens concentration measured by FCS (left). (C) Fluorescence signal intensity (RFU) from imaged cells was converted into concentration (nM) by multiplying with a particular conversion factor. Cell coordinates were then mapped in space and color-coded according to their expected cell category (defined in (B)). Contours of S-fated cells were identified from raw microscopy images and are shown in yellow. Three conversion factors were tested, as shown at top. The factors differed from one another across a nine-fold range. Only a conversion factor of 10 nM/RFU (center column) was able to recapitulate the expected pattern of S-fated cells. Three representative discs are shown. Decreasing the factor to 3.3 (left) eliminated S-fated cells. Increasing the factor to 30 (right) produced a continuous row of S-fated cells. Thus, we estimate the conversion factor to be within an order of magnitude of the estimated conversion of 1 RFU equivalent to 10 nM.
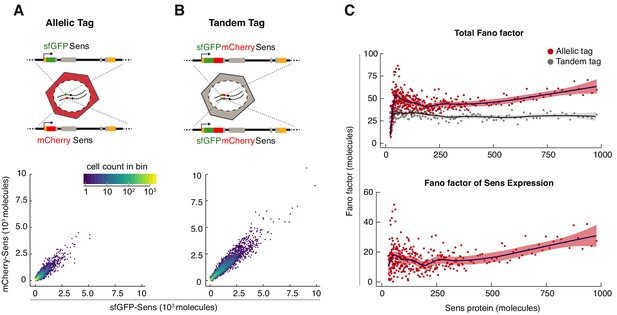
Sens Fano factor relative to protein copy number in single cells.
(A) sfGFP- and mCherry-Sens protein numbers measured from single cells in sfGFP-sens/mCherry-sens wing discs. 12,000 cells were plotted for comparison in (A,B). Colors represent cell count in each hexagonal plot region. (B) Estimated sfGFP and mCherry numbers generated from the tandem-tagged sfGFP-mCherry-sens gene in single cells of wing discs. Note that these numbers are not perfectly correlated with one another due to noise in the measurement process (see also Figure 2—figure supplement 1). As expected, median expression of the tandem tag was 2.1 ± 0.03 fold higher than allelic tag expression. (C) Top panel: The Fano factor was calculated in bins of cells expressing either tandem-tagged Sens or the singly-tagged allelic pairs of Sens (Figure 2—figure supplement 2). Bottom panel: The Fano factor of Sens expression was calculated by subtracting out the Fano factor from tandem-tagged cells. Plotted region encompasses data from 88% of tagged cells (Sens < 1000 molecules). Shading demarcates 95% confidence intervals.
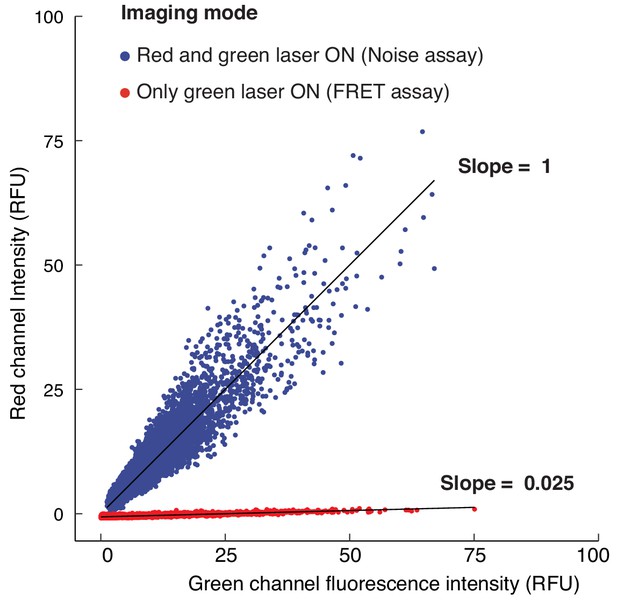
Fluorescence Resonance Energy Transfer (FRET) from sfGFP to mCherry molecules is negligible under experimental imaging conditions.
Wing disc cells with tandem tagged sfGFP-mCherry-sens alleles were imaged under identical conditions with either both green and red lasers ‘on’ to assay stochastic noise, or with only the green laser ‘on’ to assay FRET from sfGFP to mCherry molecules. Raw single cell green channel fluorescence intensity is plotted on the x-axis. All cells in the imaging field were included for analysis. Red channel fluorescence values, on y-axis, were linearly transformed to make the slope equal to 1 and y-intercept equal to 0 for noise assay data. Single cell red fluorescence values for FRET assay data were then transformed with identical parameters for comparison.
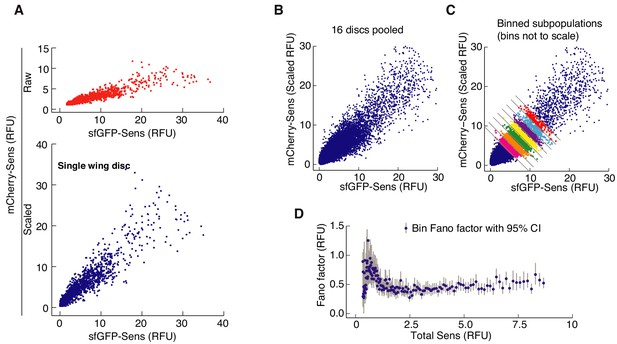
Fano factor calculation from nuclear fluorescence signals of Sens positive cells.
(A) The mCherry signal intensity is scaled relative to sfGFP signal in Sens-positive cells from individual discs. (B) Data from discs of the same genotype are pooled together. (C) Pooled data are sorted into separate bins according to total Sens output. The intrinsic noise and mean for Sens is calculated for each binned sub-population. (D) The Fano factor of each bin is plotted as a function of mean Sens output in a bin. 95% confidence intervals for the estimated Fano factor are calculated by bootstrapping with resampling within each binned sub- population.
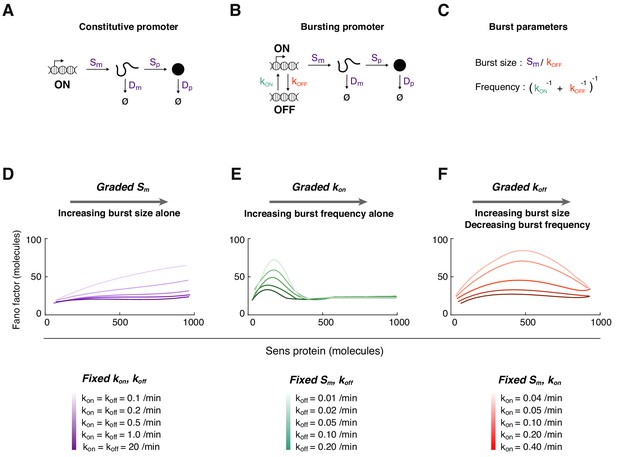
Mathematical modeling of Sens protein noise.
(A) The model of gene expression with a constitutively active promoter. (B) The two-state model with a promoter having distinct on and off states, and independent rate constants for state conversion (see also Figure 3—figure supplements 1–2). (C) The relation between transcription rate constants and transcription burst frequency and burst size. (D-F) The Fano factor derived from simulations of the two-state model. In each panel, a different transcription rate constant is varied to generate a range of Sens protein output. Trend lines were generated by smoothing the Fano factor profiles obtained from binning 5,000 simulated cells. (D) The rate constant Sm is systematically varied from 0.1 to 1 mRNA/min. Rate constants kon and koff are fixed at the values shown for each simulation curve. The promoter switches rapidly at high values of kon and koff such that the Fano factor is similar to one from a constitutively active promoter (dark purple line). (E) The rate constant kon is systematically varied from 0.025 to 10 min-1. Rate constant Sm is fixed at 0.25 mRNA/min, and koff is fixed at the values shown for each simulation curve. (F) The rate constant koff is systematically varied from 0.025 to 3 min-1. Rate constant Sm is fixed at 0.5 mRNA/min, and kon is fixed at the values shown for each simulation curve.
-
Figure 3—source data 1
Rate parameters used in modeling.
Default parameter values (indicated in bold) were used for all simulations unless noted.
- https://cdn.elifesciences.org/articles/53638/elife-53638-fig3-data1-v2.xlsx
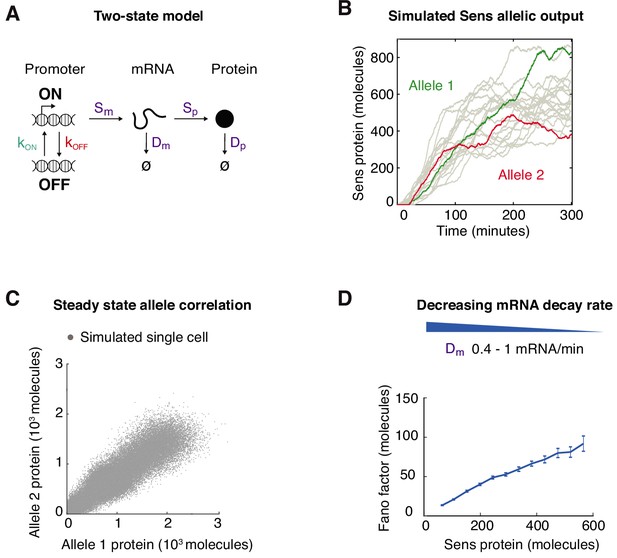
Modeling of Sens expression and noise.
(A) Simple two-state model of gene expression with six rate parameters as shown. (B) Simulated trajectories of Sens protein molecule numbers over time starting from zero molecules. Two randomly paired trajectories are shown in green and red, which mimic two independently acting alleles within the same cell. (C) Sens protein output from randomly paired virtual alleles. Sampling was taken when all simulations had reached steady state. The kon rate parameter was varied to generate a range of steady-state protein output, and pairing was only done between simulations with identical rate parameters. The full range of Sens expression can be recapitulated by varying any of the six gene expression rate parameters in the model. (D) Simulations were performed by systematically varying the mRNA decay rate constant Dm and the Fano factor was calculated for bins of randomly paired simulations according to their protein output. Error bars are 95% confidence intervals as calculated by bootstrapping. The resulting Fano factor slowly rises as protein levels increase. Similar profiles were obtained if the rate parameters Sp and Dp were systematically varied (not shown).
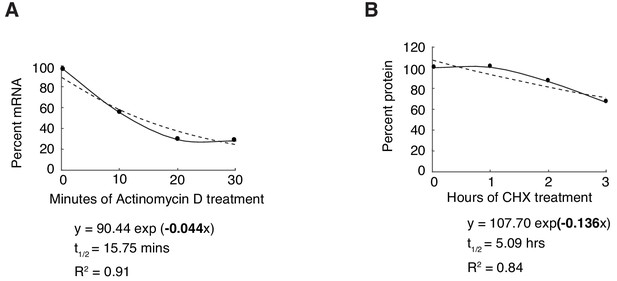
Measurement of sens mRNA and protein decay in the wing disc.
(A) Developing wing discs were treated with Actinomycin D to block mRNA synthesis. sens mRNA was measured at 10 min time intervals by RT-qPCR, and an exponential decay rate was estimated from multiple time points. Dashed line shows the fitted decay curve. (B) Developing wing discs were treated with cycloheximide (CHX) to block protein synthesis. Sens protein was measured at 1 hr time intervals by indirect enzyme linked immunosorbent assay (ELISA), and an exponential decay rate was estimated from multiple time points. Dashed line shows the fitted decay curve.
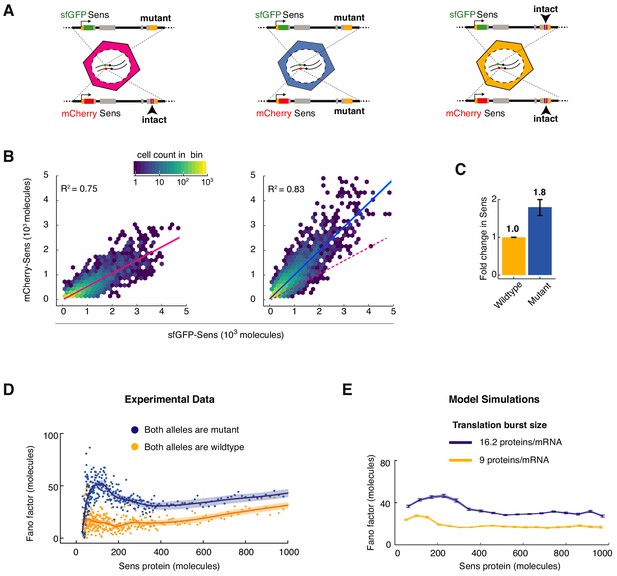
MicroRNA regulation uniformly decreases Sens protein output and noise.
(A) The sens transgenes were modified to mutate the two miR-9a binding sites in the 3’UTR. Wing disc cells were generated that had different combinations of mutant and wildtype alleles. (B) Protein output measured from the mCherry-sens allele with intact (left plot) or mutated (right plot) miR-9a sites. These were measured relative to protein output from the sfGFP-sens allele with mutated sites. A sample of 6,000 cells is shown for each plot, and colors represent cell density in the plotted hexagonal bins. Lines represent the linear regression model fit and shaded regions are the 95% confidence intervals. Dotted red line in right plot shows fit from the left plot. Since the linear model fits both genotypes equally well, it argues that the strength of miR-9a repression is equivalent in all cells, regardless of Sens output. (C) Sens protein output increases with loss of miR-9a binding sites. Error bars are standard error of the mean. Shown is the comparison of mutant sfGFP-sens to wildtype mCherry-sens but the same result was observed if the mutant mCherry-sens was compared to wildtype sfGFP-sens (Figure 4—figure supplement 1). (D) Loss of miR-9a regulation leads to an increase in Fano factor across the entire range of Sens protein output. Shaded regions are 95% confidence intervals. (E) Model simulations with a 1.8-fold increase in translation burst size (defined as Sp/Dm) reproduces the experimentally observed Fano profile. Error bars are 95% confidence intervals.
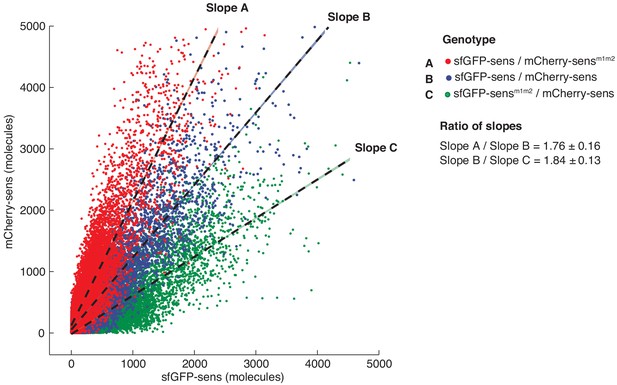
Fluorescent protein tags sfGFP and mCherry behave interchangeably in vivo.
Single wing disc cells of the indicated genotypes were imaged and their nuclear sfGFP and mCherry fluorescence intensity was measured. Cells from individual discs were pooled separately to calculate the mCherry:sfGFP linear slope. Slopes were compared across genotypes (with relative errors propagated) to sensitively assay if the nature of the fluorescent protein tag interferes with quantitative protein measurements. No statistical difference was observed when fold-repression was calculated by mCherry-Sens ratios or by sfGFP-Sens ratios between wild type and mutant sens alleles.
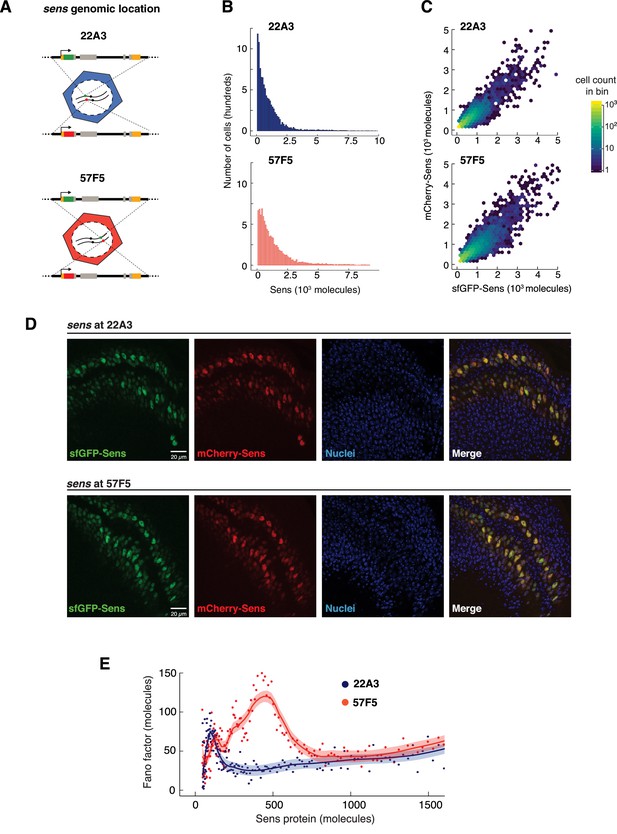
Genome location of the sens gene dramatically affects Sens noise.
(A) The sens transgenes were inserted into one of two locations on chromosome II - 22A3 or 57F5. (B) Histograms depicting the frequency distribution of Sens protein number per cell. 10,000 cells were randomly sampled from 22A3 and 57F5 datasets each for comparison (see also Figure 5—figure supplement 1). (C) Scatter plot with hexagonal binning of single-cell sfGFP-Sens and mCherry-Sens protein numbers from sens genes inserted at 22A3 or 57F5. Randomly chosen subsets of 6,000 cells were plotted for each genotype for ease of comparison. Colors represent cell counts in respective hexagonal bins. (D) Confocal micrographic images of sfGFP- and mCherry-Sens fluorescence in wing discs expressing the sens gene inserted at 22A3 or 57F5. Nuclei are counterstained with DAPI (blue). Scale bars are 20 μm. Image brightness is enhanced (identically across 22A3 and 57F5) for ease of visualization. (E) The Fano factor profiles from cells expressing sens at 57F5 or 22A3. Cells with more than 800 molecules have identical Fano values at both genomic positions. The Fano peaks at lower Sens levels are very different between the genomic locations. Shaded regions are 95% confidence intervals.
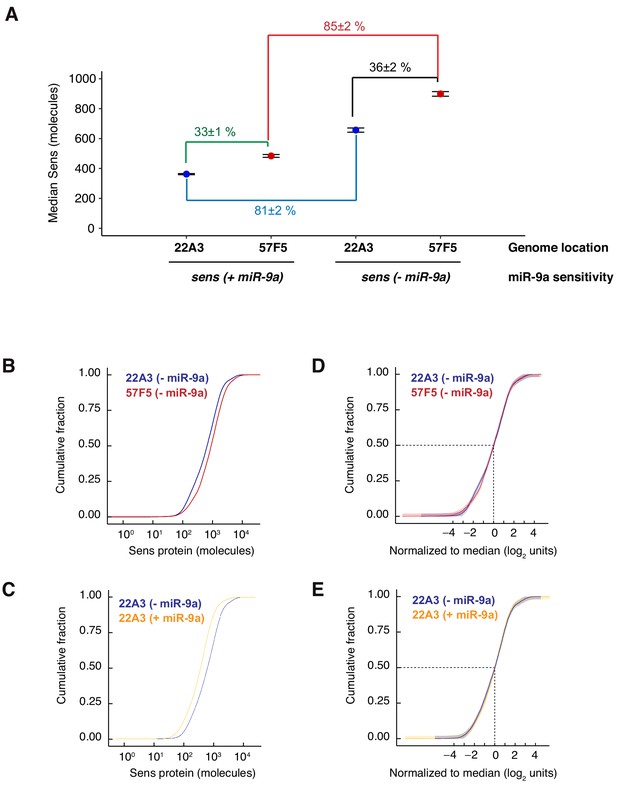
Comparison of Sens expression profiles for various allelic pairs.
(A) Median protein number per cell for total Sens (sfGFP-Sens + mCherry-Sens) expressed from allelic pairs inserted at either 22A3 or 57F5; with or without the miR-9a binding sites in the 3’UTR. Error bars are 95% confidence intervals. Percentages represent the percent increase in median number from lower to higher expressing conditions. (B,C) Empirical cumulative density profiles (CDFs) of Sens protein number per cell for the conditions comparing (B) insertion site and (C) miR-9a regulation. (D,E) CDFs in B and C were normalized by their respective median protein numbers and log2 transformed. Dashed lines mark the normalized median number per cell, corresponding to log2(1)=0. Shaded regions depict the upper and lower 95% confidence intervals computed for each CDF by Kolmogorov-Smirnov’s D statistic.
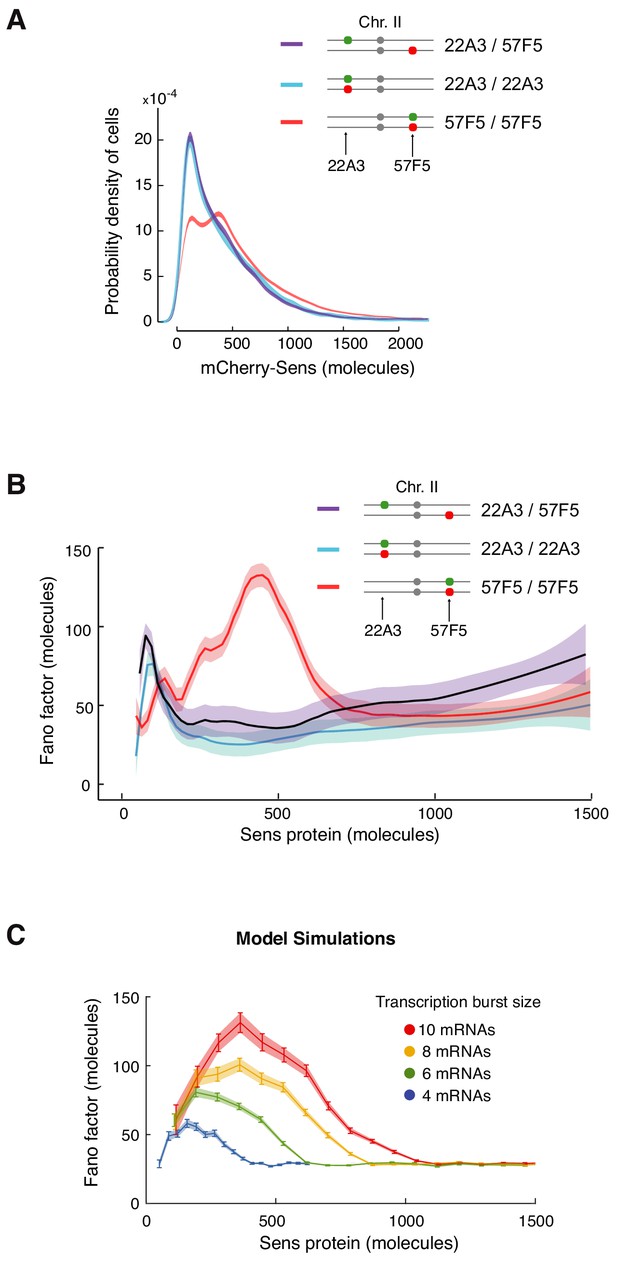
Enhanced protein noise requires trans regulation at 57F5.
(A) Frequency distribution of mCherry-Sens protein number in cells. The mCherry-sens allele is located at either 22A3 or 57F5 as indicated. The sfGFP-sens allele is either paired or unpaired with the mCherry-sens allele as indicated. Lines are 95% confidence intervals of moving averages (see also Figure 6—figure supplement 1). (B) The Fano factor of Sens from cells expressing Sens protein either from paired alleles (22A3/22A3 and 57F5/57F5) or unpaired alleles (22A3/57F5). Moving line averages are shown, and shaded regions are 95% confidence intervals. (C) Model simulations in which transcription burst size (defined as Sm/koff) is set to different values as shown. The Fano peak amplitude and position change as burst size varies, but all relax to a constant basal level. These trends are similar to those observed in (B). Error bars and shaded regions are 95% confidence intervals (see also Figure 6—figure supplement 2).
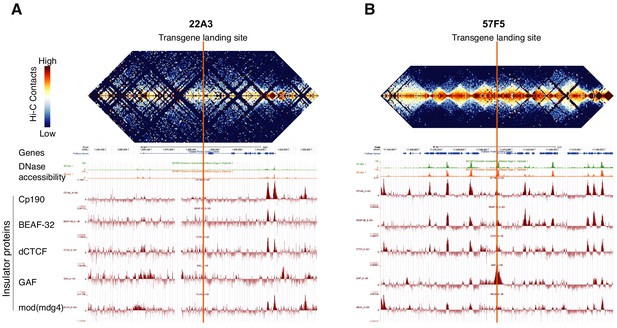
Chromatin landscape of genomic loci 22A3 and 57F5 determined from Hi-C data.
(A, B) Heat maps of aggregate Hi-C data used to calculate chromosomal contact frequency for the embryonic nc14 genome. Vertical orange lines denote the precise locations of the two transgenic landing sites at 22A3 (A) and 57F5 (B). Below the heat maps are shown tracks for annotated genes, DNase accessibility, and ChIP-seq of the insulator proteins CP190, BEAF-32, dCTCF, GAF and mod(mdg4). The plots are adapted from Stadler et al. (2017), Liu et al. (2008) and Nègre et al. (2010). Although these data were derived from embryonic genomes, Stadler et al. (2017) showed that the embryonic TAD boundaries correspond to the locations of interband regions of larval polytene chromosomes. This strongly suggests that TAD organization is largely maintained during the development of embryos into third-instar larvae.
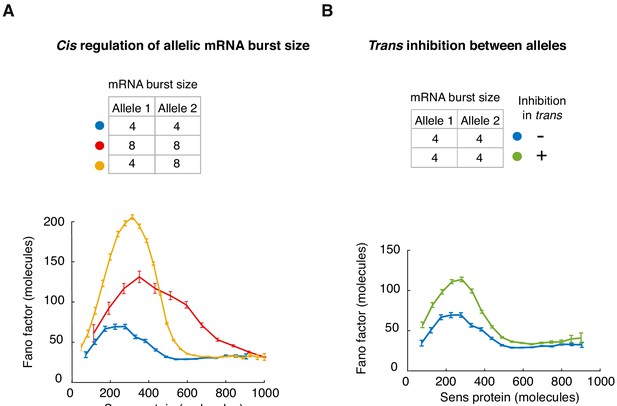
Fano factor profiles from model simulations in which sens alleles at 57F5 are regulated independently in cis or there is promoter inhibition in trans.
(A) In order to mimic a cis-only mode of regulation, one set of simulations was run with an average burst size (Sm/koff) of 4 mRNAs (blue). Another set of simulations was run with an average burst size of 8 mRNAs (red). We assumed these different burst sizes corresponded to 22A3 and 57F5 alleles, respectively. For each set, the kon rate parameter was systematically varied to generate simulations with a range of steady-state protein output. To mimic cis-only regulation in 57F5/22A3 cells, each allele’s expression in the virtual cell was simulated independent of the other allele. This was done by simulating an average burst size of 4 mRNAs for one allele and 8 mRNAs for the other allele (yellow). Thousands of such cells were simulated for each value of kon and then binned according to total protein output. Fano factor of bins is plotted for each virtual allelic combination. (B) To simulate inhibitory interactions between paired 57F5 alleles (green), the kon of one allele was downregulated by 90% if the other allele was turned on to transcribe, and vice-versa. Non-interacting alleles (blue) were simulated independent of each other as in (A). Both sets of simulations were run with average burst size of 4 mRNAs. Thousands of cells were simulated across a spectrum of kon rates. Fano factor for binned cell populations is plotted. All error bars are 95% confidence intervals.
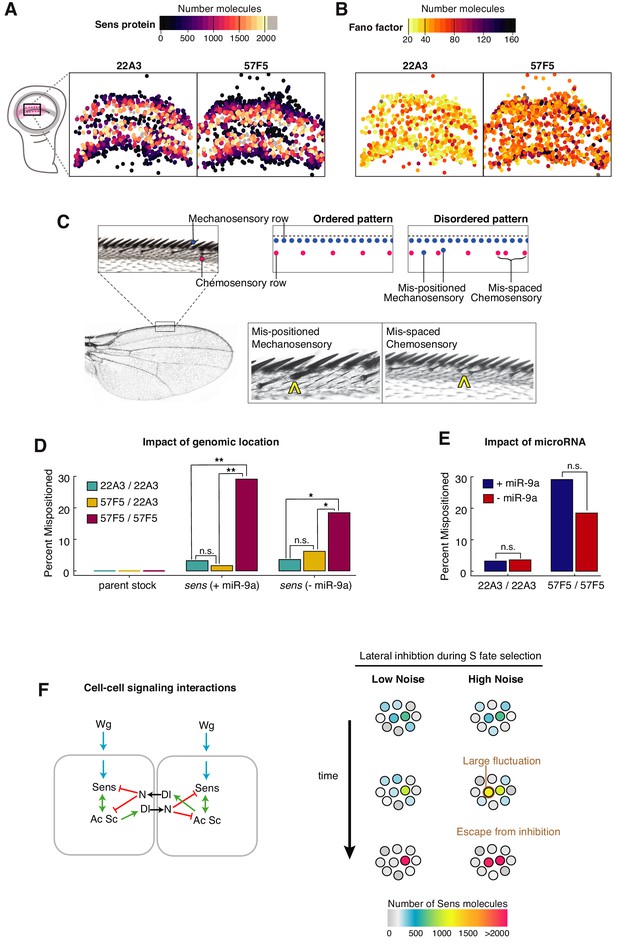
Self-organized sensory patterning is disrupted by stochastic gene expression.
(A, B) The centroids of Sens-positive cells in 22A3/22A3 and 57F5/57F5 wing discs are mapped and color coded according to Sens protein number (A), and Fano factor level (B). Cells exhibiting high Fano factor values are distributed throughout the proneural zone of the 57F5/57F5 disc, where S fate determination occurs (see also Figure 7—figure supplements 1 and 2). (C) The dorsal surface of the adult wing margin displays two ordered rows of sensory organs - an outer continuous row of thick mechanosensory bristles (blue) and an inner periodic row of thin chemosensory bristles (magenta). Disorganized patterns are observed when bristles are incorrectly positioned. Instances of ectopic (mis-positioned) mechanosensory bristles in the chemosensory row (center) and ectopic chemosensory bristles, which disrupt periodic spacing (right), were observed and counted. (D) Percentage of adult wings with one or more patterning error. Pattern disorder is much greater when sens alleles are expressed from 57F5/57F5 relative to alleles at 22A3/22A3 or 57F5/22A3. This result is observed regardless of whether miR-9a regulates sens or not. Genotypes were compared by calculating the odds ratio of mispatterning, and determined to be significantly different from one if p<0.05 using a Fischer’s exact test: n.s., not significant; *p<0.05; **p<.005 (Figure 7—figure supplement 3). (E) Uniformly increasing Sens protein number 1.8-fold by removing miR-9a regulation does not lead to greater pattern disorder. Genotypes were compared by calculating the odds ratio of mis-patterning, and determined to be significantly different from one if p<0.05 using a Fischer’s exact test: n.s., not significant (Figure 7—figure supplement 3). (F) Left: A schematic outlining the induction of sens by Wg (blue) followed by sens auto-activation through feedback with Ac Sc (green) and through mutual inhibition between neighboring cells (red). Right: When expression noise is sufficiently small, Sens levels progressively increase or decrease in neighboring cells. However, when noise is enhanced, a large fluctuation will alter the trajectory of Sens expression towards an erroneous outcome.
-
Figure 7—source data 1
Odds ratio of wings with mispositioned mechanosensory bristles.
For each genotype, the proportion of wings containing at least one ectopic mechanosensory bristle was calculated. To compare between two genotypes, the odds ratio (of wings with ectopic bristles) was calculated from these proportions. Test column indicates the variable being compared (genomic locus of sens alleles, or presence of miR-9a binding sites in the sens 3’UTR) across each subgroup. Fisher’s exact test was used to determine if the odds ratio was not equal to 1. Odds ratios significantly different from one are highlighted in bold (*p-value<0.05, ** p-value<0.01, *** p-value<0.005, p-value>0.05, not significant).
- https://cdn.elifesciences.org/articles/53638/elife-53638-fig7-data1-v2.xlsx
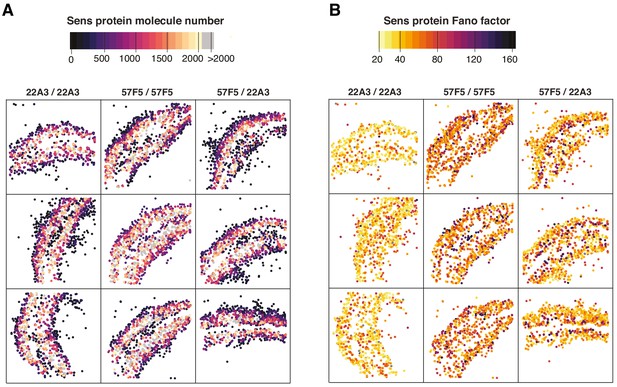
Maps of Sens protein number and Fano factor in replicate wing discs.
(A, B) Spatial maps of cell coordinates from individual wing discs. Three representative wing discs with homologous allele pairs for sens at 22A3 (left), 57F5 (center) and the non-homologous pair 57F5/22A3 (right) are shown. Cell centroids are color coded according to their corresponding (A) Sens protein number or (B) Fano factor. Color scales on top. Only cells with fewer than 2000 molecules of Sens are plotted for ease of visualization.
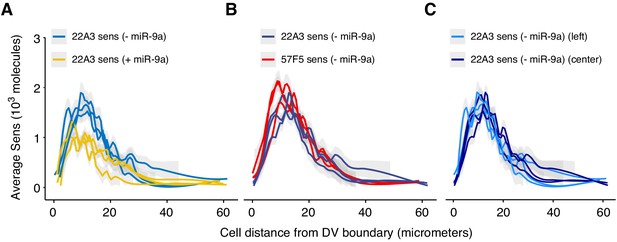
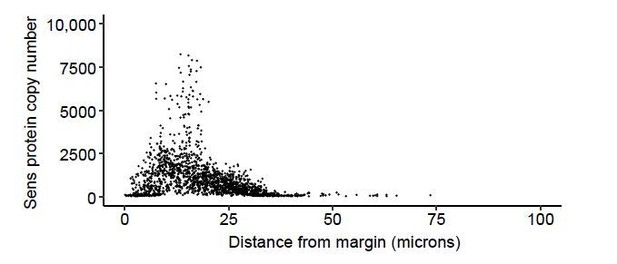
Average Sens expression as a function of cell position relative to the DV boundary.
The DV boundary was manually drawn and Sens protein levels were measured as a function of the shortest distance from a cell to the boundary. Cells in both dorsal and ventral compartments were used in this analysis. Each line represents a moving line average from one wing disc. Shaded regions are 95% confidence intervals around the estimated average. (A) Effect of loss of miR-9a repression on sens located at 22A3. (B) Effect of transgene location at 22A3 or 57F5. (C) Effect of experimental replication on sens. Sets of discs were collected in separate experimental runs.
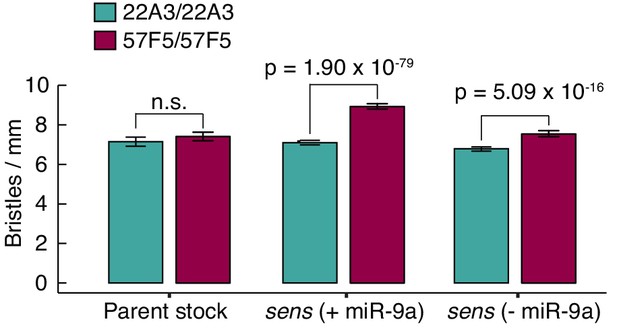
Chemosensory bristle spacing is dependent on genomic location of the sens gene.
If ectopic chemosensory bristles arise due to Sens protein fluctuations, they would decrease the average distance between adjacent chemosensory bristles. Therefore, we measured chemosensory bristle density on the anterior wing margin of adult females. When sens is expressed from 57F5, chemosensory bristle density is greater relative to 22A3. This result is observed irrespective of the nature of the sens allele compared that is with or without miR-9a regulation of sens. In contrast, bristle density is not significantly different between the parental 22A3 and 57F5 stocks which express sens from the endogenous locus on chromosome III. Error bars are 95% confidence intervals and p values are from a student’s t-test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | white1118 | BloomingtonDrosophilaStock Center | BDSC: 3605 Flybase: FBst0003605 | |
| Gene (Drosophila melanogaster) | sensE1 | Nolo et al. (2001) | Flybase: FBal0098024 | From Hugo Bellen |
| Gene (Drosophila melanogaster) | sensE2 | BloomingtonDrosophilaStock Center | BDSC: 5311 Flybase: FBal0098023 | |
| Strain, strain background (Drosophila melanogaster) | y1 w1118; PBac{y+-attP-3B} VK00037 | BloomingtonDrosophilaStock Center | BDSC: 9752 Flybase: FBst0009752 | 22A3 |
| Strain, strain background (Drosophila melanogaster) | y1 w1118; PBac{y+-attP-9A} VK00022 | BloomingtonDrosophilaStock Center | BDSC: 9740 Flybase: FBst0009740 | 57F5 |
| Genetic reagent (Drosophila melanogaster) | Wildtype sfGFP-sens [22A3] | Venken et al. (2006). From Hugo Bellen | Pacman construct containing sens gene with N-terminal 3xFlag-TEV-StrepII-sfGFP-FlAsH fusion tag inserted at 22A3 | |
| Genetic reagent (Drosophila melanogaster) | Mutant sfGFP-sens [22A3] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-sfGFP-FlAsH fusion tag and two miR-9a binding sites mutated inserted at 22A3 | |
| Genetic reagent (Drosophila melanogaster) | Wildtype mCherry-sens [22A3] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-mCherry-FlAsH tag inserted at 22A3 | |
| Genetic reagent (Drosophila melanogaster) | Mutant mCherry-sens [22A3] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-mCherry-FlAsH fusion tag and two miR-9a binding sites mutated inserted at 22A3 | |
| Genetic reagent (Drosophila melanogaster) | Tandem tag sfGFP-mCherry -sens [22A3] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-mCherry-sfGFP-FlAsH fusion tag inserted at 22A3 | |
| Genetic reagent (Drosophila melanogaster) | Wildtype sfGFP-sens [57F5] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-sfGFP-FlAsH fusion tag inserted at 57F5 | |
| Genetic reagent (Drosophila melanogaster) | Mutant sfGFP-sens [57F5] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-sfGFP-FlAsH fusion tag and two miR-9a binding sites mutated inserted at 57F5 | |
| Genetic reagent (Drosophila melanogaster) | Wildtype mCherry-sens [57F5] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-mCherry-FlAsH tag inserted at 57F5 | |
| Genetic reagent (Drosophila melanogaster) | Mutant mCherry -sens [57F5] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-mCherry-FlAsH fusion tag and two miR-9a binding sites mutated inserted at 57F5 | |
| Genetic reagent (Drosophila melanogaster) | Tandem tag sfGFP-mCherry -sens [57F5] | This paper | sens transgene with N-terminal 3xFlag-TEV-StrepII-mCherry-sfGFP-FlAsH fusion tag inserted at 57F5 | |
| Antibody | Guinea Pig polyclonal anti-Sens | Nolo et al. (2000). From Hugo Bellen | IF(1:1000) | |
| Antibody | Goat anti-guinea pig IgG Alexa 488 | Invitrogen | Cat# A-11073, RRID: AB_2534117 | IF(1:250) |
| Antibody | Mouse monoclonal anti-Flag, clone M2 | Sigma | Cat# F1804, RRID: AB_262044 | Elisa (1:100) |
| Antibody | Rabbit polyclonal anti-GFP | Molecular Probes | Cat# A-11122, RRID: AB_221569 | Elisa (1:5000) |
| Antibody | Goat polyclonal anti-rabbit IgG – HRP conjugated | GE Healthcare | Cat# RPN4301, RRID: AB_2650489 | Elisa (1:5000) |
| Recombinant DNA reagent | P[acman] wild type sfGFP-sens | Venken et al. (2006). Gift of Koen Venken | ||
| Recombinant DNA reagent | P[acman] mutant sfGFP-sens | This paper | 19.2 kb D. melanogaster genomic fragment including sens gene with miR-9a binding sites mutated and N-terminal fusion to sfGFP | |
| Recombinant DNA reagent | P[acman] wild type mCherry-sens | This paper | 19.2 kb D. melanogaster genomic fragment including sens gene with N-terminal fusion to mCherry | |
| Recombinant DNA reagent | P[acman] miR-9a binding site mutant mCherry-sens | This paper | 19.2 kb D. melanogaster genomic fragment including sens gene with miR-9a binding sites mutated and N-terminal fusion to mCherry | |
| Recombinant DNA reagent | P[acman] wild type tandem tag sfGFP-mCherry-sens | This paper | 19.2 kb D. melanogaster genomic fragment includingsens gene with miR-9a binding sites mutated and N-terminal fusion to mCherry and sfGFP | |
| Sequence-based reagent | 18S - Forward | This paper | PCR primers | CTGAGAAACGGCTACCACATC |
| Sequence-based reagent | 18S - Reverse | This paper | PCR primers | ACCAGACTTGCCCTCCAAT |
| Sequence-based reagent | Rpl21 - Forward | This paper | PCR primers | CTTGAAGAACCGATTGCTCT |
| Sequence-based reagent | Rpl21 - Reverse | This paper | PCR primers | CGTACAATTTCCGAGCAGTA |
| Sequence-based reagent | Sens - Forward | This paper | PCR primers | CAGGAATTTCCAGTGCAAACAG |
| Sequence-based reagent | Sens - Reverse | This paper | PCR primers | CGCCGGTATGTATGTACGTG |
| Sequence-based reagent | Hsp70Ba - Forward | This paper | PCR primers | AGTTCGACCACAAGATGGAG |
| Sequence-based reagent | Hsp70Ba - Reverse | This paper | PCR primers | GACTGTGGGTCCAGAGTAGC |
| Commercial assay or kit | 1-Step Ultra TMB-ELISA | Thermo Fisher | Cat #34028 | For Elisa assays |
| Chemical compound, drug | Paraformaldehyde (powder) | Polysciences | 00380–1 | |
| Chemical compound, drug | Triton X-100 | Sigma Aldrich | T9284-500ML | |
| Chemical compound, drug | VectaShield | Vector Labs | H-1000 | |
| Chemical compound, drug | 4′,6-diamidino-2-phenylindole (DAPI) | Life Technologies | D1306 | |
| Software, algorithm | MATLAB script to automatically segment disc nuclei | Peláez et al. (2015) | https://github.com/ritika-giri/stochastic-noise | |
| Software, algorithm | MATLAB scripts for modeling simulations | This paper | https://github.com/ritika-giri/stochastic-noise/tree/master/MATLAB%20scripts%20for%20gene%20expression%20simulation | |
| Software, algorithm | R scripts for imaging data analysis | This paper | https://github.com/ritika-giri/stochastic-noise/tree/master/R%20script%20for%20data%20analysis | |
| Other | WM1 medium | Restrepo et al. (2016) | Growth medium for organ culture |