Circulating myeloid cells invade the central nervous system to mediate cachexia during pancreatic cancer
Figures
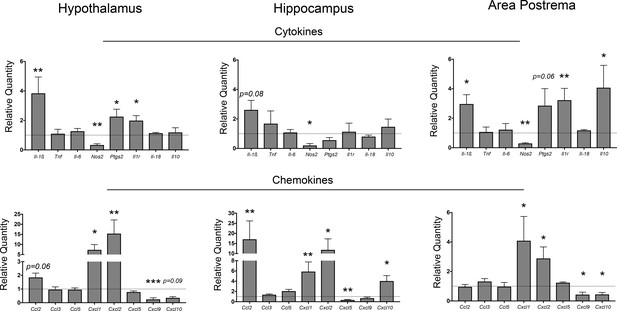
Neuroinflammation in the CNS during PDAC.
qRT-PCR analysis of cytokine and chemokine transcripts in the hypothalamus, hippocampus, and area postrema in PDAC-bearing animals at 10 d.p.i. Values are relative to sham group. All analyses are from 10 d.p.i. n = 4–5/group, *p<0.05, **p<0.01, ***p<0.001 compared to sham group in one-way ANOVA analysis of ΔCT values. Results are representative of at least two independent experiments. For all figures, data are presented as mean ± s.e.m.
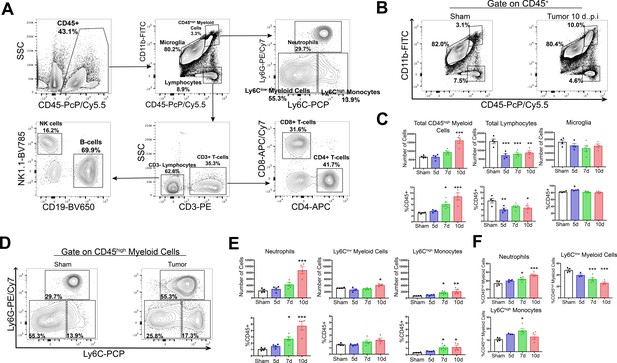
Circulating myeloid cells infiltrate the brain during PDAC.
(A) Flow cytometry plots of immune cells isolated from whole brain homogenate, showing gating strategy to identify different immune cell populations. (B) Representative flow cytometry plots displaying CD45 and CD11b fluorescent intensities of immune cells isolated from brains of tumor and sham animals, gated on live, singlet, CD45+ cells. (C) Quantification of different immune cell populations in the brain at different time points throughout PDAC course. d = days post inoculation. Populations were identified as shown in (A). (D) Representative flow cytometry plots displaying Ly6C and Ly6G fluorescent intensities of immune cells isolated from brains of tumor and sham animals, gated on CD45highCD11bhigh cells. (E) Quantification of different CD45high myeloid cell populations in the brain at different time points during PDAC progression. (F) Relative amounts of different CD45high myeloid cell populations as a percentage of total CD45high myeloid cells, throughout the course of PDAC. Populations identified as described for (E). n = 4–5/group, *p<0.05, **p<0.01, ***p<0.001 compared to sham group in one-way ANOVA Bonferroni post hoc analysis, and results are representative of three independent experiments.
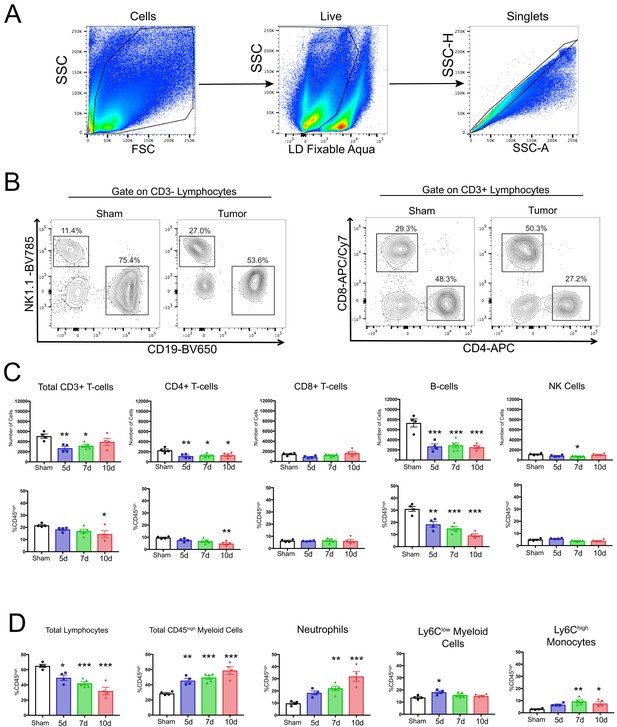
Decreased lymphocytes in the brain during PDAC cachexia.
(A) Gating strategy to identify live single cells from whole brain homogenate. (B) Representative plots of different lymphocyte populations from brain homogenate from sham and tumor (10 d.p.i.) animals. For CD3- cells, NK cells = NK1.1+CD19-, B-cells = CD19+NK1.1-. For CD3+ cells, CD4+ and CD8+ T-cells were identified. (C) Quantification of different lymphocyte populations throughout the course of cachexia. *p<0.05, **p<0.01, ***p<0.001 compared to sham one-way ANOVA Bonferroni post hoc analysis. (D) Quantification of different immune cell populations in the brain throughout the course of cachexia, as a percentage of CD45high cells. *p<0.05, **p<0.01, ***p<0.001 compared to sham. n = 4–5/group. Results are representative of three independent experiments.
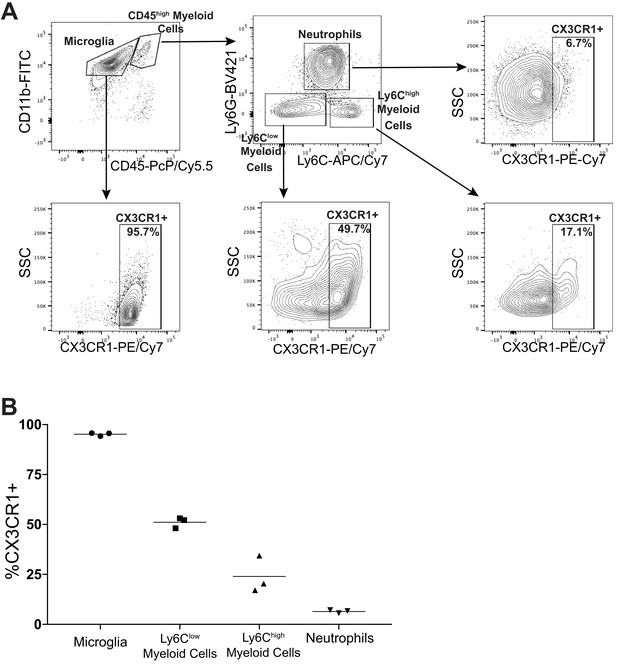
Infiltrating Ly6G+ cells are not microglia.
(A) Gating strategy to identify different immune cell populations isolated from brains of tumor-bearing animals, 14 d.p.i., gated on live, singlet, CD45+ cells. (B) Percentage of cells CX3CR1+. n = 3/group.
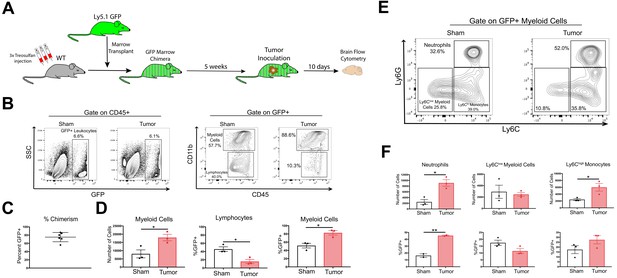
GFP BMT confirms peripheral origin of infiltrating myeloid cells in the CNS during PDAC.
(A) Diagram of bone marrow transplant protocol to generate GFP+ bone marrow chimeras. (B) Gating strategy for CD45+GFP+ cells isolated from brains of tumor and sham GFP chimera animals. (C) Percent chimerism, identified as percentage of CD45+ cells in the blood that were GFP+. (D) Quantification of GFP+ myeloid cells and lymphocytes in the brains of tumor and sham mice, 10 d.p.i. (E) Representative flow cytometry plot of different GFP+ myeloid cell populations in the brains of tumor and sham GFP bone marrow chimera animals, 10 d.p.i. (F) Quantification of different GFP+ myeloid cell populations in the brains of tumor and sham GFP bone marrow chimera animals, 10 d.p.i. n = 3/group, *p<0.05, **p<0.01 in student’s t-test.

Infiltrating immune cells accumulate at CNS interfaces during PDAC cachexia.
(A) Picture of sagittal mouse brain section to illustrate different regions analyzed. (B-D) 20X images of velum interpositum (B), mediobasal hypothalamus (C), and area postrema (D) of brain from sham animal and tumor animal at 10 d.p.i., with 60X inset shown on the right, along with quantification of MPO+ and total CD45+ cells. For B, dashed line denotes VI borders. For C, dashed line denotes borders of meninges adjacent to ME. Scale bar for 20X images = 100 μm. Scale bar for 60X insets = 10 μm. Data are presented as mean ± s.e.m., n = 5/group, *p<0.05, **p<0.01 in student’s t-test.
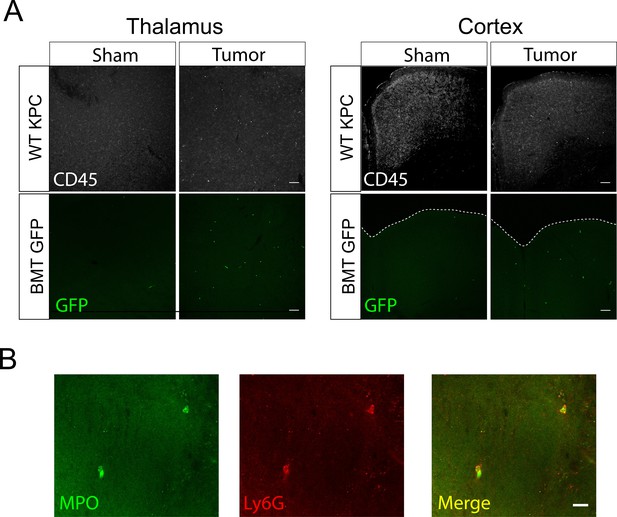
Immunofluorescence analysis of infiltrating immune cells during PDAC.
(A) 10X confocal images of thalamus and cortex from sham and tumor mouse brains, 10 d.p.i.WT KPC = WT animals, BMT GFP = Ly5.1 eGFP marrow transplanted into WT recipient after treosulfan conditioning to ablate marrow (see Materials and methods). Scale bar = 100 μm. (B) 40X confocal image of thalamus from tumor mouse, 12 d.p.i. Scale bar = 20 μm.
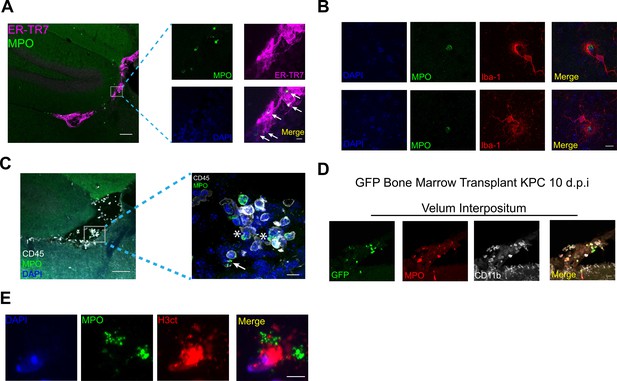
Characteristics of brain-infiltrating immune cells during PDAC.
(A) Representative 10X image of VI from the brain of a tumor animal 10 d.p.i., showing ER-TR7 staining to label meninges and MPO staining to label neutrophils. Scale bar = 100 μm. Inset = 60X showing neutrophils (indicated by arrows) within the meninges of the VI. Scale bar = 10 μm. (B) Representative 60X images of microglia phagocytosing neutrophils in the thalamus of animals with KPC tumor, 10 d.p.i. Scale bar = 10 μm. (C) 20X image a VI with 60X inset showing neutrophils degranulating. Asterisk = myeloperoxidase ‘blebs’ coming off neutrophil. Arrow = extracellular myeloperoxidase. (D) Representative 60X images of VI from BMT GFP mice at 10 d.p.i. Scale bars = 20 μm. (E) 60X image of neutrophil extracellular trap in the VI of a tumor animal, 10 d.p.i. Scale bar = 5 μm.
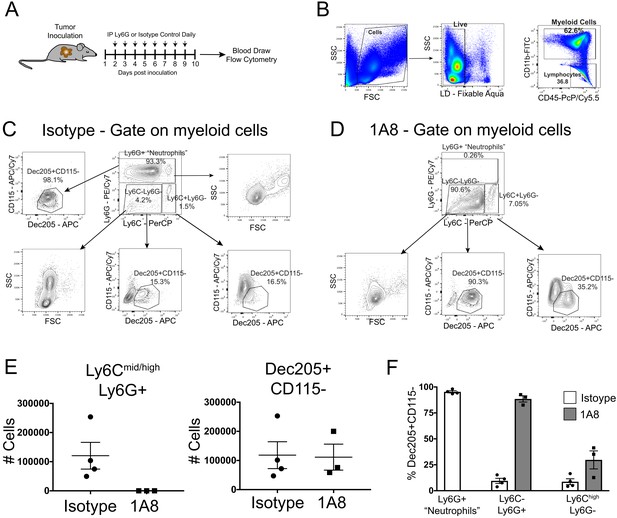
Neutrophils are not depleted by Ly6G antibody administration.
(A) Schematic of experimental setup. (B) Representative plots of gaiting strategy for myeloid cells in isolated blood leukocytes. (C) Representative plots of gating strategy used to identify neutrophils in isotype-treated tumor-bearing animals at 10 d.p.i. Cells were initially gated on myeloid cells identified in B. (D) Representative plots of gating strategy used to identify different cell types in anti Ly6G-treated tumor-bearing animals at 10 d.p.i. 1A8 = clone 1A8 anti-Ly6G antibody group. Note large population of Ly6C-Ly6G- cells that is not present in representative plot from isotype-treated animal. Also note that this population has similar forward FSC and SSC properties as the Ly6Cmid/highLy6G+ population in C, rather than the LyC-Ly6G- population in C. (E) Number of the Ly6Cmid/highLy6G+ and Dec205+CD115- myeloid cells in isotype- and 1A8-treated animals. # Cells = number of cells per 200 μl blood. (F) Percentage of different myeloid cell populations that are Dec205+CD115- in isotype- and 1A8-treated tumor-bearing animals. Ly6G+ ‘Neutrophils’=Ly6Cmid/highLy6G+. Ly6Cmid/highLy6G+ population omitted for 1A8-treated animals since this was only 100–200 cells in each animal.
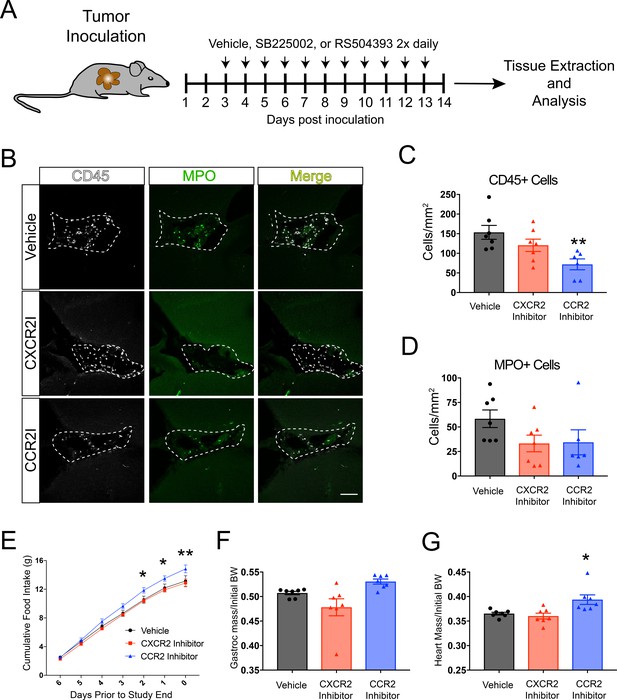
CCR2 signaling is important for cachexia and immune cell infiltration into the brain during PDAC.
(A) Diagram depicting treatment schedule after OT tumor inoculation with PDAC cells. (B) Representative images of the VI from brains of vehicle-, SB225002-, or RS504393-treated tumor-bearing animals at 14 d.p.i. CXCR2I = SB225002. CCR2I = RS504393. Dashed line denotes VI borders. Scale bar = 100 μm. (C) Quantification of CD45+ globoid cells in the VI at 14 d.p.i. n = 7/group. **p<0.01 compared to vehicle-treated in Bonferroni post-hoc analysis in one-way ANOVA. (D) Quantification of MPO+ cells in the VI at 14 d.p.i. One RS504393-treated animal was excluded from quantification analysis due to meeting Grubbs outlier criterion. (E) Cumulative food intake starting when animals develop cachexia, at 7 d.p.i. *p<0.05, **p<0.01, comparing RS504393-treated tumor vs. vehicle-treated tumor in Bonferroni post hoc analysis in two-way ANOVA. n = 7/group. (F) Mass of dissected gastrocnemius, normalized to initial body weight, at 14 d.p.i. (G) Mass of dissected heart, normalized to initial body weight, at 14 d.p.i. *p<0.05 compared to vehicle-treated in Bonferroni post-hoc analysis in one-way ANOVA.
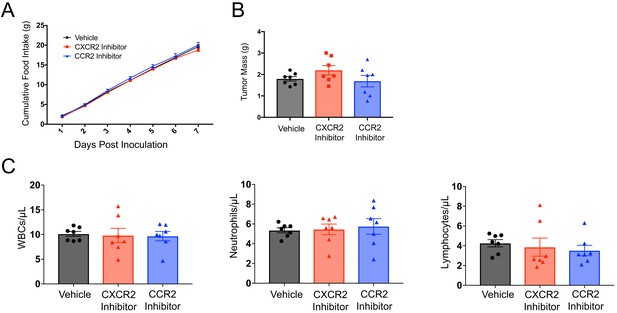
No differences in pre-cachexia food intake, tumor mass, or circulating immune cells in SB225002-, or RS504393-treated tumor-bearing animals.
(A) Cumulative food intake from tumor inoculation to 7 d.p.i.in vehicle-, SB225002-, RS504393-treated tumor-bearing animals. CXCR2I = SB225002. CCR2I = RS504393. (B) Tumor mass in vehicle-, SB225002-, RS504393-treated tumor-bearing animals. (C) Quantification of circulating immune cells using Hemavet analyzer in vehicle-, SB225002-, RS504393-treated tumor-bearing animals. WBC = white blood cell.
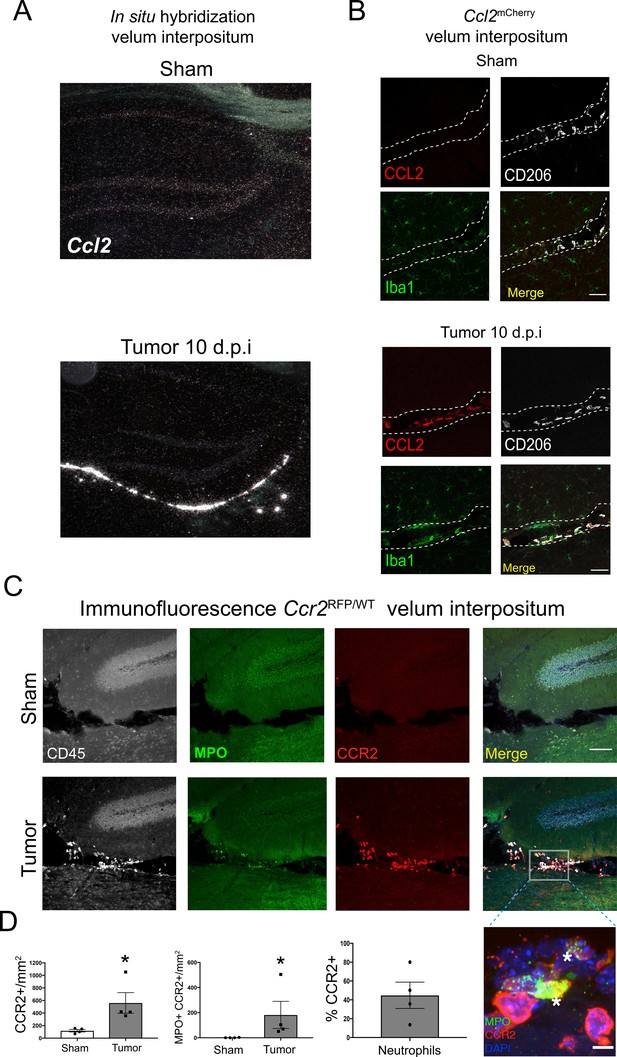
The CCR2-CCL2 axis is activated in the CNS during PDAC.
(A) Representative darkfield microscopy image of in situ hybridization for Ccl2 in sham and tumor (10 d.p.i) mouse brains. (B) Representative 40X confocal microscopy images of the VI from Ccl2mCherry sham and tumor mouse brains, 10 d.p.i., demonstrating that CCL2 protein expression is confined to meningeal macrophages, identified by CD206 labeling. Scale bar = 20 μm. (C) Representative 20X confocal microscopy image of brain from Ccr2RFP/WT tumor (10 d.p.i.) and sham mouse brain. Scale bar = 100 μm. Inset = 60X image identifying CCR2+ neutrophils in the VI of a tumor animal, indicated by asterisks. Scale bar = 5 μm. (D) Quantification of different RFP+ cell populations in the VI of Ccr2RFP/WT tumor (10 d.p.i.) and sham animals. n = 4/group. *p<0.05, Mann-Whitney U-test comparing sham to tumor. Results are representative of two independent experiments.
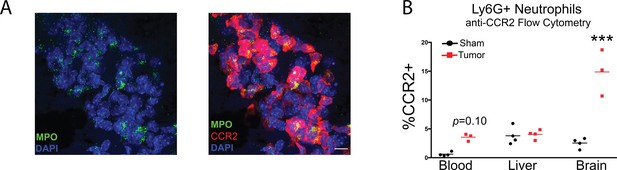
Neutrophils in the velum interpositum express CCR2 during PDAC.
(A) 60X image identifying cluster of CCR2+ neutrophils in the VI of a Ccr2RFP tumor animal. Scale bar = 10 μm. (B) Flow cytometry analysis of CCR2+ neutrophils in the blood, brain, and liver of sham and tumor animals, 10 d.p.i. Neutrophils defined as live, CD45highCD11b+Ly6G+ cells. CCR2+ neutrophils identified by AF647 anti-CCR2 labeling compared to AF647 isotype control. n = 3–4/group. ***p<0.001 in repeated measures one-way ANOVA compared to sham. Bars denote mean. Results are representative of two independent experiments.

Chemokine transcripts are upregulated in microglia in vitro in response to PDAC-conditioned media.
(A) Schematic representation of in vitro PDAC-conditioned media treatment system. (B) qRT-PCR analysis of chemokine transcripts after PDAC-conditioned media treatment. Values are relative to those from control media-treated primary microglia. ***p<0.001 compared to control media-treated in repeated measures one-way ANOVA. n = 3/group. Note: one Cxcl9 PDAC-conditioned sample was not amplified after 45 cycles, so was not included. Therefore, no statistics were performed for Cxcl9. (C) qRT-PCR analysis of chemokine transcripts after 10 ng LPS treatment. Values are relative to those from control media-treated primary microglia. ***p<0.001 compared to control media-treated in repeated measures one-way ANOVA. n = 3/group.
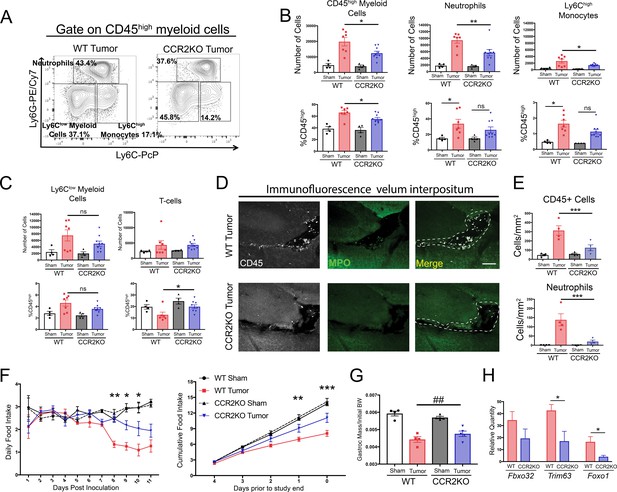
The CCR2-CCL2 axis in the CNS is critical for brain inflammation, anorexia, and muscle catabolism during PDAC.
(A) Representative plot of different CD45high myeloid cell populations from WT and CCR2KO tumor animal brains, 11 d.p.i. Cells are gated on live, singlet, CD45+, CD45highCD11b+ cells. (B and C) Flow cytometry analysis of immune cells isolated from whole brain homogenate. *p<0.05, **p<0.01, WT tumor vs. CCR2KO tumor, or tumor vs. sham in the same genotype in Bonferroni post hoc analysis in two-way ANOVA. ns = not significant. n = 4–9/group. Data consist of two independent experiments pooled (n = at least 2/group in each experiment). (D) Representative 20X confocal microscopy images of the VI from WT tumor and CCR2KO tumor brain, 10 d.p.i. Scale bar = 100 μm. Dashed lines represent VI boundary. (E) Quantification of total CD45+ globoid cells and MPO+ neutrophils in the VI of WT and CCR2KO animals, 10 d.p.i. ***p<0.001, WT tumor vs. CCR2KO tumor in Bonferroni post hoc analysis in two-way ANOVA. n = 4/group. (F) Daily food intake (left) and cumulative food intake for final 5 days of the study (right, starting when animals develop cachexia) in WT and CCR2KO tumor and sham mice. *p<0.05, **p<0.01, ***p<0.001 comparing WT tumor vs. CCR2KO tumor in Bonferroni post hoc analysis in two-way ANOVA. n = 4/5 per group. Results are representative of three independent experiments. (G) Left = mass of dissected gastrocnemius, normalized to initial body weight, at 11 d.p.i. ##p<0.01 for interaction effect between genotype and tumor status in two-way ANOVA analysis. (H) qRT-PCR analysis of Fbxo32, Trim63, and Foxo1 from RNA extracted from gastrocnemii dissected at 11 d.p.i. Values normalized to those from WT sham. *p<0.05, WT tumor vs. CCR2KO tumor dCt values. n = 3–5/group.
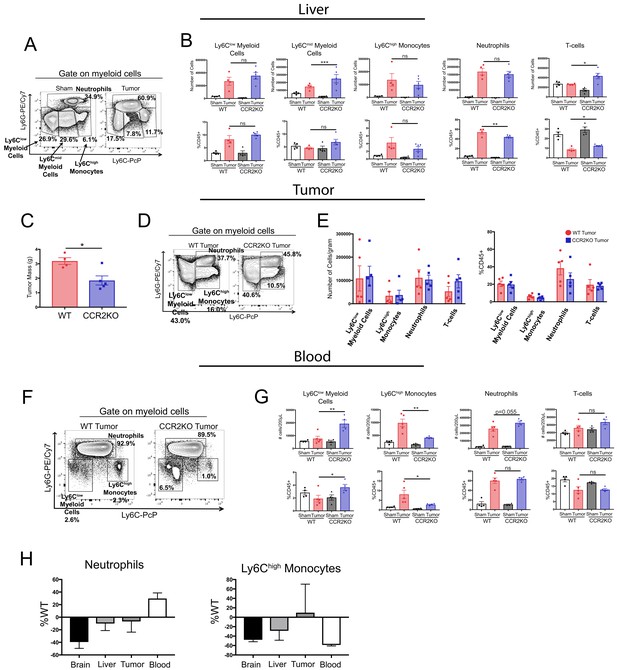
The CCR2-CCL2 axis is of selective importance for the brain in PDAC cachexia.
(A) Representative flow cytometry plot of different myeloid cell populations from WT sham and tumor livers, 11 d.p.i., in order to illustrate different myeloid cell populations identified based on Ly6C and Ly6G expression. Cells are gated on live, singlet CD45+CD11b+ cells. (B) Quantification of flow cytometry analysis of different immune cell populations in the liver from WT and CCR2KO sham and tumor animals, 11 d.p.i. *p<0.05, **p<0.01, WT tumor vs. CCR2KO tumor, or tumor vs. sham in the same genotype in Bonferroni post hoc analysis in two-way ANOVA. ns = not significant. n = 4–9/group. (C) Tumor mass from WT and CCR2KO animals, 11 d.p.i. Data are representative of three independent experiments. Data are presented as mean ± s.e.m. (D) Representative flow cytometry plot of different myeloid cell populations from WT and CCR2KO tumors, 10 d.p.i. Cells are gated on live, singlet CD45+CD11b+ cells. (E) Quantification of flow cytometry analysis of different immune cell populations isolated from tumor from WT and CCR2KO tumor animals, 10 d.p.i. Data consist of two independent experiments pooled (n = at least two per group per experiment). Data are presented as mean ± s.e.m. (F) Representative plot of different myeloid cell populations from WT and CCR2KO tumor animal blood, 10 d.p.i. Cells are gated on live, singlet CD45+CD11b+ cells. (G) Quantification of flow cytometry analysis of different immune cell populations in the blood from WT and CCR2KO sham and tumor animals, 10 d.p.i. *p<0.05, **p<0.01, WT tumor vs. CCR2KO tumor, or tumor vs. sham in the same genotype in Bonferroni post hoc analysis in two-way ANOVA. ns = not significant. n = 4–5/group. Data are representative of two independent experiments. (H) Analysis of neutrophils and Ly6Chigh monocytes in brain, liver, tumor, and blood in CCR2KO tumor mice, normalized to number in WT tumor mice. n = 5–9/group.
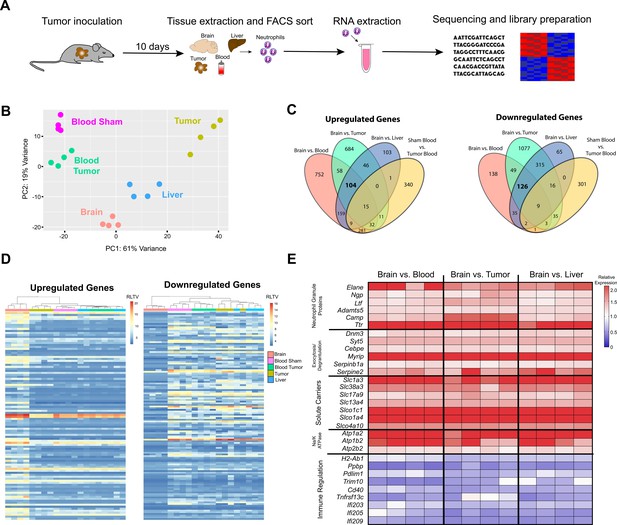
RNASeq of neutrophils in different organs during PDAC.
(A) Workflow for neutrophil isolation, RNA extraction, and RNAseq analysis. (B) Principal component analysis of 500 most varying genes in neutrophils isolated from blood, tumor, liver, and brain from mice with PDAC at 10 d.p.i., as well as blood from sham mice. (C) Venn diagram of different comparisons of transcripts expressed in neutrophils from different organs. (D) We identified putative ‘brain-specific’ transcripts by comparing the transcriptome of brain-infiltrating neutrophils to that of liver- and tumor-infiltrating neutrophils, as well as circulating neutrophils (all from tumor animals). In order to control for the nonspecific effects of malignancy on circulating neutrophils, we any excluded transcripts that were upregulated in circulating neutrophils from tumor animals compared to circulating neutrophils from sham animals. Using this approach, we identified 104 upregulated and 126 downregulated ‘brain-specific’ transcripts RLTV = regularized logarithm transformed value. (E) Heatmap of select brain-specific transcripts showing relative expression, comparing average of brain neutrophils to neutrophils in different organs. Functional enrichment analysis (based on Gene Ontology curation) of brain-specific transcripts identified enrichment for the term ‘extracellular space’ (GO:0005615) in upregulated genes and enrichment for the terms ‘external side of plasma membrane’ (GO:0009897), ‘immune response’ (GO:0006955), and”response to interferon-gamma’ (GO: 00034341) in downregulated genes. Several brain-specific upregulated transcripts encoded neutrophil granule components and enzymes, such as neutrophil granule protein (Ngp), the metalloproteinase ADAMTS5 (Adamts5) neutrophil elastase (Elane), lactoferrin (Ltf), cathelicidin antimicrobial peptide (Camp), and transthyretin (Ttr), as well as proteins important for granule secretion and NET formation such as dynamin 3 (Dnm3), synaptotagmin 15 (Syt15), Serpinb1a (Serpinb1a), Serpin Family E Member 2 (Serpine2), C/EBPε (Cebpe) (Gombart et al., 2003), and Myosin VIIA And Rab Interacting Protein (Myrip) (Desnos et al., 2003). We also observed an increase in genes for solute carriers (Slc gene family) and components of the Na/K ATPase, suggesting brain-infiltrating neutrophils are highly metabolically active. Many brain-specific downregulated transcripts encoded proteins important for immune function and responsiveness to T-cell-derived cytokines, such as MHC II (H2-Ab1), CXCL7 (Ppbp), PDLIM1 (Pdlim1, a negative regulator of NFκβ signaling), and the interferon-inducible genes Ifi203, Ifi205, and Ifi209.
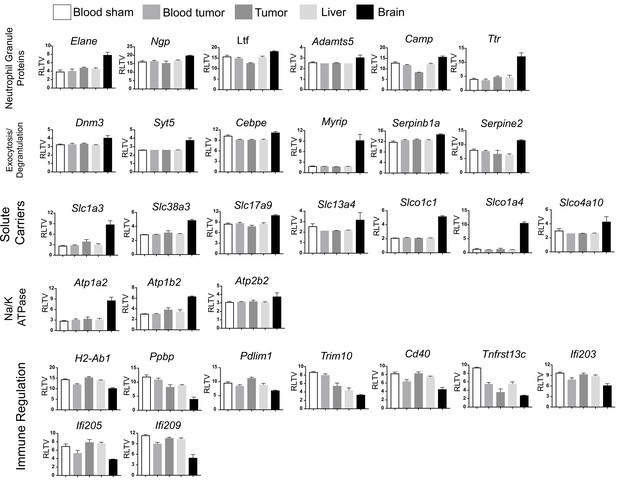
Expression of neutrophil ‘brain-specific’ transcripts.
Normalized expression values of transcripts depicted in heatmap in Figure 7e. RLTV = regularized logarithm transformed value.
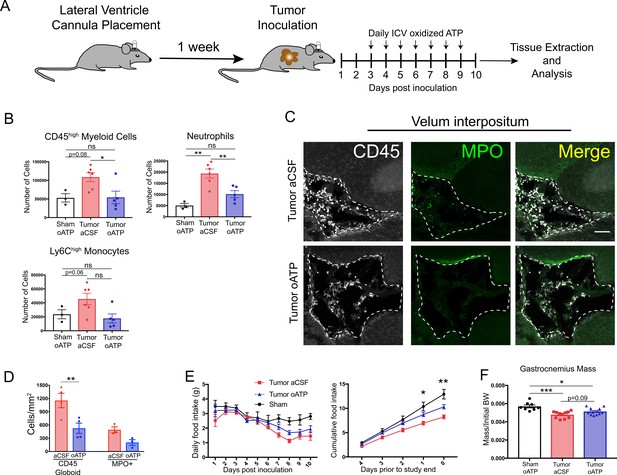
Intracerebroventricular administration of oxidized ATP prevents immune cell recruitment to the brain and attenuates anorexia during PDAC.
(A) Diagram depicting workflow for lateral ventricle cannulation and ICV oATP treatment during PDAC. ICV = intracerebroventricular. (B) Quantification of immune cells isolated from whole brain homogenate. *p<0.05, **p<0.01, in Bonferroni post hoc analysis in two-way ANOVA. ns = not significant. n = 4–7/group. (C) Representative 20X confocal microscopy images of the VI from aCSF-treated and oATP-treated tumor animals. Dashed lines denote VI border. Scale bar = 100 µm. (D) Quantification of VI-infiltrating CD45+ globoid and MPO+ cells, comparing tumor aCSF to tumor oATP-treated animals. n = 3–4/group, 4 VI images per animal. **p<0.01, in Bonferroni post hoc analysis in one-way ANOVA. (E) Daily food intake (left) and cumulative food intake for the final 5 days of the study (right, starting when animals develop symptoms) *p<0.05, **p<0.01, comparing aCSF tumor vs. oATP tumor in Bonferroni post hoc analysis in two-way ANOVA. n = 8–12/group. Results consist of two independent experiments pooled (n = 4–7/group in each experiment). (F) Mass of dissected gastrocnemius, normalized to initial body weight, at 10 d.p.i. *P < 0.05, **P < 0.01, in Bonferroni post hoc analysis in two-way ANOVA. BW = body weight.
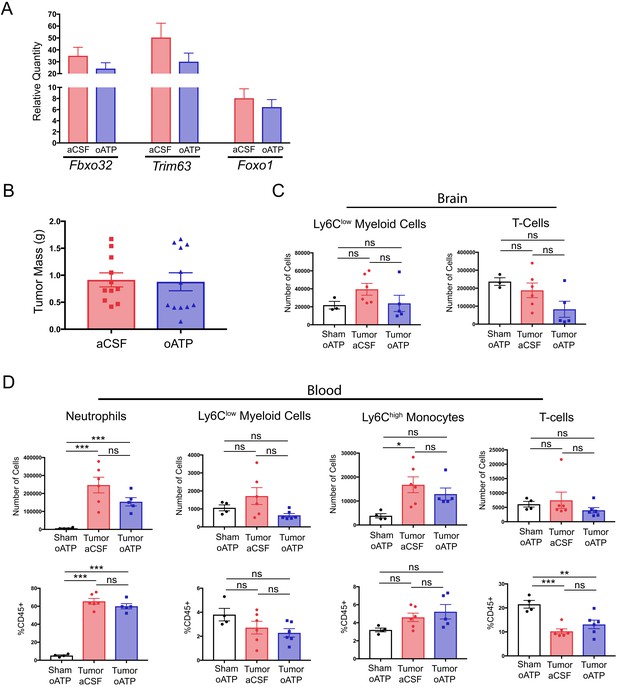
Intracerebroventricular antagonism of P2RX7 does not affect systemic inflammation or tumor size during PDAC.
(A) qRT-PCR analysis of Mafbx, Murf1, and Foxo1 from RNA extracted from gastrocnemii dissected at 8–10 d.p.i. Values normalized to those from sham oATP. n = 4–7/group. (B) Tumor mass from aCSF- and oATP-treated tumor-bearing mice, 8–10 d.p.i. n = 11–12/group. Results consist of two independent experiments pooled (n = 5–7/group in each experiment). (C) Quantification of immune cells isolated from whole brain homogenate. ns = not significant in Bonferroni post hoc analysis in two-way ANOVA. n = 4–7/group. (D) Flow cytometry of immune cells isolated from blood, per 200 µL of blood. *p<0.05, **p<0.01, ***p<0.001 in Bonferroni post hoc analysis in two-way ANOVA. ns = not significant. n = 4–6/group.
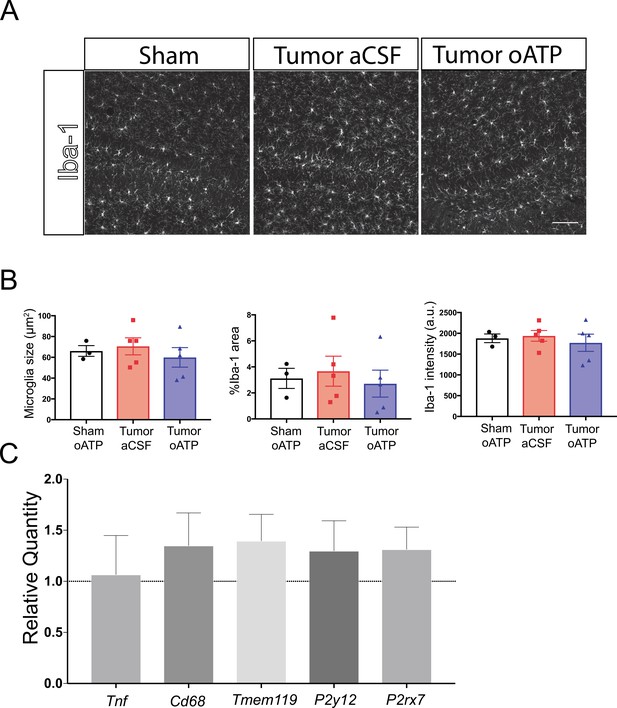
Intracerebroventricular administration of oxidized ATP does not affect microglia activation during PDAC.
(A) Representative 20X images of Iba-1 immunofluorescence in the dentate gyrus, 10 d.p.i. Scale bar = 100 µm. (B) Quantification of microglia morphology in the dentate gyrus 10 d.p.i., showing mean microglia size (left), percent area covered by Iba-1 immunofluorescence (middle), and mean Iba-1 fluorescent intensity per microglia (right). a.u. = arbitrary units. n = 3–5/group. (C) qRT-PCR analysis of transcripts associated with microglia activation in the hippocampus. Values are compared those from oATP-treated tumor animals relative to aCSF-treated tumor animals, at 10 d.p.i. n = 7/group.
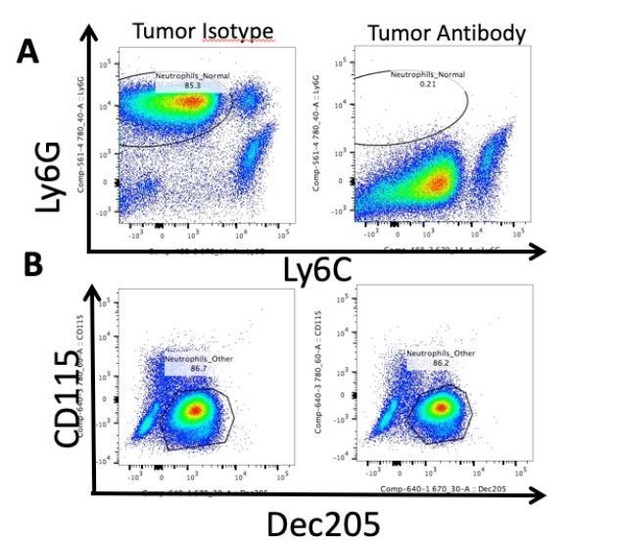
Neutrophils are not depleted by Ly6G antibody administration.
Representative flow cytometry analysis of isolated circulating leukocytes from PDAC-bearing mice, 10 d.p.i. Mice received daily IP injections of 500 ug Ly6G antibody (1A8, Biolegend) starting 2 d.p.i. or isotype control. (A) Gating on live, CD45+CD11b+ myeloid cells. Note Ly6GlowLy6Cmid population in “Tumor Antibody” plot. (B) CD115 and Dec205 expression on same CD45+CD11b+ myeloid cells as in A. Population drawn = neutrophils.





