TMEM95 is a sperm membrane protein essential for mammalian fertilization
Figures
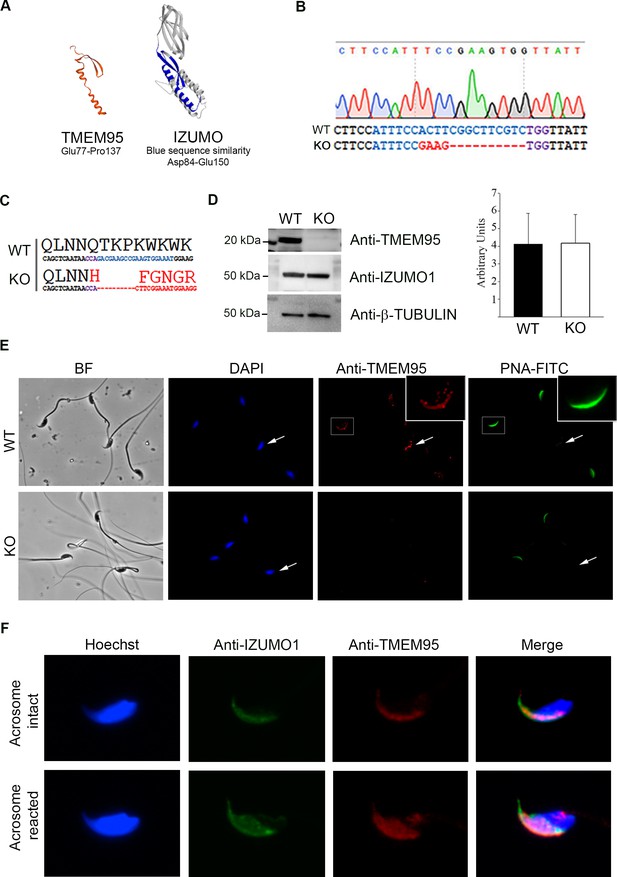
Generation of TMEM95 deficient mice.
(A) Structure prediction of TMEM95 protein (left) using IZUMO1 (right) as template, created by SWISS-MODEL software. (B) Tmem95 KO allele generated following CRISPR-mediated edition. CRISPR target sequence and PAM are depicted in blue and purple letters, respectively. (C) The deletion of 10 bp altered Tmem95 ORF. Large letters indicate the aminoacid sequence corresponding to the codons (DNA sequence) shown in smaller letters below. (D) Western Blot images for TMEM95, IZUMO1 and β-tubulin proteins from protein extracts from WT or KO sperm. Graph on right indicates the abundance of IZUMO1 in WT and KO extracts. (E) Immunocytochemistry images of KO and WT sperm stained with an antibody against TMEM95 and the acrosomal stain PNA. TMEM95 localized to the acrosomal cap in acrosome intact sperm and in the equatorial segment after acrosome reaction. (F) Immunocytochemistry images of acrosome intact (upper images) or reacted (lower images) WT sperm stained against IZUMO1 and TMEM95. Both proteins relocalize to the equatorial segment following acrosome reaction.
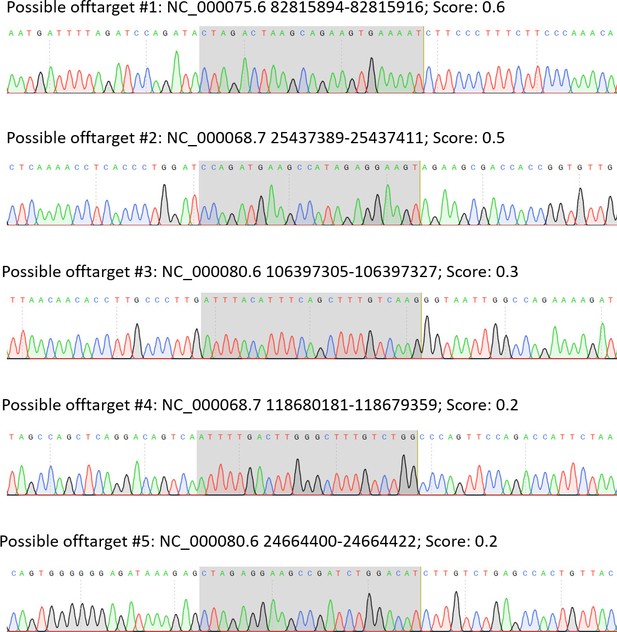
Offtarget analysis.
None of the 5 most probable off-target sites for the sgRNA used were edited, showing the same sequence as the WT. Location of the possible off-target is coloured in gray.
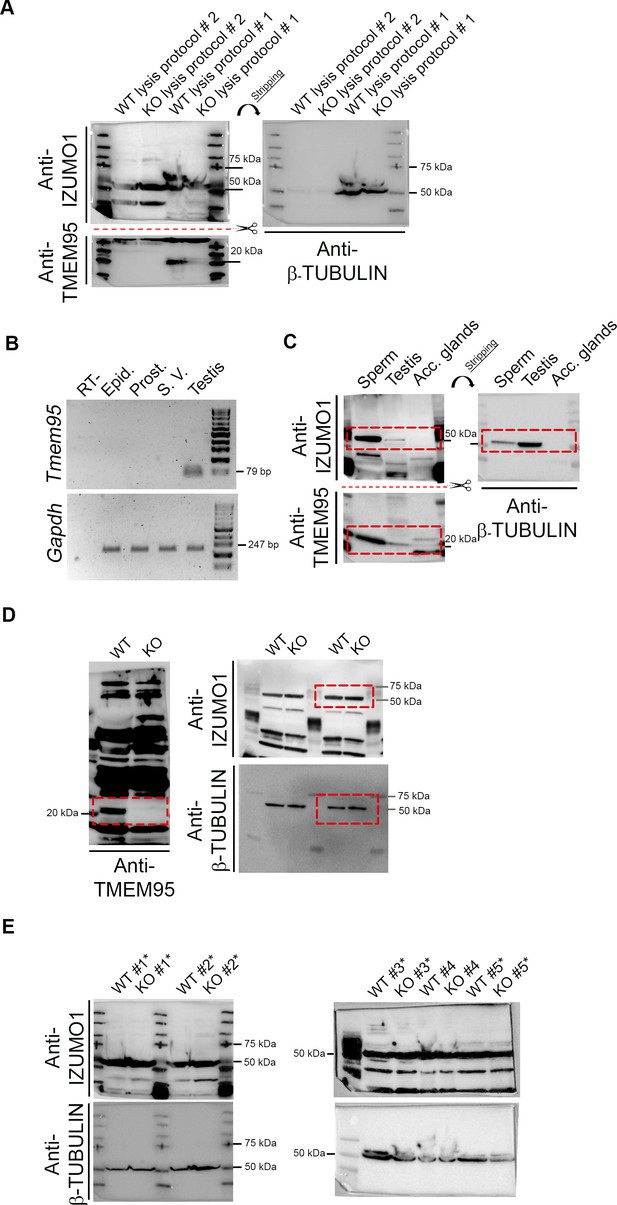
WB images related to Figure 1D.
(A) Immunoblotting of protein extracts obtained from WT and KO epididymal sperm samples following two lysis protocols. In lysis protocol #1, sperm were re-suspended in 4X reducing SDS Sample Buffer and boiled for 10 min. In lysis protocol #2, sperm were re-suspended in 1% Octyl β-D-glucopyranoside solution in PBS and incubated on ice for 30 min (Nishimura et al., 2011). Supernatants were probed with anti-TMEM95 (MyBiosource MBS7004333), anti-IZUMO1 (Abcam ab211623) or anti-β-TUBULIN (Sigma T8328) antibodies. (B) Gel electrophoresis of PCR products amplified from cDNA obtained from testis, seminal vesicle (S.V.) prostate (prost.), epididymis (epid.) or a negative control testis RNA not retrotranscribed (RT-) to detect Gapdh and Tmem95 transcripts. (C) Immunoblotting of protein extracts obtained from WT epididymal sperm, testis or accessory glands (seminal vesicle and prostate). Same antibodies than (A). (D) Uncropped images of the WB used to generate Figure 1D. (E) WB images used for the quantification of IZUMO1 and β-TUBULIN shown in Figure 1D. Four samples were used for quantification (marked with asterisk), as β-TUBULIN band on samples #4 was dispersed, leading to inaccurate quantification.
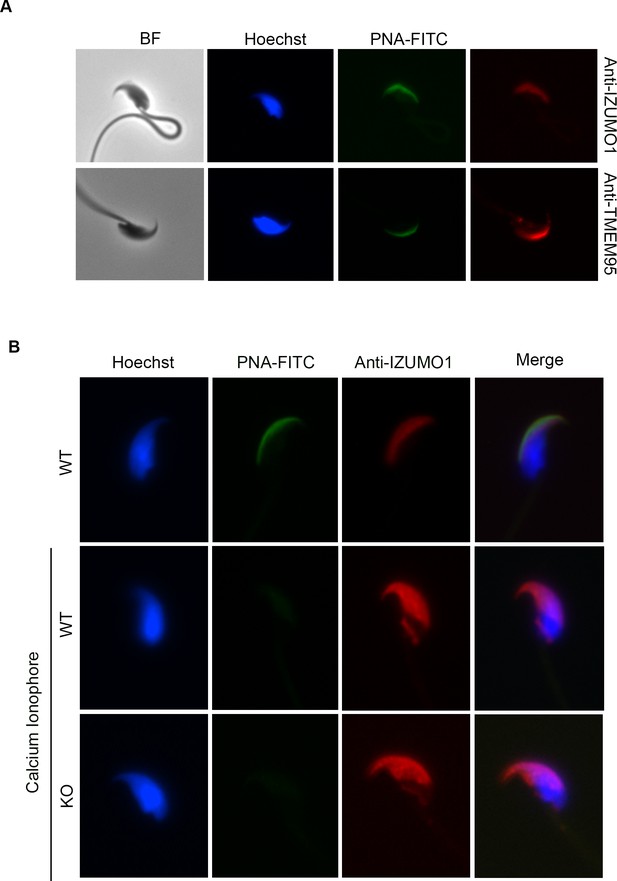
Acrosomal cap localization of TMEM95 and IZUMO1 and IZUMO1 relocalization in WT and KO sperm.
(A) Immunocytochemistry images of WT sperm stained with PNA and IZUMO1 antibody (upper row) or PNA and TMEM95 antibody (lower row) in the absence of permeabilizing agents. Despite the absence of permeabilizing agents (30 min fixation in 4% PFA without Triton X-100), TMEM95 and inner acrosomal (PNA) and acrosomal membrane (IZUMO1) markers were detected. (B) IZUMO1 relocates to the equatorial segment following acrosome reaction in TMEM95 KO sperm. Upper row shows one acrosome intact spermatozoon. Lower rows show WT and KO acrosome reacted sperm where IZUMO1 has relocated to the equatorial segment.
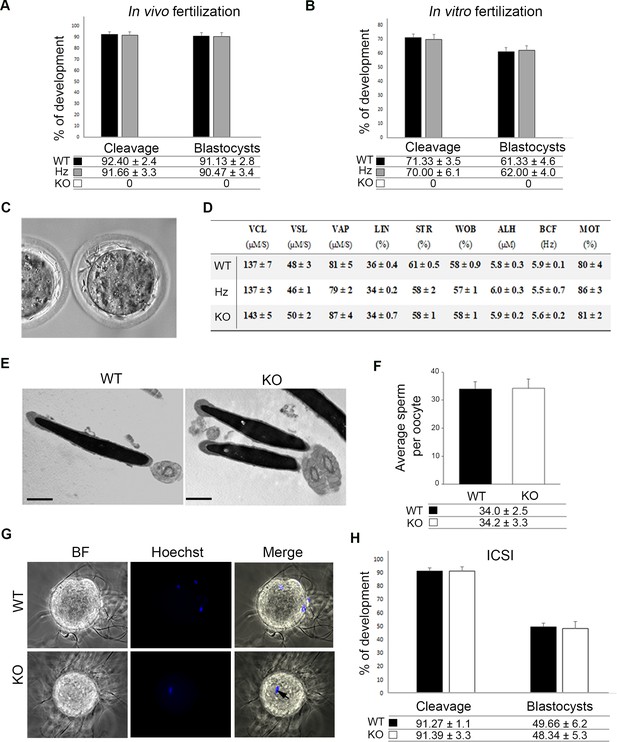
Reproductive performance of TMEM95-null male mice.
(A) Developmental outcomes following in vivo fertilization of WT eggs with WT, Hz or KO males. TMEM95-deficient males were unable to fertilize eggs, whereas no statistical differences were found in developmental rates following fertilization with sperm obtained from Hz or KO males (ANOVA p>0.05). (B) Developmental outcomes following in vitro fertilization of WT eggs with WT, Hz or KO males. TMEM95-deficient males were unable to fertilize eggs, whereas no statistical differences were found in developmental rates following fertilization with sperm obtained from Hz or KO males (ANOVA p>0.05).(C) Representative image of a non-fertilized egg following co-incubation with TMEM95-disrupted sperm. Egg penetration failure prevents zona hardening leading to the accumulation of sperm in the perivitelline space. (D) Motility parameters of WT, Hz or KO sperm analysed by CASA; no differences were found between groups in any of the parameters analysed (ANOVA p>0.05). (E) Transmission Electron Microscopy images of WT or KO sperm showing an overtly normal morphology in TMEM95-deficient sperm. (F) Average number of WT or KO sperm bound to each oocyte following binding assay. TMEM95 disruption did not impaired sperm binding to the egg membrane (ANOVA p>0.05). (G) Sperm-egg fusion assay. WT sperm fused with Hoechst pre-loaded zona-free eggs, which transferred the stain to them upon membrane fusion. In contrast TMEM95-depleted sperm were unable to fuse, exhibiting no Hoechst staining (only the egg DNA, marked by an arrow, is stained). (H) Development of WT eggs microinjected with WT or KO sperm following Intracytoplasmic Sperm Injection (ICSI). Similar developmental rates were obtained using WT or KO sperm (ANOVA p>0.05), indicating that TMEM95 null sperm were able to fertilize eggs when the sperm-egg membrane fusion step is bypassed by ICSI.
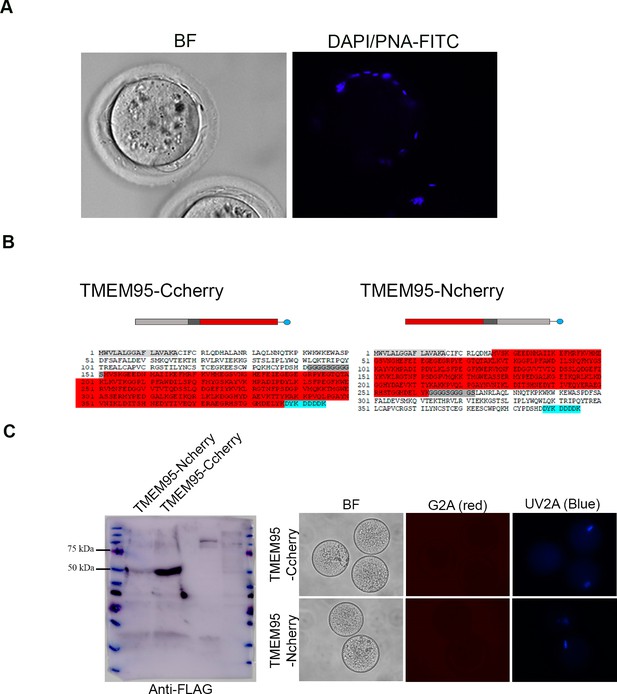
TMEM95 null sperm undergo acrosome reaction and bind to the egg membrane.
(A) Unfertilized eggs co-incubated with TMEM95 null sperm and stained with DAPI and PNA. All sperm that penetrated into the perivitelline space underwent the acrosome reaction (PNA negative) but were still unable to penetrate into the ooplasm. (B) Schematic representation of TMEM95:Cherry fusion proteins. Cherry (highlighted in red) was inserted near the C-terminus of TMEM95 (TMEM95-Ccherry) or just downstream of the signal peptide of TMEM95 (TMEM95-NCherry). (C) Immunoblotting of TMEM95:Cherry fusion proteins expressed in CHO cells (left). Media was separated by SDS-PAGE and analysed by WB using an anti-FLAG antibody. Zona-free eggs were incubated with purified proteins at 0.9 mg/ml for 1 hr to determine protein binding. No protein signal was detected at the egg membrane.
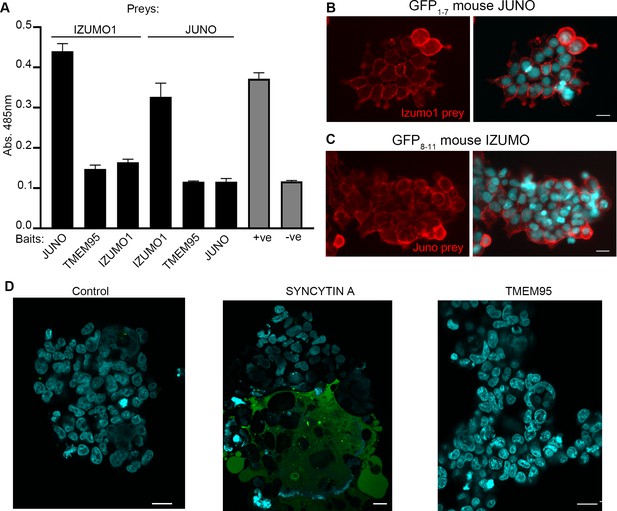
TMEM95 does not interact with JUNO nor IZUMO1.
(A) Binding analysis using the AVEXIS assays shows that the soluble recombinant TMEM95 ectodomain does not interact with JUNO nor with IZUMO1. The entire ectodomains of the named proteins were expressed in HEK293-6E cells either as biotinylated baits or as pentameric beta-lactamase-tagged preys. Bait proteins were immobilised on streptavidin-coated plates and captured prey proteins quantified by measuring the absorbance of a colorimetric reaction product of the beta-lactamase substrate, nitrocefin. The CD200R (bait)-CD200 (prey) binding pair was used as positive control. The same prey, CD200R, was tested against TMEM95 and is shown as negative control. Bars represent means + s.d.; n = 3. (B) HEK293 cells stably expressing the N-terminal half of GFP (GFP1-7) and mouse JUNO stained with a highly avid IZUMO1 probe. (C) HEK293 cells stably expressing the C-terminal half of GFP (GFP8-11) and mouse IZUMO1 stained with a highly avid JUNO. (D) TMEM95 does not induce fusion when expressed in HEK293T cells in the presence of JUNO and IZUMO1 using a GFP-complementation cell fusion assay. HEK293T cells expressing either half of GFP and either JUNO or IZUMO1 were mixed and their fusogenic ability visualized by GFP fluorescence. The IZUMOI-expressing cells were either mock transfected prior to mixing (Control), transfected with Syncitin a, as a positive fusion control, or Tmem95. By contrast to the cells transfected with Syncytin a, Tmem95 did not induce cell fusion. Cell nuclei are stained with DAPI and scale bar represents 20 µm.
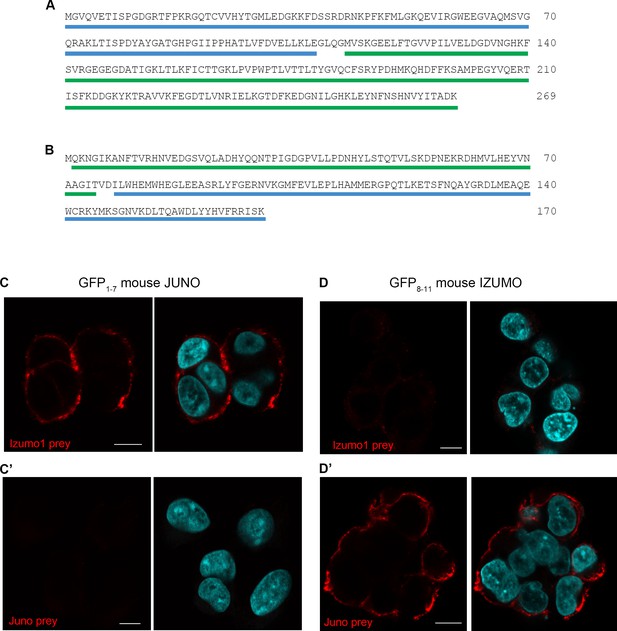
Split GFP fusion assay.
(A) Amino acid sequence of FKBP1A underlined in blue fused via a short linker to the N-terminal region of GFP (GFP1-7) (green underline). (B) Amino acid sequence of the C-terminal region of GFP (GFP8-11) highlighted in green, fused to RB, underlined in blue. (C, C´, D and D´) The cells expressing the GFP1-7 stably express mouse Juno and the cells expressing the GFP8-11 stably express mouse Izumo1. To establish functional cell surface expression of both Juno and Izumo1 on the cells, noth cell lines were stained with the corresponding pentameric FLAG-tagged Izumo1 and Juno preys. The cells expressing mouse Juno were stained with the Izumo1 prey but not Juno, and the cells expressing mouse Izumo1 stained with the Juno prey but not Izumo1. Images are representative of at least three independent replicates. Nuclei are stained with DAPI (blue) and scale bars represents 10 µm.
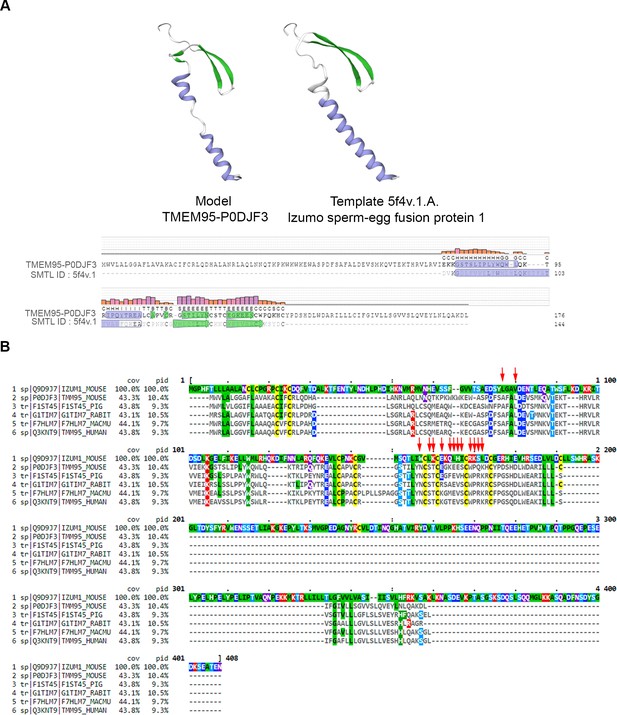
Sequence alignments of TMEM95 and IZUMO1.
(A) Structure prediction of TMEM95 created by SWISS-MODEL web-based integrated service. Model TMEM95-P0DJF3 (left) was built using Template 5f4v.1.A (right) from Izumo1 sperm-egg fusion protein 1 (Ohto et al., 2016). Secondary structure alignment shows coloured residues by secondary structure elements: green for β-hairpin and blue for α-helix. (B) Sequence alignment of IZUMO1 from Mus musculus (Q9D9J7) (top) and TMEM95 from Mus musculus (P0DJF3), Sus scrofa (F1ST45), Oryctolagus cuniculus (G1TIM7), Macaca mulatta (F7HLM7) and Homo sapiens (Q3KNT9) are shown. Residues are colored by identity to top sequence (IZUMO1) and to indicate physicochemical properties: dark-gray for mismatch, yellow for cysteine, green for hydrophobic, dull-blue for small alcohol, bright-blue for negative charge, red for positive charge, and purple for polar. The residues of the IZUMO1-JUNO interface binding (Ohto et al., 2016) are indicated by red arrows. cov, percentage of coverage; pid, percentage of identity with top sequence. The sequence identity between mouse and human TMEM95 is 67%.
Videos
Confocal z-sections of an egg fixed in PFA following IVF with TMEM95-null sperm.
Multiple sperm are found in the perivitelline space, unable to fuse with the egg membrane.
Tables
Details of peptides identified by MS/MS from 20 kDa gel band.
| Protein name | Score | Spi % | Sequence |
|---|---|---|---|
| TMEM95 | 5.33 | 68.8 | LLLCIFGIVLLsGVVSLQ |
| TMEM95 | 5.17 | 59.8 | LLLCIFGIVLLsGVVSLQ |
| TMEM95 | 4.12 | 46.1 | LLSGVVSLQVEY |
| TMEM95 | 3.42 | 53 | KTRYP |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Tmem95 KO | This article | Generated by CRISPR (M and M), available upn request | |
| Antibody | Rabbit Anti-TMEM95 | MyBioSource | MBS7004333 | |
| Antibody | Rabbit Anti-IZUMO1 | AbCam | Ab211623; RRID:AB_2650506 | |
| Antibody | Rat Anti-Izumo | Gift from Dr. Ikawa | Clone KS139-34 | |
| Antibody | Mouse Anti-Tubulin | Sigma | T8328, RRID:AB_1844090 | |
| Antibody | Rabbit anti-Flag | Sigma | F7425, RRID:AB_439687 | |
| Recombinant DNA reagent | Plasmid pMJ920 | (Jinek et al., 2013) | RRID:Addgene_42234 | |
| Recombinant DNA reagent | Plasmid px330 vector | (Yang et al., 2014) | RRID:Addgene_42230 | |
| Sequence-based reagent | Primers | This article | Detailed on M and M (ordered from Sigma) |





