CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication
Figures
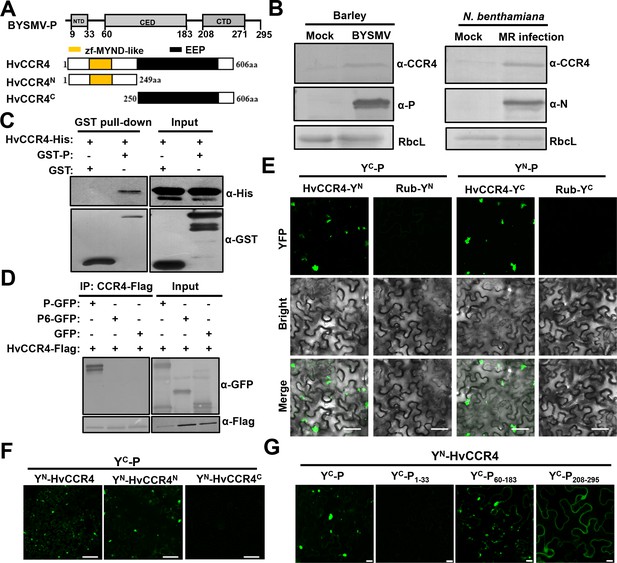
The barley HvCCR4 protein interacts with the BYSMV P protein in vitro and in vivo.
(A) Modular organization of BYSMV P, HvCCR4 and schematic presentation of deletion mutants. The BYSMV P protein consists of three structural domains, including an N terminal domain (NTD), a central domain (CED), and a C terminal domain (CTD). The yellow and black boxes indicate the HvCCR4 zf-MYND-like and EEP nuclease domains. (B) Western blotting analyses showing accumulation of the CCR4 (α-CCR4), P (α-P) and N (α-N) proteins in BYSMV-infected barley plants, BYSMV-MR-infected N. benthamiana leaves, and mock inoculated plants. Rubisco complex large subunit (RbcL) detected by Stain-Free technology was used as a loading control. (C) GST pull-down assays showing HvCCR4–P interactions in vitro. His-tagged HvCCR4 was incubated with GST-tagged P or GST and immunoprecipitated with glutathione-Sepharose beads. The pull down and input proteins were detected by western blotting assays with anti-GST (α-GST) and anti-His (α-His) antibodies. (D) Co-IP analysis of the interaction between P-GFP and HvCCR4-Flag in vivo. N. benthamiana leaves were agroinfiltrated with A. tumefaciens cells expressing various proteins as indicated. At three dpi, leaf extracts were incubated with anti-Flag beads, and then the IP and input proteins were analyzed by western blotting with anti-GFP (α-GFP) and anti-Flag (α-GFP) antibodies. (E) BiFC analysis of the HvCCR4–P interaction in epidermal cells of N. benthamiana leaves infiltrated with Agrobacterium strains expressing proteins tagged with the YN or YC halves of YFP. Rubisco protein (Rub) served as a negative control. Bar = 50 μm. (F) BiFC analysis of BYSMV P interactions with the HvCCR4N or HvCCR4C domains. Bar = 50 μm. (G) BiFC analysis of the interaction between HvCCR4 and BYSMV P domains indicated in panel A. Bar = 10 μm. In (E), (F), and (G), images were taken at three dpi with a Leica laser scanning microscope.
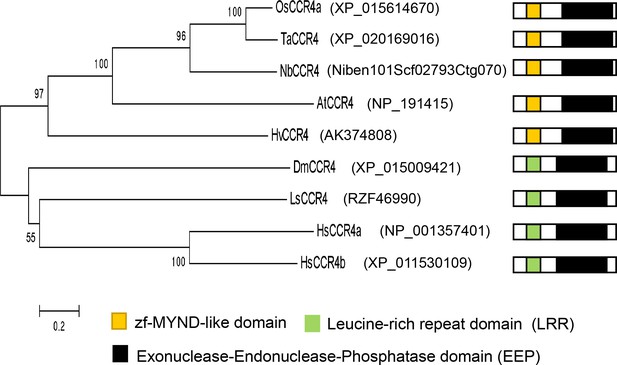
Phylogenetic tree of plant and animal CCR4 orthologues.
Plant CCR4 orthologues are from rice (Os), wheat (Ta), barley (Hv), N. benthamiana (Nb) and Arabidopsis (At). Animal CCR4 orthologues are from human (Hs), fruit fly (Dm), and planthopper (Ls). The phylogenetic tree was constructed by mega 5.0, bootstraps values (>50) indicated in each node. Accession numbers of the genes were listed. Organization of CCR orthologues including the zf-MYND-like domain (yellow boxes), Leucine-rich repeat domain (LRR) (green boxes), and Exonuclease-Endonuclease-Phosphatase domain (EEP) (Black boxes) were shown.

Sequence alignments of the zf-MYND-like, LRR, and EEP domains of various CCR4 protein.
(A) Sequence alignments of the zf-MYND-like domains of rice, wheat, barley, N. benthamiana and Arabidopsis CCR4 proteins (B) Sequence alignments of human, fruit fly, and planthopper CCR4 protein LRR domains. (C) Sequence alignments of the EEP domains of both plant and animal CCR4 proteins.
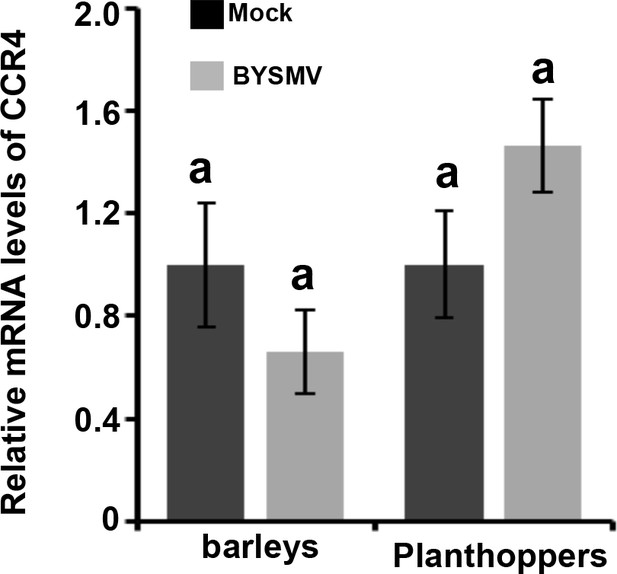
qRT-PCR analysis of the expression of CCR4 in BYSMV-infected and mock-treated barley at 15 dpi or planthoppers at seven dpi.
The values of CCR4 accumulation in mock-treated barley and planthopper were set to 1. Error bars indicate standard errors of three independent experiments. Letters above the bars indicate statistical significance (p<0.01) evaluated by Turkey’s Multiple Comparison Test analysis.
BiFC assays showing negative Rub controls with HvCCR4 and LsCCR4.
Leaves were infiltrated with Agrobacterium strains harboring pairs of YN and YC tagged proteins. Images were taken at three dpi with a confocal scanning laser microscope. Bar = 50 μm.
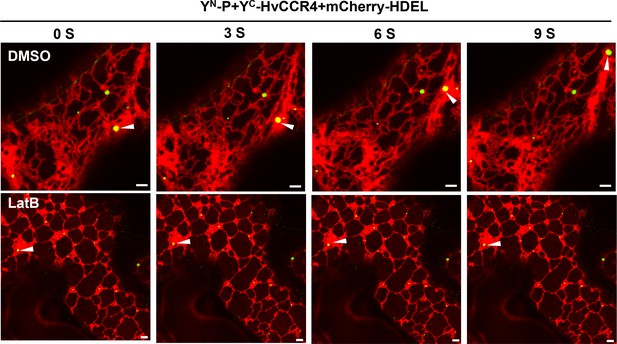
Time-lapse confocal images of YN-P and YC-HvCCR4 in N.
benthamiana leaves. YN-P and YC-HvCCR4 were co-expressed in N. benthamiana leaves following treatment with DMSO or latrunculin B (LatB) at two dpi to disrupt actin assembly. Images were taken 3 hr after DMSO or 10 μM LatB treatments. Bar = 2 μm.
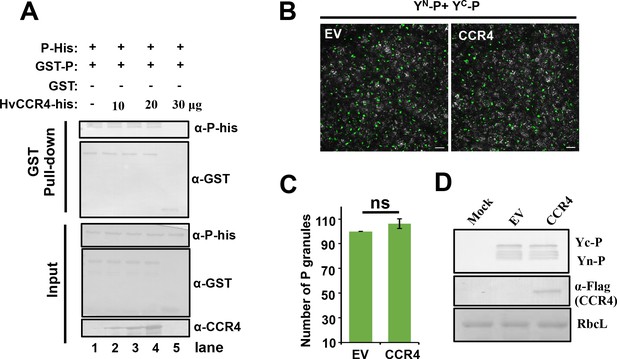
CCR4 does not affect self-interaction of BYSMV P in vitro and in vivo.
(A) Competitive GST pull-down in vitro. GST-P and P-his were pulled down by glutathione sepharose beads incubated with increasing concentrations of HvCCR4-His. Input and GST pull down proteins were analyzed with anti-P, anti-CCR4 and anti-GST, respectively. (B) Competitive BiFC assays. N. benthamiana leaves were infiltrated with Agrobacterium strains expressing YN-P and YC-P with overexpression HvCCR4, empty vector as negative control. (C) Number of P granules shown in the panel B. (D) Western-blotting analysis showing accumulation of YN-P, YC-P and HvCCR4 shown in the panel B.
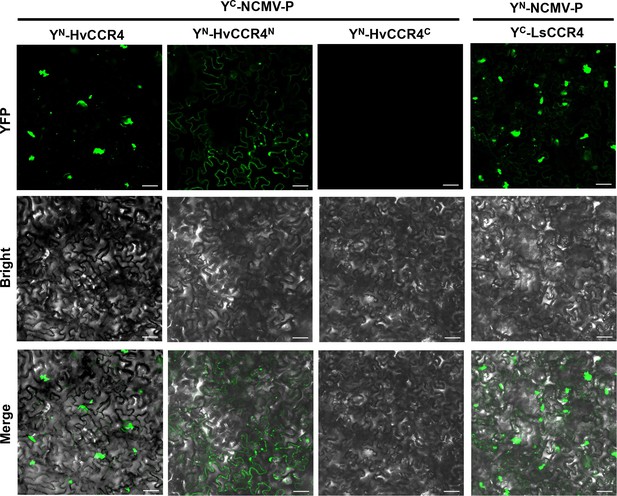
BiFC assays of NCMV-P with HvCCR4 or LsCCR4.
N. benthamiana leaves were infiltrated with Agrobacterium strains expressing YC-NCMV-P and YN-HvCCR4, YN-N-HvCCR4, YN-C-HvCCR4 or YN-NCMV-P and YC-LsCCR4. YFP fluorescence was photographed by microscope at two dpi. Bar = 50 µm.
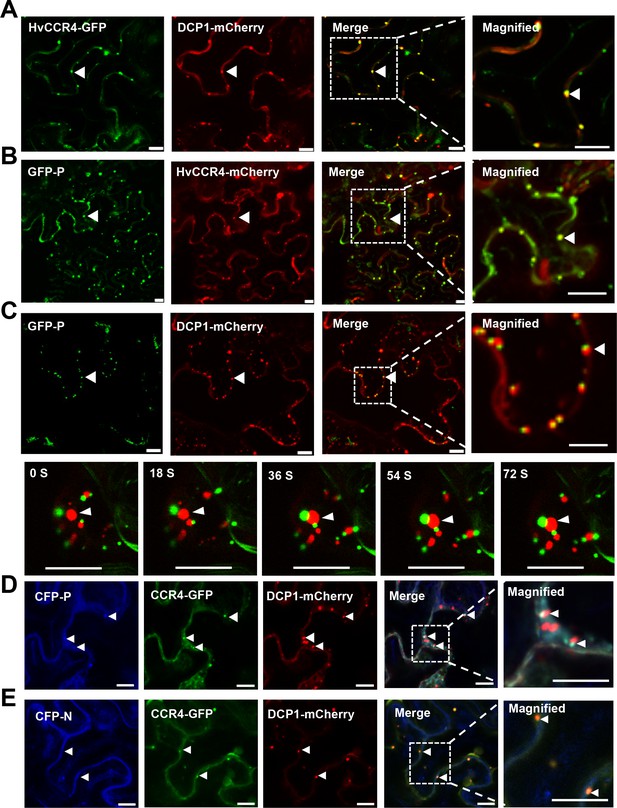
BYSMV P sequestration of HvCCR4 from the PBs.
(A–C) Confocal micrographs showing subcellular co-localization of HvCCR4-GFP and DCP1-mCherry (A), GFP-P and HvCCR4-mCherry (B), GFP-P and DCP1-mCherry (C) co-expressed in N. benthamiana epidermal cells. In the bottom panel (C), time-lapse confocal images of GFP-P and DCP1-mCherry. Arrow heads indicate association of different bodies. Magnified images (in white boxes) are shown in the right panels. Bar = 10 μm. (D and E) Transient co-expression of HvCCR4-GFP and DCP1-mCherry with CFP-P (D) or CFP-N (E) were monitored at three dpi. Bar = 10 μm.
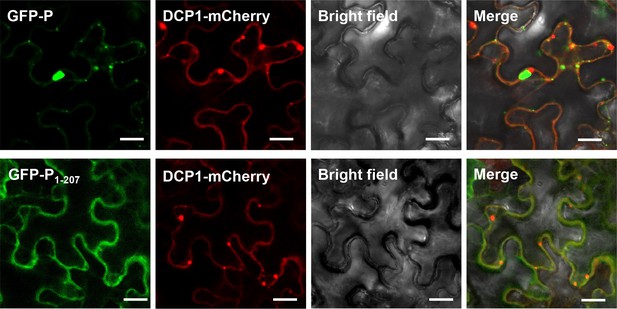
Co-localization of GFP-P or GFP-P1-207 with DCP1-mCherry.
N. benthamiana leaves were infiltrated with Agrobacterium strains expressing DCP1-mCherry with GFP-P or GFP-P1-207 were photographed by microscope at two dpi. Bar = 50 µm.
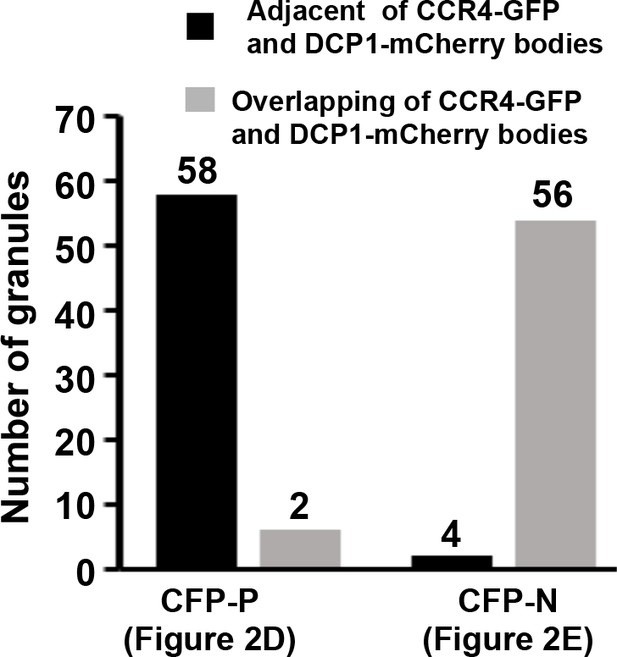
Quantitative data showing localization of CCR4-GFP and DCP1-mCherry bodies in the absence and presence of P protein.
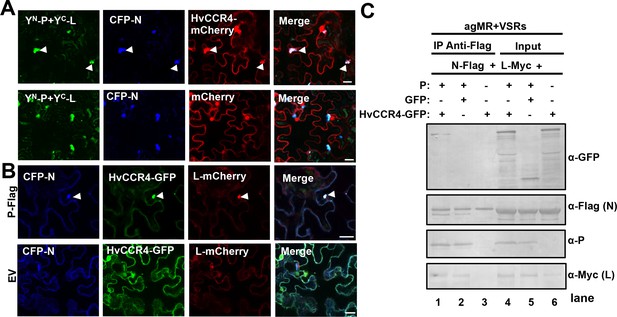
BYSMV P recruitment of HvCCR4 into viroplasm-like bodies.
(A) Confocal micrographs showing subcellular localization of the YN-P and YC-L BiFC combination and CFP-N together with HvCCR4-mCherry or free mCherry in N. benthamiana leaves. Bar = 20 μm. (B) Confocal micrographs showing subcellular localization of CFP-N, HvCCR4-GFP, and L-mCherry in the presence of P-Flag or empty vector (EV). Bar = 20 μm. Representative images in panel A and B were taken at three dpi. Arrowheads indicated viroplasm-like bodies. (C) Co-IP analysis of the HvCCR4-GFP association with N-Flag, P, and L-Myc in vivo. N. benthamiana leaves were agroinfiltrated with A. tumefaciens cells expressing various proteins as indicated. At three dpi, leaf extracts were incubated with anti-Flag beads, and then the IP and input proteins were analyzed by western blotting with anti-GFP, -Flag, -P, and -Myc antibodies, respectively.
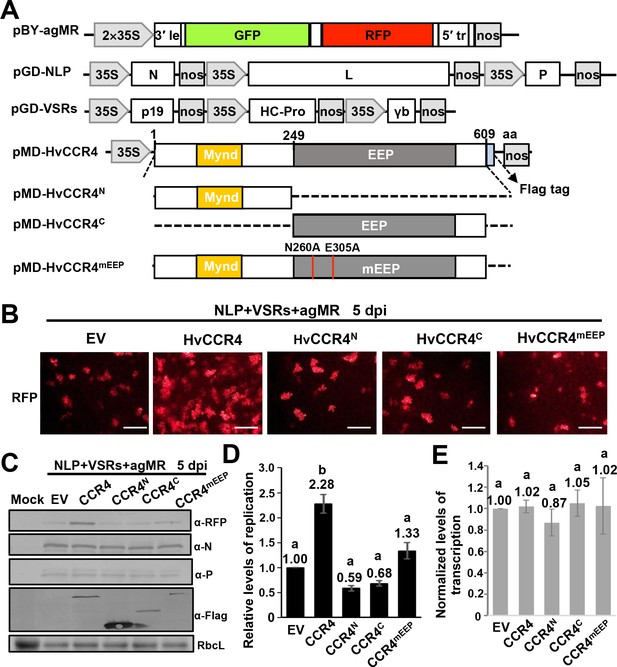
HvCCR4 enhancement of BYSMV minigenome replication.
(A) Schematic diagrams of pBY-agMR, pGD-NLP, pGD-VSRs, pMD-HvCCR4, and derivative plasmids. Mixtures of these plasmids transiently express anti-genomic minireplicon RNA, BYSMV N, P, and L proteins, as well as suppressors of RNA silencing (VSRs) in N. benthamiana leaves. The HvCCR4, HvCCR4N, HvCCR4C and HvCCR4mEEP (N260A/E305A) ORFs were cloned into pMDC32 for transient overexpression. (B) RFP foci in N. benthamiana leaves agroinfiltrated with Agrobacterium mixture harboring BYSMV-agMR combinations containing an empty vector (EV), HvCCR4, HvCCR4N, HvCCR4C, or HvCCR4mEEP. Representative images were taken at five dpi. Bar = 1 mm. (C) Western blotting analysis showing accumulation of RFP, N, P, and HvCCR4 proteins in the leaves shown in panel (B) with rabbit α-RFP, α-N, α-P, or α-Flag protein antibodies, respectively. N. benthamiana leaves infiltrated with the pGD vector were mock controls. (D) qRT-PCR analysis of minigenome RNA replication in the samples shown in panel (B). (E) qRT-PCR analysis of normalized levels of RFP transcription by comparing the relative levels of mRNA versus minigenome RNA in the samples shown in panel (B). EF1A served as an internal control gene. The values of viral replication and transcription in leaf samples agroinfiltrated with the EV plasmid were set to 1. Error bars indicate standard errors of three independent experiments. Letters above the bars indicate statistical significance (p<0.01) evaluated by Turkey’s Multiple Comparison Test analysis.
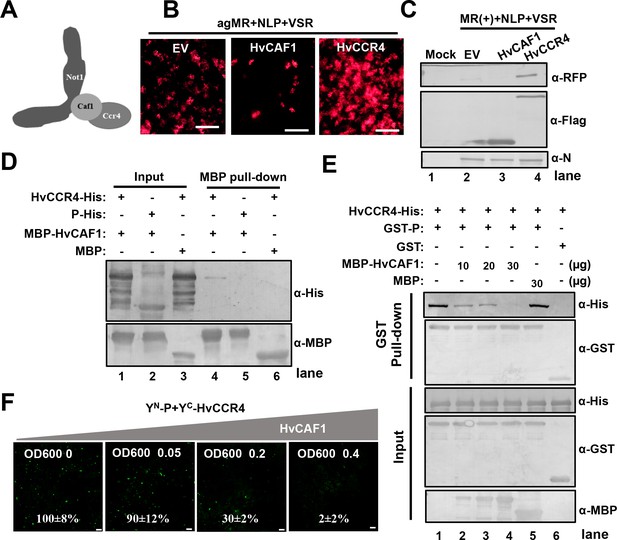
HvCAF1 negatively regulates minigenome RNA replication.
(A) Schematic representation of CAF1 linking CCR4 and the Not1 scaffold in the CCR4-NOT complex. (B) RFP foci in N. benthamiana leaves agroinfiltrated with Agrobacterium mixture harboring BYSMV-agMR combinations or the empty vector (EV), HvCAF1, or HvCCR4. Representative images were taken at five dpi. Bar = 1 mm. (C) Western blotting analysis showing accumulation of RFP, HvCAF1, and HvCCR4 proteins in the leaves shown in panel (B) with rabbit antibodies against RFP and Flag proteins, respectively. (D) MBP pull-down assays showing the in vitro interaction of HvCAF1 with HvCCR4, but not with BYSMV P. CCR4-His or P-His were incubated with MBP-HvCAF1 or MBP with anti-MBP beads. Pull down or input proteins were detected by western blotting with anti-His or anti-MBP antibodies. (E) Competitive GST pull-down assays in vitro. GST-P and HvCCR4-His were pull-downed with glutathione-Sepharose beads incubated with increasing concentrations (10, 20, and 30 μg) of MBP-HvCAF1 or free MBP (30 μg). Input and GST pull down proteins were analyzed with anti-His, -GST, or -MBP antibodies. Free GST and HvCCR4-His served as negative controls. (F) Competitive BiFC assays in vivo. N. benthamiana leaves were infiltrated with Agrobacterium strains expressing YN-P and YC-HvCCR4 with increasing concentrations (OD600 0, 0.05, 0.2, 0.4) of Agrobacterium expressing HvCAF1. The bottom values represent relative granule numbers of different treatments. Values in leaf samples agroinfiltrated with HvCAF1 (OD600 0) were set to 100%. Error bars indicate standard errors of three independent experiments. Images were taken at three dpi by a Leica laser scanning microscope. Bar = 50 μm.
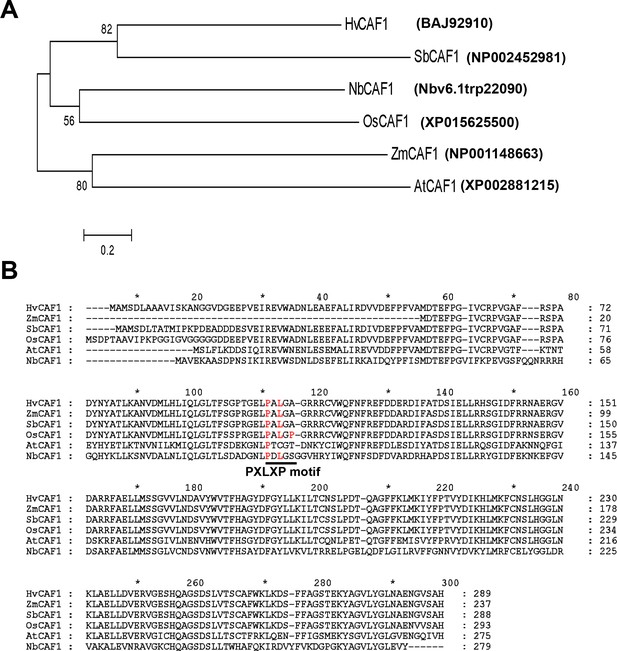
Phylogenetic tree (A) and sequence alignments (B) of plant CAF1 orthologues.
Plant CCR4 orthologues from sorghum (Sb), rice (Os), maize (Zm), barley (Hv), N. benthamiana (Nb) and Arabidopsis (At). The phylogenetic tree was constructed by mega 5.0. Accession numbers of genes were listed. The conserved PXLXP motifs are indicated by a solid line beneath the motifs.
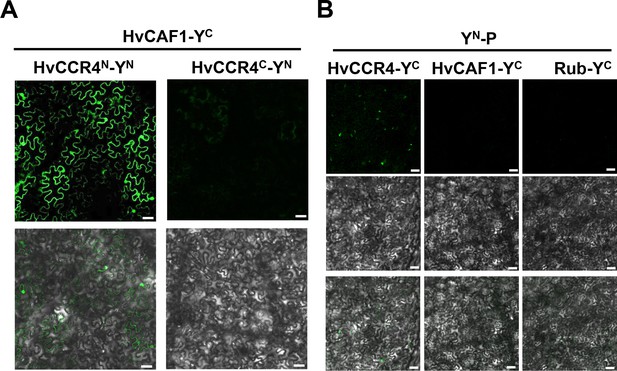
BiFC assays examining interactions between the P, HvCCR4 and HvCAF1 proteins.
(A) BiFC analysis of interactions between HvCAF1-YC and HvCCR4N-YN or CCR4C-YN in coinfiltrated N.benthamiana leaves. Bar = 50 μm. (B) BiFC analysis of the interactions between YN-P and HvCCR4-YC, HvCAF1-YC or Rub-YC in co-infiltrated N. benthamiana leaves. Bar = 50 μm.
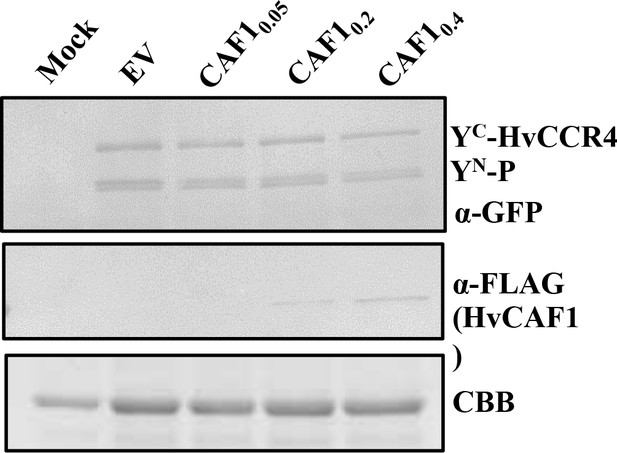
Western blotting analysis of accumulation of expressed proteins in competitive BiFC assays.
The accumulation of YN-P and YC-HvCCR4 with anti-GFP, HvCAF1-Flag with anti-Flag, CBB represents the quantify of protein.
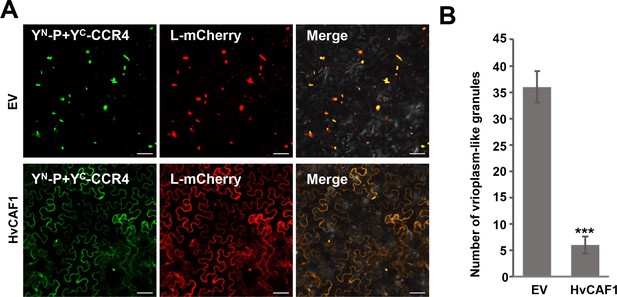
HvCAF1 negatively affect viroplasm-like structure formation.
(A) Co-localization of YN-P and YC-CCR4 with L-mCherry. N. benthamiana leaves were infiltrated with Agrobacterium strains expressing YN-P, YC-CCR4 and L-mCherry with empty vector (EV) or HvCAF1. YFP and mCherry fluorescences were photographed by microscope after two dpi. Bar = 50 µm. (B) Number of viroplasm-like granules when overexpression CAF1 and empty vector in the same area. Error bars indicate the standard error of the mean values of three independent experiments. Statistical significance was determined by Student’s t test (*** p-value<0.001).
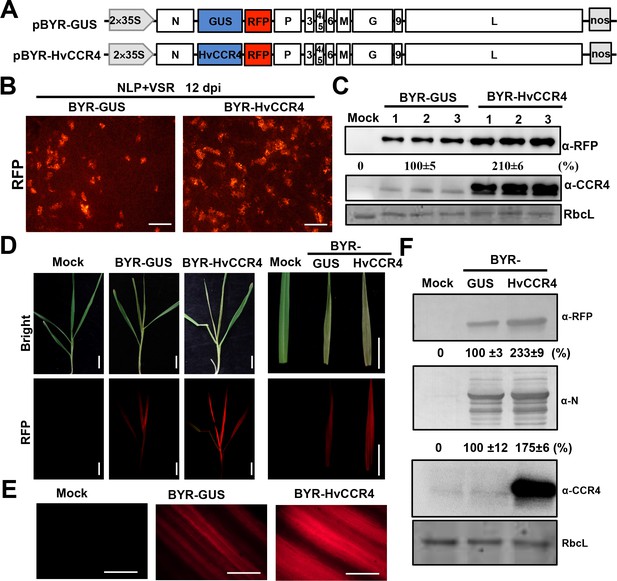
BYSMV-mediated HvCCR4 overexpression enhances virus pathogenesis in barley plants.
(A) Schematic diagrams of BYSMV pBYR-GUS and pBYR-HvCCR4 derivatives harboring GUS and HvCCR4 ORFs. (B) RFP foci in N. benthamiana leaves infiltrated with Agrobacterium strains containing pGD-NLP, pGD-VSRs, pBYR-GUS or pBYR-HvCCR4 plasmids. RFP fluorescence was photographed after 12 dpi with a fluorescence microscope. Bar = 1 mm. (C) Western blotting analysis showing accumulation of RFP and CCR4 with anti-RFP and -CCR4 antibodies. Relative accumulation of RFP from three repetitions are shown at the bottom of RFP panels. (D) Disease symptoms and RFP fluorescence of barley plants infected with BYR-GUS or BYR-HvCCR4 at 15 dpi. Bar = 1 cm. (E) RFP fluorescence of systemically infected barley leaves with BYR-GUS or BYR-HvCCR4 at 15 dpi. Bar = 1 mm. (F) Western blotting analysis showing accumulation of RFP, N, and HvCCR4 proteins in the leaves shown in panel D. The mean relative values of three experiments were shown under the results. The mean values in BYR-GUS infected samples were set as 100%.
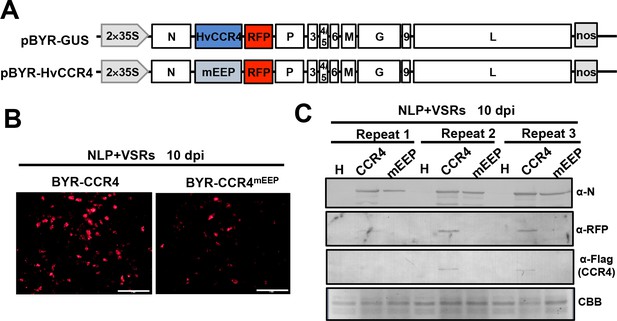
HvCCR4mEEP overexpression did not enhances virus pathogenesis in N. benthamiana leaves.
(A) Schematic diagrams of BYSMV pBYR-GUS and pBYR-HvCCR4 derivatives harboring GUS and HvCCR4 ORFs. (B) RFP foci in N. benthamiana leaves infiltrated with Agrobacterium strains containing pGD-NPL, pGD-VSRs, with pBYR-HvCCR4 or pBYR-HvCCR4mEEP plasmids. RFP fluorescence was photographed after 10 dpi with fluorescence microscope. Bar = 1 mm. (C) Western blotting analyses showing accumulation of N, RFP, Flag-HvCCR4 and Flag-HvCCR4mEEP with anti-N, anti-RFP, and anti-Flag, respectively.
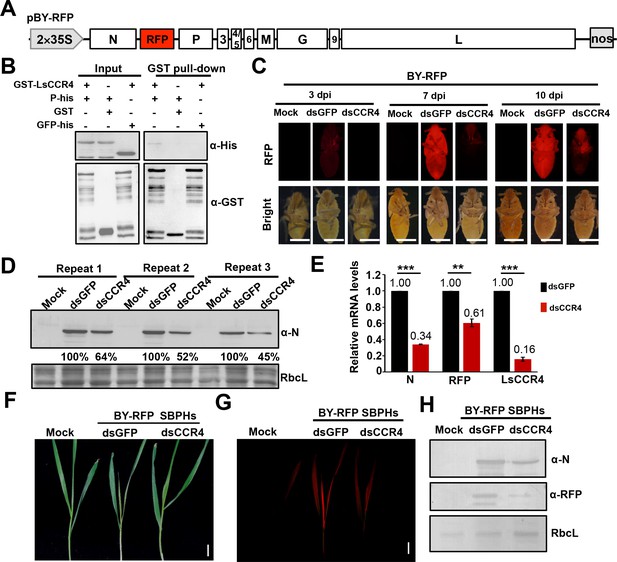
Requirement of the planthopper LsCCR4 protein for efficient BYSMV infection in SBPHs.
(A) Schematic diagrams of the BYSMV pBY-RFP derivative containing an RFP gene between the N and P genes of the antigenome cDNA. (B) GST pull-down analysis of interactions between BYSMV P and LsCCR4. GST and GFP-His served as negative controls. (C) RFP fluorescence of SBPHs microinjected with crude extracts of BY-RFP-infected barley leaves and mock buffer, dsGFP, or dsCCR4. At 3-, 7-, and 10- dpi, the insects were photographed with a fluorescence microscope. Representative confocal images from 30 insects are shown. Bar = 1 mm. (D) Western blotting analysis of BYSMV N protein accumulation in the samples shown at 10 dpi in panel (C) with the anti-N polyclonal antibody. Protein accumulation in the dsGFP-injected samples was set to 100%. Three independent repeats from 30 insects were shown. (E) qRT-PCR analysis of N, RFP and CCR4 mRNA accumulation in the samples shown in panel (C) at 10 dpi. EF1A acted as an internal control gene and the values in the dsGFP-injected insects were set to 1. Error bars indicate the standard error of the mean values of three independent experiments. Statistical significance was determined by Student’s t test (** p-value<0.01; *** p-value<0.001). (F) Symptoms in systemically infected barley leaves at 15 days after feeding with BY-RFP infected SBPHs treated with dsGFP or dsCCR4. Bar = 2.0 cm. (G) RFP fluorescence in the barley leaves shown in panel (F). Bar = 2.0 cm. (H) Western blotting analysis of accumulation of BYSMV N and RFP in the samples shown in panel (F). Rubisco complex large subunit (RbcL) was detected with Stain-Free technology as equal loading controls.
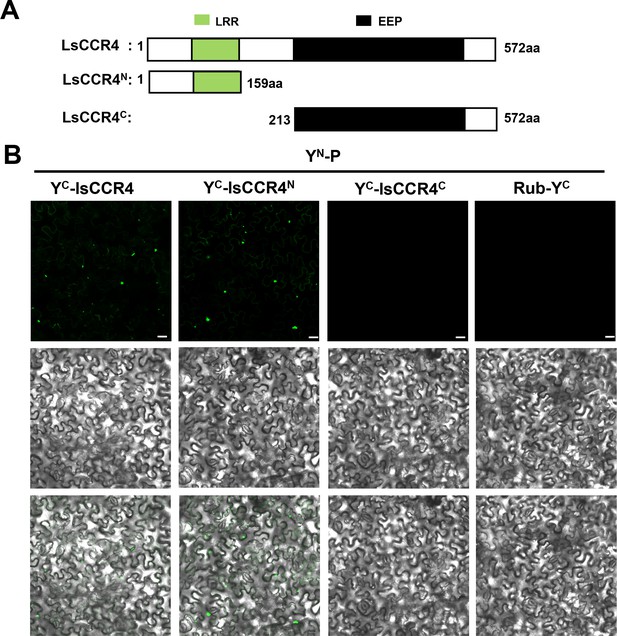
BiFC assays examining interactions between BYSMV P and LsCCR4.
(A) Schematic diagrams of LsCCR4 and different domains for BiFC assays. (B) BiFC analysis of the interactions of YN-P with LsCCR4-YC, LsCCR4N-YC, LsCCR4C-YC, or Rub-YC in coinfiltrated N. benthamiana leaves at three dpi. Bar = 20 μm.
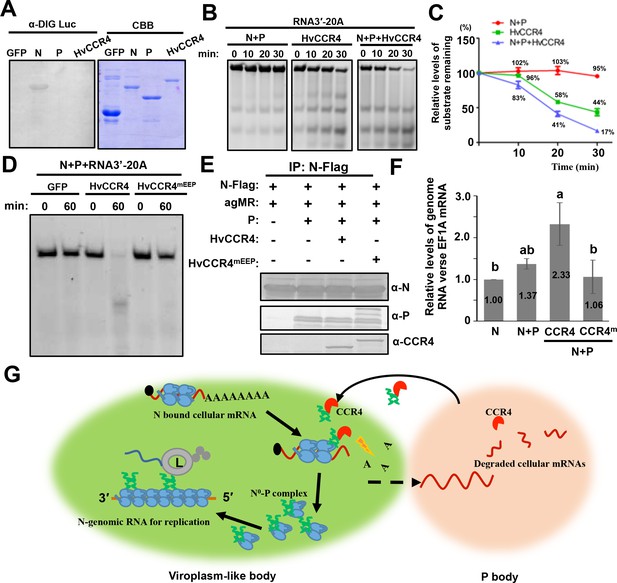
CCR4 facilitates binding of BYSMV N with viral genome RNAs by triggering N-bound cellular mRNA decay.
(A) Northwestern blotting assays detecting non-specific RNA-binding abilities of HvCCR4, BYSMV N, P protein by digoxigenin-labeled Luciferase mRNA (See Materials and methods for details). (B) BYSMV N and P in vitro facilitation of deadenylase activity of purified HvCCR4 protein. The 5'-fluorescein isothiocyanate-labeled RNA substrate (RNA 3′−20A) was incubated with different protein combinations as indicated. At different time after incubation, the RNA substrate was analyzed by denaturing PAGE. Relative accumulation of undegraded RNA substrate was examined by Image J. The 0 min after incubation with proteins was set as one unit. (C) Degradation of the RNA 3′−20A substrate. The graphs plot the relative accumulation of remaining substrates estimated by electronic autoradiography in the gels of panel D. Data points represent mean values from three independent repeats. Error bars indicate Error bars indicate standard errors of three independent experiments. The curve diagrams were drawn with GraphPad Prism. (D) The N260 and E305 residues of HvCCR4 are required for the deadenylase activity of purified HvCCR4 protein in vitro. (E) and (F) HvCCR4 increases the binding specificity of the N protein to minigenome RNA. N. benthamiana leaves were infiltrated with Agrobacterium strains for expression of agMR, N-Flag, and other protein combinations as indicated. At three dpi, the N-Flag protein was immunoprecipitated from the infiltrated leaves with the anti-Flag M2 affinity gel. Western-blot analysis of N, P and CCR4 protein accumulation in the IP products (D). Analysis of N-Flag bound RNA from IP products by qRT-PCR to determine the ratios of N bound gRNA and host EF1A mRNA (E). Numbers above the graph show the mean values of three independent experiments. Statistical significance (p<0.05) was evaluated by two-way ANOVA with Tukey’s test. (G) CCR4-mediated mRNA decay model for optimal BYSMV replication. Host CCR4 proteins hijacked by the BYSMV phosphoprotein trigger degradation of N-bound cellular mRNAs to release RNA-free N protein for enhanced encapsidation and replication of viral genomic RNA.
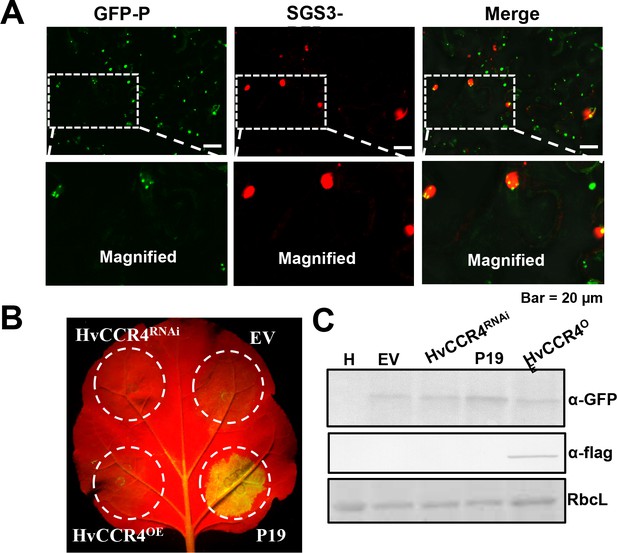
HvCCR4 did not exhibit obvious activities involved in RNAi.
(A) Confocal micrographs showing subcellular co-localization of GFP-P and SGS3-RFP. (B) Transgene GFP-induced silencing co-expressed with HvCCR4OE and HvCCR4RNAi, TBSV P19 suppressor, and empty vector. were photographed by microscope at three dpi. (C) Western blotting analysis of GFP and Flag-CCR4 with anti-GFP and anti-flag.
Videos
Motile cytoplasmic BiFC bodies of YN-P and YC-CCR4 in the epidermal cells of agroinfiltrated N. benthamiana leaves.
Images were taken at three dpi with a Leica laser scanning microscope. Bar = 50 μm.
Intracellular movement of YN-P and YC-CCR4 inclusion bodies in epidermal cells of agroinfiltrated N. benthamiana leaves expressing mCherry-HDEL after DMSO treatment.
Scale bar = 2 μm.
Intracellular movement of YN-P and YC-CCR4 inclusion bodies in epidermal cells of agroinfiltrated N. benthamiana leaves expressing mCherry-HDEL with LatB (10 μM) treatment.
Scale bar = 2 μm.
High mobility of GFP-P and CCR4-mCherry colocalized bodies in epidermal cells of agroinfiltrated N. benthamiana leaves.
Scale bar = 20 μm.
High mobility of GFP-P bodies and DCP-mCherry bodies in epidermal cells of agroinfiltrated N. benthamiana leaves.
Scale bar = 2 μm.
A representative movie showing CFP-N, L-mCherry, and CCR4-GFP in epidermal cells of agroinfiltrated N. benthamiana leaves expressing BYSMV P.
Scale bar = 20 μm.
Additional files
-
Supplementary file 1
List of the BYSMV P protein interacting barley proteins obtained in IP-MS assays.
- https://cdn.elifesciences.org/articles/53753/elife-53753-supp1-v2.docx
-
Supplementary file 2
Primers used in this study.
- https://cdn.elifesciences.org/articles/53753/elife-53753-supp2-v2.docx
-
Supplementary file 3
Key Resources Table.
- https://cdn.elifesciences.org/articles/53753/elife-53753-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53753/elife-53753-transrepform-v2.pdf





