A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila
Figures

Conditional CRISPR mutagenesis with pCFD6 is robust across target genes and tissues.
(A) Schematic overview of the workflow. To perform tissue-specific targeted mutagenesis flies transgenic for a specific Gal4 driver (X-Gal4) and UAS-Cas9 are crossed to flies with a UAS-sgRNA transgene. Offspring from this cross express Cas9 and sgRNAs in Gal4 expressing cells, leading to mutagenesis of the target gene. (B) Schematic of gene editing outcomes typically observed with a single, ubiquitous sgRNA (lower left) or a conditional array of several sgRNAs (lower right). Leaky expression, that is expression in the absence of Gal4, from conditional Cas9 transgenes gives rise to ectopic mutagenesis in combination with ubiquitous, but not conditional, sgRNAs. Gene editing in tissues typically results in genetic mosaics, which can be enriched for bi-allelic knock-out cells through sgRNA multiplexing. (C) Conditional CRISPR mutagenesis in wing imaginal discs with nub-Gal4 in the wing pouch. Gene editing with pCFD6-arm2x results in loss of Arm protein exclusively in the Gal4 expression domain in nearly all cells. Control animals express the nub-Gal4 driver and UAS-cas9.P2. Scale bar = 50 µm. (D) Conditional CRISPR mutagenesis of Notch in intestinal stem cells drives tumor formation in the midgut. esgts (esg-Gal4 tub-Gal80ts) was used to repress expression of UAS-cas9.P2 and pCFD6-N2x until adult stages. Mutagenesis was induced for 5 days at 29°C and flies were returned to 18°C to avoid Cas9.P2 mediated toxicity. Posterior midguts 15 days after induction of mutagenesis are shown. esgts UAS-cas9.P2 pCFD6-N2x tissue shows an accumulation of stem cells (DNA marked in cyan) and an increase in mitotic cells (pHistone3 in magenta). Quantification of phenotypes are shown in Figure 1—figure supplement 2. Control genotype is esgts UAS-cas9.P2 pCFD6-se2x. Scale bar = 50 µm. (E) Mutagenesis of neur in pnr-Gal4 UAS-cas9.P2 pCFD6-neur2x animals results in loss of thoracic bristles along the dorsal midline, where pnr-Gal4 is expressed. Note the tissue patch that retains bristles, reflecting mosaic mutagenesis. (F) Mutagenesis of the pigmentation gene se in the eye. GMR-Gal4 UAS-casp.P2 pCFD6-se2x animals develop a uniform dark eye coloration. Control animals in (E) and (F) express the respective Gal4 driver and UAS-cas9.P2 pCFD6-Sfp24C12x. (G) pCFD6 mediated mutagenesis in the germline. Shown is a summary of the mutational status at each sgRNA target site in individual F1 flies. nos-Gal4VP16 UAS-cas9.P1 pCFD6 flies expressing sgRNAs targeting the indicated essential genes are viable, demonstrating germline restricted mutagenesis, and transmit mutant alleles to their offspring. Shown is a summary of the mutational status at each sgRNA target site in individual flies. All lines, except the one targeting Dpp (asterisk), transmit mutant alleles to the majority of offspring. Flies expressing sgRNAs targeting Dpp in the germline produce few viable offspring and transmitted only a single, in-frame, mutation out of 11 analysed offspring. The same sgRNA construct results in highly efficient mutagenesis in somatic tissues (see Figure 4), consistent with haploinsufficiency of Dpp in the Drosophila embryo.
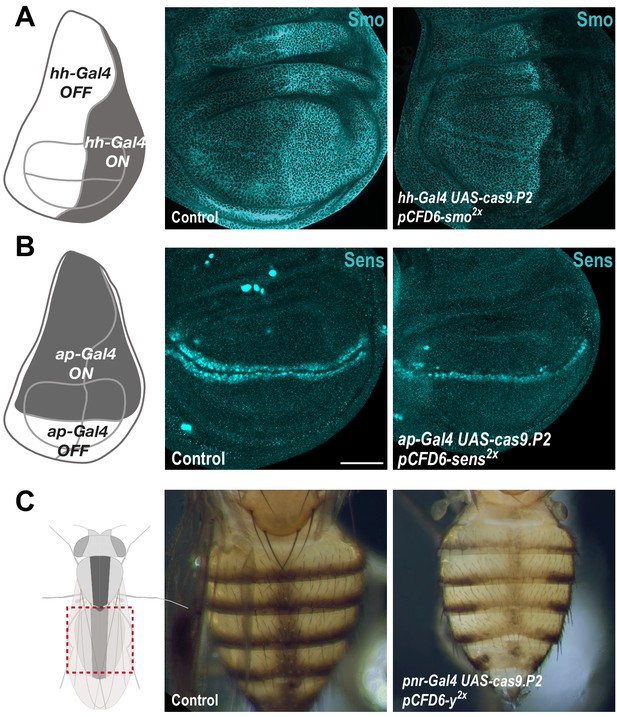
Efficient conditional CRISPR mutagenesis in various Drosophila tissues.
(A) CRISPR mutagenesis of smo in the posterior compartment of the wing imaginal disc. Smo protein was detected by immunohistochemistry. Smo is normally expressed in all wing disc cells, but protein levels are higher in the posterior compartment (see Control (hh-Gal4 UAS-cas9.P2)). In hh-Gal4 UAS-cas9.P2 pCFD6-smo2x wing disc cells in the posterior compartment express no or reduced levels of Smo, presumably reflecting cells containing only one or no functional smo alleles. (B) CRISPR mutagenesis of sens in the dorsal compartment of wing imaginal discs with ap-Gal4 leads to a loss of Sens expression in most, but not all cells. (C) Mutagenesis of y in the dorsal abdomen. In pnr-Gal4 UAS-cas9.P2 pCFD6-y2x animals, cuticle coloration is uniformly changed in a broad stripe centred around the dorsal midline, compared to control animals (pnr-Gal4 UAS-cas9.P2 pCFD6-Sfp24C12x). Note that the strong phenotype mediated by pCFD6-y2x is in line with the high levels of mutagenesis with this construct reported in Figure 3—figure supplement 1B.
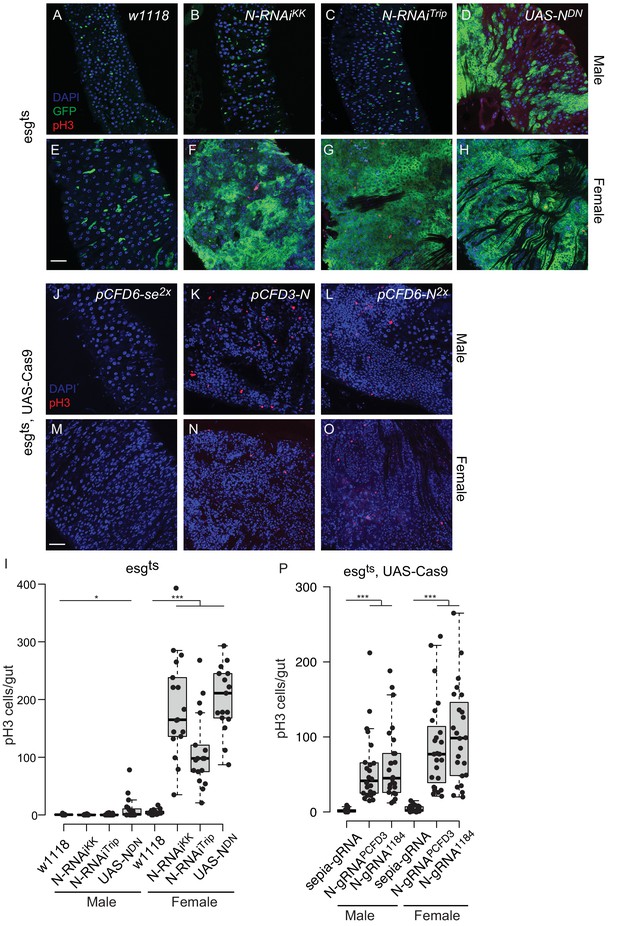
Qualitative differences between CRISPR mutagenesis and RNAi knock-down of Notch in the Drosophila midgut.
(A–H) Representative images of the posterior midgut of male or female esgts > w1118 (A, E), esgts > N RNAiKK (B, F), esgts > N RNAiTrip (C, G) and esgts > UAS NDN animals, stained with pH3 antibody in red to mark mitotic cells. esg-GFP is shown in green, nuclei are stained with DAPI (blue). Animals were raised at 18°C to adulthood and then incubated at 29°C for 15 days. (I) Quantification of pH3-positive cells per adult midgut of the indicated genotypes after 15 days at 29°C. (J–O) Representative images of the posterior midgut of male or female esgts UAS-Cas9.P2 pCFD6-se2x (J, M), esgts UAS-Cas9.P2 pCFD3-N (K, N) and esgts UAS-Cas9.P2 pCFD6-N2x (L, O) flies stained with pH3 antibody in red and nuclei are stained with DAPI (blue). Animals were raised at 18°C, after eclosion mutagenesis was induced for 5 days at 29°C, followed by 18°C for 15 days. Unlike in the RNAi condition, no GFP is visible due to repression by Gal80 at 18°C. (P) Quantification of pH3-positive cells per adult midgut of the indicated genotypes at 18°C for 30 days after inducing CRISPR/Cas9 mediated mutagenesis. Significant differences in the number of pH3-positive cells between Notch RNAi or knock-out group (esgts > N RNAiKK, esgts > N RNAitrip, esgts;UAS-Cas9 >N gRNA PCFD3 or esgts;UAS-Cas9 >N-gRNA1184) and the control groups. (esgts > w1118 or esgts;UAS-Cas9 >sepia gRNA) are indicated with asterisks (*p<0.05; **p<0.01; ***p<0.001; two-tailed T-test) Scale bars: 30 µm (A–H, J–O). Note that the total number of mitotic cells in the RNAi and CRISPR conditions cannot be directly compared, as animals were raised at different temperatures.

A transgenic series for tunable Cas9 expression to balance activity and toxicity.
(A) Principle of the UAS-uCas9 series. Translation of the downstream ORF is inversely correlated with length of the upstream ORF in bicistronic mRNAs. The UAS-uCas9 series consists of transgenes that harbor uORFs of different length to modulate expression of Cas9. (B - D) Systematic characterization of Cas9 expression, toxicity and mutagenesis efficiency of the UAS-uCas9 series. Transgenes of the UAS-uCas9 series were recombined with nub-Gal4 and crossed to the apoptosis sensor UAS-GC3Ai (B, C) or pCFD6-evi2x (D). Graphs show data as individual dots, and boxplots as a data summary, with the line representing the median and the box the interquartile range. (B) Quantification of anti-Cas9 staining intensity in wing discs of the indicated genotype. Cas9 levels gradually reduce as the size of the uORF increases. N ≥ 6 wing discs. (C) Elevated levels of apoptosis were only observed with UAS-uXSCas9. The longest uORF (uXXL) encodes EGFP, preventing visualization of dying cells with GC3Ai. Quantification of fluorescent intensity of the GC3Ai reporter in the wing pouch. N ≥ 14 wing discs. (D) All transgenes of the UAS-uCas9 series mediate evi mutagenesis, with transgenes containing the four shortest uORFs (XS-L) leading to comparable gene editing that removes Evi from nearly all cells in the Gal4 expression domain. Quantification of staining intensity for Evi protein in the wing pouch (Gal4 on), relative to Evi staining in the hinge region (Gal4 off). N ≥ 6 wing discs. (E, E’) CRISPR mutagenesis patterns reflect Gal4 expression history. (E) Fluorescence of GFP, which turns over, reflects most recent Gal4 expression pattern. (E’) CRISPR mutagenesis, visualized by activation of the CIGAR reporter, is permanent and reveals the Gal4 expression history. Images of a representative wing disc are shown to the left of each panel and average intensity projection of several discs registered to a common template are shown on the right (see Materials and methods). Areas that are CIGAR positive in many discs appear bright, while areas devoid of signal in most discs appear dark. (F, F’) Incomplete repression of CRISPR mutagenesis by temperature-sensitive Gal80. (F) Principle of the Gal80ts system. At 18°C Gal80 binds and inhibits Gal4. (F’) Mutagenesis is still observed at 18°C in 11/24 discs and observed preferentially in the Gal4 expression domain, indicating incomplete Gal4 suppression by Gal80ts. (G - G’’) Control of CRISPR mutagenesis by a flip-out cassette. (G) In the absence of FLP recombinase a FRT-flanked GFP flip-out cassette (FRT sites represented by triangles) separates Cas9 from the promoter, resulting in cells that express GFP, but no Cas9. In the presence of FLP, the GFP cassette is excised and Cas9 is expressed. (G’) Staining for the transcription factor Cut reveals a continuous stripe of cells expressing ct along the dorsal-ventral boundary in wildtype tissue. (G’’) A pulse of FLP expression was used to excise the GFP flip-out cassette in a subset of cells (marked by the absence of GFP). Cut expression (magenta) is exclusively lost in GFP negative cells. Scale bar = 50 µm.

High levels of Cas9 expression from UAS-cas9.
P2 are cytotoxic. (A) Expression of Cas9 in nub-Gal4 UAS-Cas9.P2 causes excessive cell death. Apoptotic cells were visualized by co-expression of GC3Ai, which is fluorescent in cells with activated caspase 3. Expression of GC3Ai in the wing pouch highlights a few cells undergoing apoptosis. Additional expression of UAS-cas9.P2 causes a dramatic increase in the cells undergoing programmed cell death, highlighting cytotoxicity caused by high levels of Cas9. (B) Comparison of Cas9 expression levels between act-cas9 and hh-Gal4 UAS-cas9.P2 wing discs. Cas9 was detected by antibody staining and imaged in the same session with identical setting at either low (top panel) or high (bottom panel) detector gain. The difference in Cas9 levels is such, that Cas9 expressed from act-cas9 is undetectable with low detector gain and Cas9 staining in hh-Gal4 UAS-cas9.P2 wing discs is oversaturated with high detector gain, where Cas9 from act-cas9 is visible. The act-cas9 transgene is known to mediate highly efficient mutagenesis in Drosophila (Port et al., 2014; Port et al., 2015 ). (C) Systematic characterization of Cas9 expression, toxicity and mutagenesis of the UAS-uCas9 series. Transgenes of the UAS-uCas9 series were recombined with nub-Gal4 and wing imaginal discs were stained for Cas9 protein. Cas9 levels gradually reduce as the size of the uORF increases (left panel). nub-Gal4 UAS-uCas9 flies were crossed to UAS-GC3Ai to visualize cells undergoing apoptosis. Elevated levels of apoptosis were only observed with UAS-uXSCas9. The longest uORF (uXXL) encodes EGFP, preventing visualization of dying cells with GC3Ai (middle panel). nub-Gal4 UAS-uCas9 flies were crossed to pCFD6-evi2x and mutagenesis of evi was indirectly observed by loss of Evi staining. All transgenes of the UAS-uCas9 series mediate evi mutagenesis, with transgenes containing the four shortest uORFs (XS-L) leading to comparable gene editing that removes Evi from nearly all cells in the Gal4 expression domain (right panel).
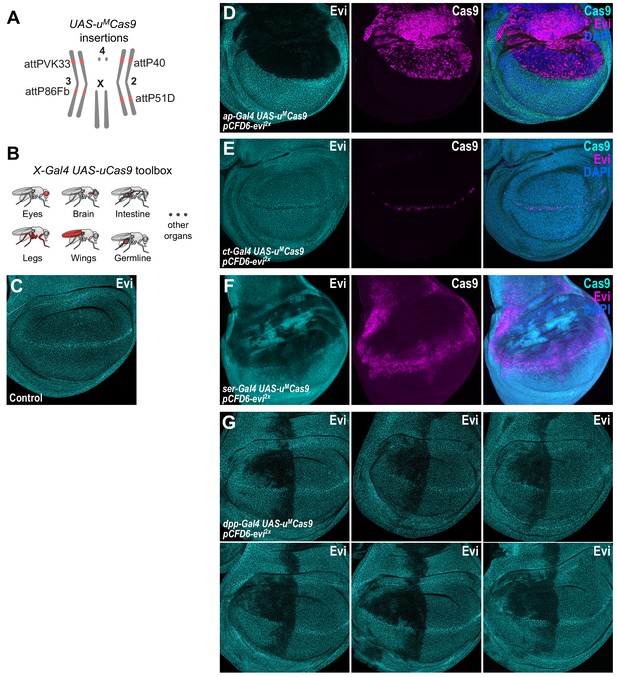
CRISPR mutagenesis patterns reflect expression patterns of Gal4 lines throughout development.
(A) Chromosomal locations of UAS-uMCas9 transgenes currently available. (B) A growing collection of Gal4 UAS-uCas9 stocks will allow tissue-specific mutagenesis in Drosophila. (C) Evi is expressed in all cells of wildtype third-instar wing imaginal discs and accumulates in cells at the dorsal-ventral boundary. (D) Mutagenesis of evi in ap-Gal4 UAS-uMCas9 pCFD6-evi2x wing discs results in loss of Evi protein exclusively in the dorsal compartment, which expresses Cas9 protein. (E) CRISPR mutagenesis of Evi with ct-Gal4 results in loss of Evi staining only in cells along the dorsal-ventral boundary, where also Cas9 protein is expressed. (F) CRISPR mutagenesis with ser-Gal4 results in loss of Evi staining in the pouch, hinge and notum region of the wing disc and includes large areas where no Cas9 protein is detectable in third instar wing discs. (G) Mutagenesis of evi with dpp-Gal4. Dpp is known to be expressed in a stripe along the anterior-posterior boundary in third instar wing imaginal discs. In addition to this domain, mutagenesis is consistently found in the anterior-dorsal region of the pouch. Six representative discs for the same genotype are shown.
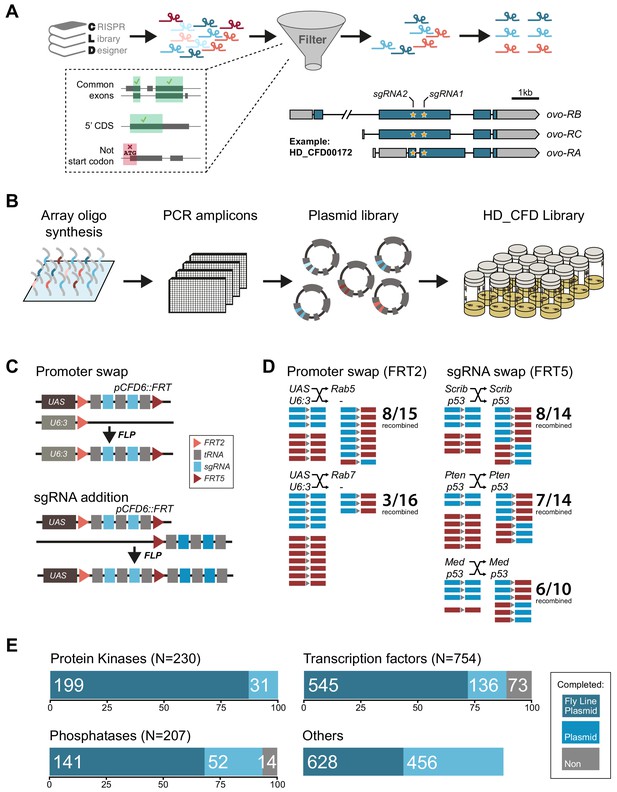
Generation of a large-scale sgRNA library.
(A) Design of the sgRNA pairs used for the HD_CFD library. sgRNAs were designed through CLD and filtered to target common exons in the 5’ORF and not overlap the start codon. sgRNAs were then paired to target two independent positions in the same gene. As an example the locations of the two target sites in ovo targeted by the two sgRNAs encoded in line HD_CFD000172 is shown. Exons are represented as boxes and regions in blue are protein coding. (B) Experimental strategy for the generation of the transgenic sgRNA library. sgRNA target sequences are encoded on oligonucleotides synthesized and cloned in pool. Individual plasmids are sequence verified and transformed into Drosophila at attP40 on the second chromosome following a pooled injection protocol followed by genotyping of individual transformants. (C) Applications of the pCFD6::FRT vector. pCFD6::FRT contains two non-compatible FRT sites either side of the sgRNA cassette. Using compatible FRT sides in trans allows to exchange sequences upstream or downstream of the sgRNAs in vivo. (D) Efficient promoter or sgRNA exchange in vivo. Summary of FLP/FRT mediated exchange of the sgRNA promoter (left) or sgRNAs (right). Each line represents a single sequenced animal. Red and blue boxes either side of the triangle (representing FRT) indicate successful recombination. (E) Summary statistics of the different functional groups present in the sgRNA library. Given is the number of genes from each category that are covered by fly lines, plasmids or against which currently no tools are available. Note that for some genes two fly lines or plasmids exist. Status in September 2019 is shown. Group ‘Others’ contains mainly genes with human orthologs associated with cancer development in humans.
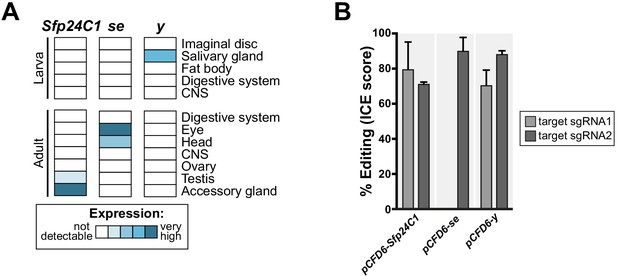
Negative control sgRNA transgenes for use with HD_CFD library.
(A) Sfp24C1, se and y are not expressed in most tissues (upper panel, data from modENCODE), making them suitable as negative controls in most circumstances. (B) sgRNA lines mediate efficient mutagenesis at both (pCFD6-Sfp24C12x, pCFD6-y2x) or one (pCFD6-se2x) target sites.

A large-scale CRISPR screen for essential genes in Drosophila.
(A) The majority of sgRNA lines mediates efficient mutagenesis on both sgRNA target sites. sgRNA transgenes were combined with act-cas9 and tub-Gal4 to induce ubiquitous mutagenesis. Mutagenesis was measured by sequencing PCR amplicons spanning the target sites followed by Inference of CRISPR Edits (ICE) analysis. Shown are mean values of 2–4 independent experiments and the standard error of the mean. (B) CRISPR screening for essential genes in Drosophila. Ubiquitous mutagenesis was induced in offspring of HD_CFD sgRNA lines crossed with act-cas9;;tub-Gal4/TM6B partners. Vials were analysed after 15–17 days (~5–7 days after eclosion) for viable act-cas9;pCFD6-sgRNA2x; tub-Gal4 offspring. Summary statistics are shown on the right. Crosses were scored as semi-lethal when flies of the correct genotype were present, but <50% of the number of TM6B flies, and dead larva, pupae or adults were evident in the vail. (C) False-negative results are rare and often occur for genes controlling early development. Summary statistics for 208 HD_CFD sgRNA lines targeting known essential genes are shown. 23 (11%) lines give rise to the incorrect (viable) phenotype. mRNA expression data for these target genes is shown below (data from modENCODE). Most genes have maternally contributed mRNA, are highly expressed in early embryonic stage or play known roles in embryonic development. (D) Low number of false-positive results caused by HD_CFD sgRNA lines. 54 HD_CFD lines in the screen target genes known to be dispensable for fly development. five lines result in lethality when crossed to act-cas9;;tub-Gal4/TM6B flies. Note that lines HD_CFD795 and HD_CFD1058 target the same gene with independent sgRNAs. (E) Tissue-specific CRISPR mutagenesis in the developing wing. Representative images of adult wing phenotypes caused by CRISPR mutagenesis of Dpp signalling components are shown. All lines give rise to the expected alterations in wing size and vein patterning with varying strength.
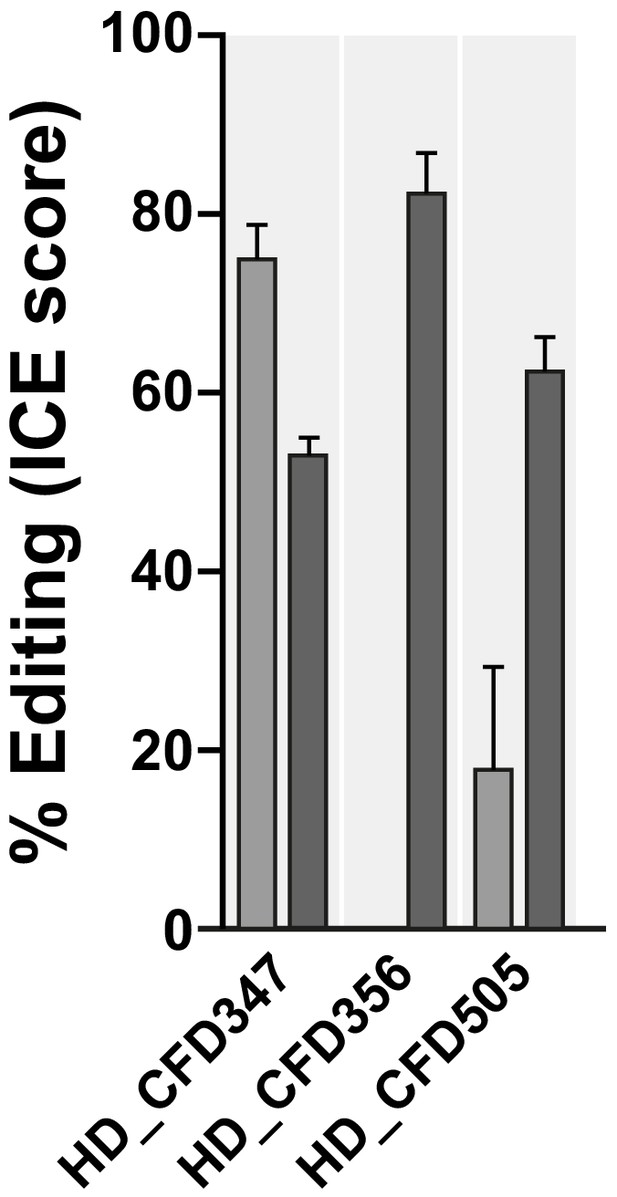
HD_CFD sgRNA line resulting in false-negative results mediate efficient on-target mutagenesis.
Lines HD_CFD347 (Target: hbn), HD_CFD356 (Target: Ets97D) and HD_CFD505 (Target: gcm) give rise to viable offspring when crossed to act-cas9;;tub-Gal4/TM6B. Failure to result in lethality does not reflect inactive sgRNAs. Target locus was amplified by PCR and editing efficiency was measured by Sanger sequencing followed by ICE analysis. Bars represent mean and error bars show standard deviation.
Additional files
-
Supplementary file 1
HD_CFD Drosophila lines 2019.
- https://cdn.elifesciences.org/articles/53865/elife-53865-supp1-v2.xlsx
-
Supplementary file 2
Results of the lethality screen.
- https://cdn.elifesciences.org/articles/53865/elife-53865-supp2-v2.xlsx
-
Supplementary file 3
Uncharacterized essential genes.
- https://cdn.elifesciences.org/articles/53865/elife-53865-supp3-v2.xlsx
-
Supplementary file 4
Oligonucleotide sequences.
- https://cdn.elifesciences.org/articles/53865/elife-53865-supp4-v2.xlsx
-
Supplementary file 5
Drosophila strains used.
- https://cdn.elifesciences.org/articles/53865/elife-53865-supp5-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53865/elife-53865-transrepform-v2.docx





