Human DECR1 is an androgen-repressed survival factor that regulates PUFA oxidation to protect prostate tumor cells from ferroptosis
Figures
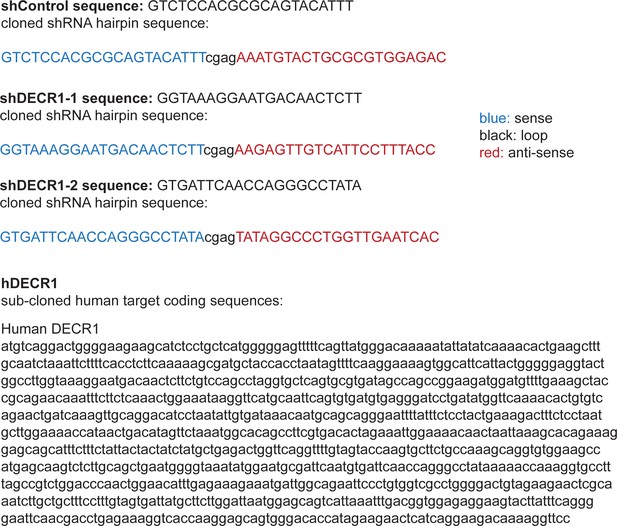
Fatty acid β-oxidation genes are overexpressed in prostate cancer and targeting this process is effective in patient-derived human prostatic ex vivo tumor explants.
(A) Etomoxir reduced cell proliferation in patient-derived human prostatic ex vivo tumor explants. Tissues were treated with 100 µM etomoxir for 72 hr, sections were fixed in formalin, paraffin embedded and stained against the proliferative marker Ki67 (n = 13) (scale bar = 50 µm). (B) Etomoxir (100 µM) decreased acylcarnitine species, the products of CPT1 activity. Acylcarnitines secreted to the conditioned medium were measured after 72 hr treatment of PDEs (n = 9). (C) A meta-analysis of fatty acid oxidation genes using four clinical datasets with malignant and matched normal RNA-sequencing data (n = 122). Genes were rank-ordered on the basis of their meta effect size scores in PCa malignant tissues versus matched normal tissues. (D) Violin plots demonstrate DECR1 mRNA overexpression in PCa primary/malignant tissues compared to normal/benign tissues in three independent datasets. (E) DECR1 mRNA expression is associated with PCa primary Gleason score, total Gleason score (>7) and diseases stage (T-stage). Data were extracted from TCGA PCa dataset. (F) Histogram displaying DECR1 mutation and copy-number amplification frequency across 13 PCa genomic datasets, and (G) across 28 tumor types. Histograms were obtained from CbioPortal platform. (H) DECR1 mRNA expression is associated with shorter relapse-free survival in TCGA PCa, Glinsky et al., 2005 and GSE21032 datasets, and shorter overall survival rates in GSE16560 dataset. (I) DECR1 copy number amplification frequency is associated with shorter relapse-free survival in TCGA PCa dataset. Data in (A) are represented as as the mean ± s.e.m and were statistically analysed using a Wilcoxon matched-pairs signed rank test. Data in (B) are represented as the mean ± s.e.m and were statistically analysed using two-tailed Student’s t-test. Data in (D) and (E) are represented as violin plots in GraphPad prism: the horizontal line within the violin represents the median, and were statistically analysed using a Mann-Whitney two-tailed t-test. Data in (H) and (I) were statistically analysed using a two-sided log-rank test. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
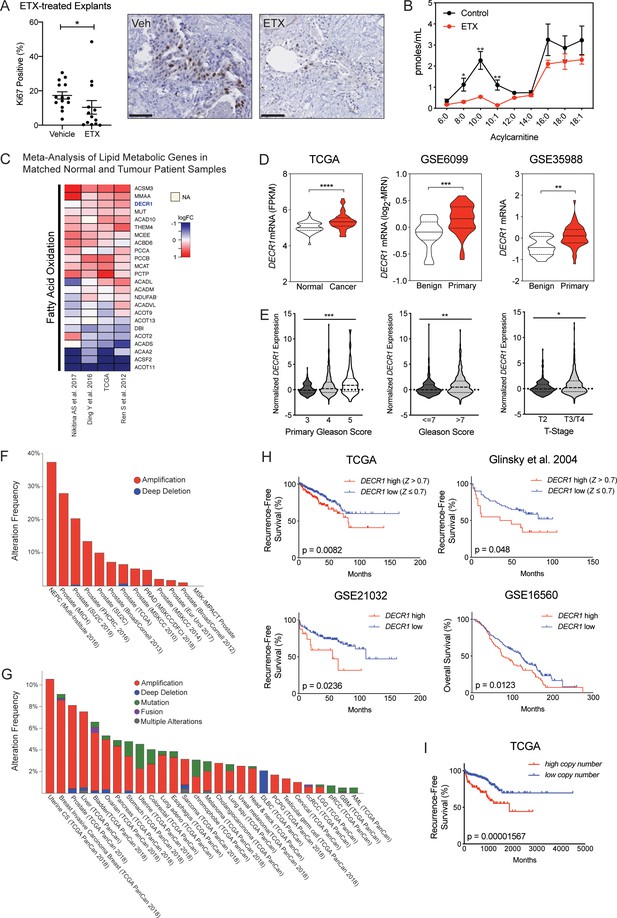
Fatty acid metabolism is consistently altered in clinical prostate tumors.
(A) Cell viability of LNCaP cells treated with indicated concentrations of etomoxir for 72 hr. Cell viability was determined using CyQUANT Cell Assay. (B) A meta-analysis of 735 lipid metabolism genes using four clinical datasets with malignant and matched normal RNA-sequencing data (n = 122). Genes were rank-ordered on the basis of their meta effect size scores in PCa malignant tissues versus matched normal. (C) Fold change of DECR1 mRNA expression in malignant tissues compared to benign/normal tissues. Data were obtained from Oncomine.
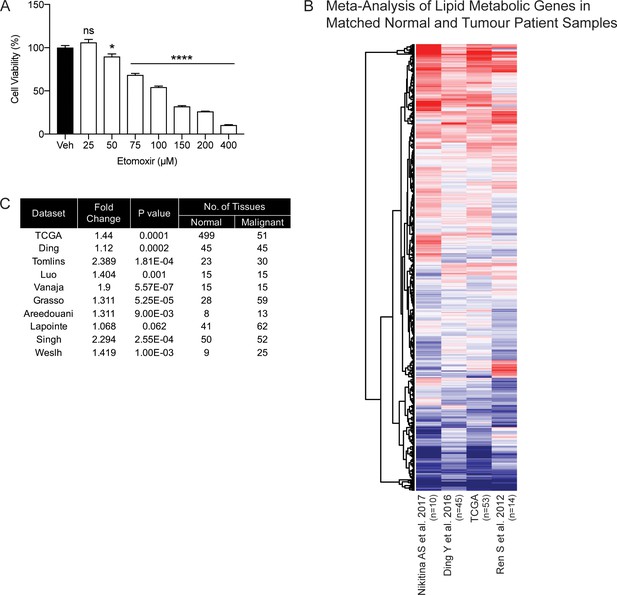
DECR1 protein in overexpressed in malignant prostate cells/tissues.
(A) Violin plots of DECR1 protein overexpression in primary PCa tissues compared to benign prostate tissues in two independent datasets. (B) Representative DECR1 IHC staining of benign prostate tissues and PCa tissues (negative control stain included in bottom right box). Scale bar, 50 µm. (C) Violin plot of DECR1 protein expression in a validation cohort consisting of benign prostate tissues (n = 3) and PCa tissues (n = 8) (top panel). Intra-tissue IHC analysis of DECR1 expression in PCa tissues (n = 8) (bottom panel). (D) DECR1 protein expression in non-malignant prostate cell lines (PNT1 and PNT2) and PCa cell lines (LNCaP, VCaP, 22RV1, C42B and PC3). (E) Immunocytochemistry staining of LNCaP cells to determine the subcellular localization of DECR1: nuclei were labelled using DAPI; mitochondria were labelled using MitoTracker Red; and DECR1 proteins were labelled using Alexa Fluor 488 secondary antibody, (Scale bar = 10 μm). Immunoblot of PNT1 and LNCaP cells separated into cytosolic, mitochondrial and nuclear fractions and incubated with poly (ADP-ribose) polymerase (PARP) and cytochrome-C antibodies to mark nuclear and mitochondrial fractions. Data are represented as violin plots in GraphPad prism: the horizontal line within the violin represents the median. Statistical analysis was performed using a Mann-Whitney two-tailed t-test (A and C top panel), or two-tailed paired t-test (C bottom panel): *p<0.05, **p<0.01 and ***p<0.001.
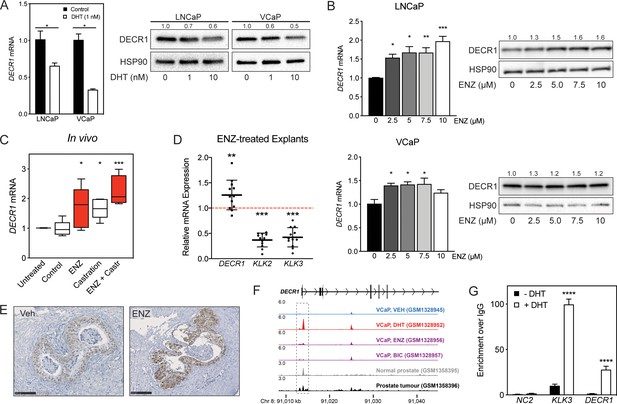
DECR1 is an androgen-repressed gene.
(A) DECR1 mRNA and protein was measured by qRT-PCR and western blot analysis after PCa cell treatment with dihydrotestosterone (DHT), or (B) enzalutamide (ENZ). Relative mRNA expression of DECR1 was calculated using comparative CT method, where the cells treated with vehicle (control) were set to one and normalised to the geometric mean CT value of GUSB and L19 (housekeeping genes). Densitometry quantification of relative DECR1 protein expression was normalized to the HSP90 internal control. n = 3 independent experiments. (C) qRT-PCR analysis of DECR1 mRNA expression in LNCaP-derived tumors treated with enzalutamide (ENZ) and/or castration (Castr). (D) qRT-PCR analysis of DECR1 and androgen regulated genes KLK2 and KLK3 mRNA expression in a cohort of 10 patient-derived human prostatic ex vivo tumor explants treated with enzalutamide (ENZ, 10 µM). Relative mRNA expression was calculated using comparative CT method, where the matched untreated tissue from the same patient was set to one and normalized to the geometric mean CT value of TUBA1B, PPIA and GAPDH. (E) Representative DECR1 IHC staining of patient-derived human prostatic ex vivo tumor explants treated with enzalutamide (ENZ, 10 µM). Scale bar, 100 µm. (F) AR ChIP-sequencing data from VCaP cells (top panel), normal human prostate and primary human prostate tumor specimens (bottom panel). Data from GSE55064 and GSE56288. (G) ChIP-qPCR analysis demonstrates AR binding at DECR1 locus in LNCaP cells after treatment with DHT. Data in bar graphs (A, B and G) are represented as the mean ± s.e.m. Data in (C) are represented as box plots using the Tukey method in GraphPad prism. Statistical analysis was performed using two-tailed Student’s t-test (A, C, D and G) or one-way ANOVA, followed by Dunnett's multiple comparisons test (B): *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
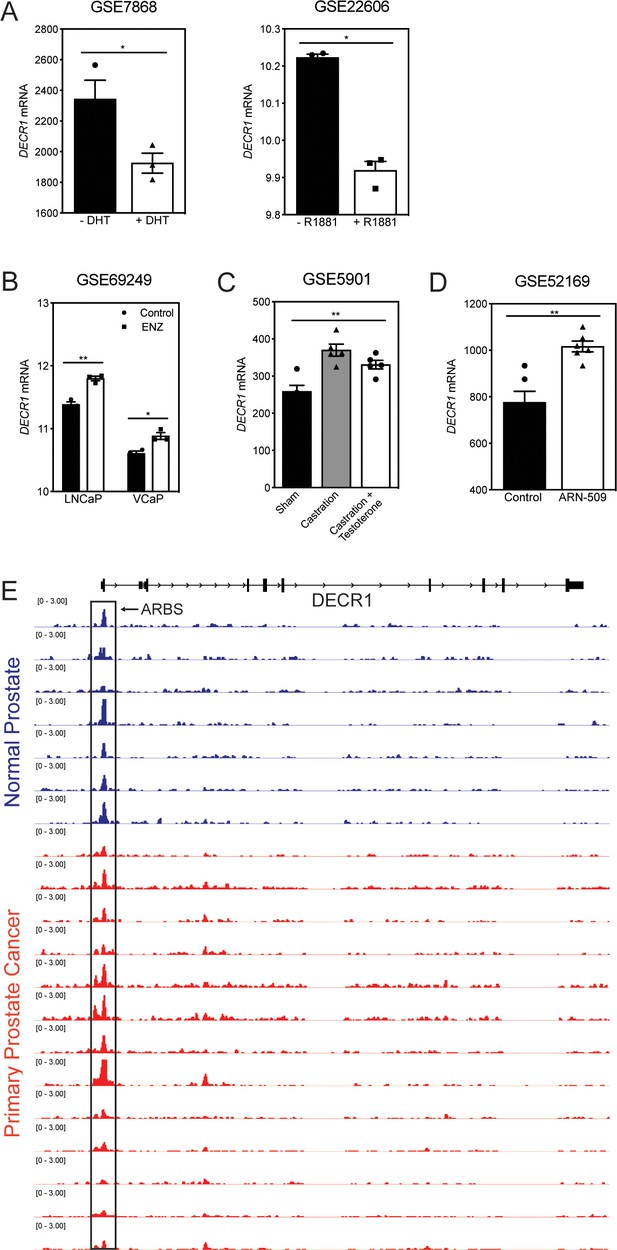
Androgenic regulation of DECR1 expression.
(A) Bar graphs of DECR1 mRNA expression in two publically available datasets shows DECR1 mRNA expression decreased after LNCaP treatment with DHT (GSE7868) or R1881 (GSE22606). (B) DECR1 expression increased in LNCaP and VCaP upon treatment with Enzalutamide (GSE69249). (C) DECR1 mRNA expression increased in mouse prostate gland after castration but decreased after testosterone administration. (D) ARN-509 (Apalutamide) treatment of LNCaP/AR xenograft increased DECR1 mRNA expression (GSE52169). (E) AR ChIP-sequencing data from normal human prostate and primary human prostate tumor specimens (normal, n = 7; tumor, n = 13). Data from GSE56288. Statistical analysis was performed using two-tailed Student’s t-test: *p<0.05, **p<0.01 and ****p<0.0001.
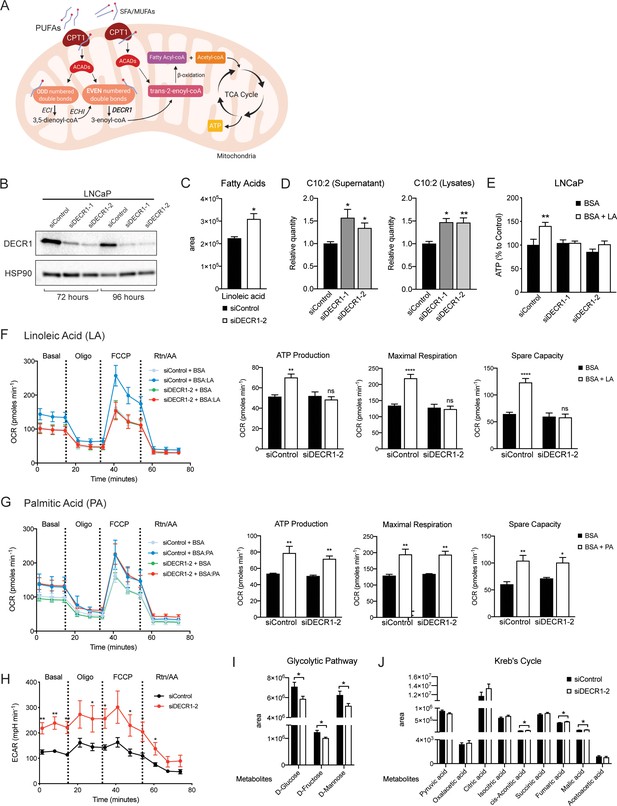
DECR1 knockdown interrupts PUFA β-oxidation in PCa cells.
(A) Schematic of DECR1 function in fatty acid (FA) β-oxidation. In order to translocate FAs into the mitochondria, CPT1 converts long-chain acyl-CoA species to their corresponding long-chain acylcarnitine species. This is followed by a dehydrogenation step mediated by acyl CoA dehydrogenase (ACAD) to generate trans-2-enoyl-CoA, the only intermediate that can be processed by downstream enzymes in the β-oxidation process. Many FAs have unsaturated bonds either on an odd-numbered carbon or in the cis-configuration, resulting in the generation of enoyl-CoA intermediates that cannot be directly processed via the downstream β-oxidation enzymes. These FAs require the activity of 3 auxiliary enzymes, ECI1, ECH1 and DECR1 in order to form trans-2-enoyl-CoA before undergoing β-oxidation. DECR1 catalyzes the conversion of either 2-trans,4-cis-dienoyl or 2-trans,4-trans-dienoyl-CoA to 3-trans-enoyl-CoA. A complete cycle of β-oxidation results in the release of the first two carbon units as acetyl-CoA, and a fatty-acyl-CoA minus two carbons. The acetyl-CoA enters the TCA cycle to produce energy (ATP). The shortened fatty-acyl-CoA is processed again starting with the ACADs to form trans-2-enoyl-CoA either directly or with the aid of the auxiliary enzymes depending on the presence of double bonds. This process continues until all carbons in the fatty acid chain are turned into acetyl-CoA. (B) DECR1 protein expression after 72 hr or 96 hr siRNA transfection. Densitometry quantification of relative DECR1 protein expression was normalized to the HSP90 internal control. (C) Linoleic acid level in LNCaP cells quantified in following 96 hr DECR1 knockdown using GC QQQ targeted metabolomics. (D) Relative quantities of the C10:2 acylcarnitine species in LNCaP cell conditioned medium (left) or cell lysates (right) (n = 3). (E) Quantification of ATP levels in LNCaP cell lysates. LNCaP cells were transfected with DECR1 siRNAs for 48 hr and then starved in no-glucose medium and treated with the lipolysis inhibitor DEUP (100 µM) in the presence (BSA-LA) or absence (BSA) of the PUFA linoleic acid for 48 hr before measuring ATP levels. (F) Oxygen consumption rate (OCR) was assessed in LNCaP cells supplemented with the PUFA linoleic acid (LA) or (G) the saturated fatty acid palmitic acid (PA). Each data point represents an OCR measurement. ATP production, maximal mitochondrial respiration and mitochondrial spare capacity were assessed. (H) Extracellular acidification rate (ECAR) was assessed in LNCaP cells. Each data point represents an ECAR measurement. For experiments (F-H) LNCaP cells were transfected with DECR1 siRNAs for 72 hr, then starved in substrate limited medium for 24 hr; the assay was run in FAO assay medium. (I and J) Metabolites were quantified in LNCaP cells following 96 hr DECR1 knockdown using GC QQQ targeted metabolomics. Data in bar graphs are represented as the mean ± s.e.m (n = 3). Statistical analysis was performed using two-tailed Student’s t-test: *p<0.05, **p<0.01 and ****p<0.0001.
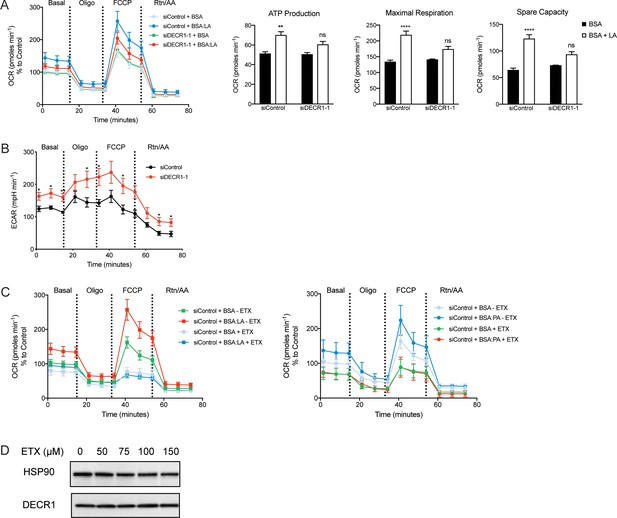
Effects of DECR1 on prostate cancer cellular metabolism.
(A) Oxygen consumption rate (OCR) was assessed in LNCaP cells supplemented with the PUFA linoleic acid (LA). Each data point represents an OCR measurement. ATP production, maximal mitochondrial respiration and mitochondrial spare capacity were assessed. (B) Extracellular acidification rate (ECAR) was assessed in LNCaP cells. Each data point represents an ECAR measurement. (C) OCR of LNCaP cells supplemented with PUFA LA (top) or palmitic acid (PA, bottom) treated with etomoxir (ETX, 100 µM). Each data point represents an OCR measurement. (D) DECR1 protein expression after 48 hr treatment with varying concentrations of etomoxir. Statistical analysis was performed using two-tailed Student’s t-test: *p<0.05, **p<0.01 and ****p<0.0001.
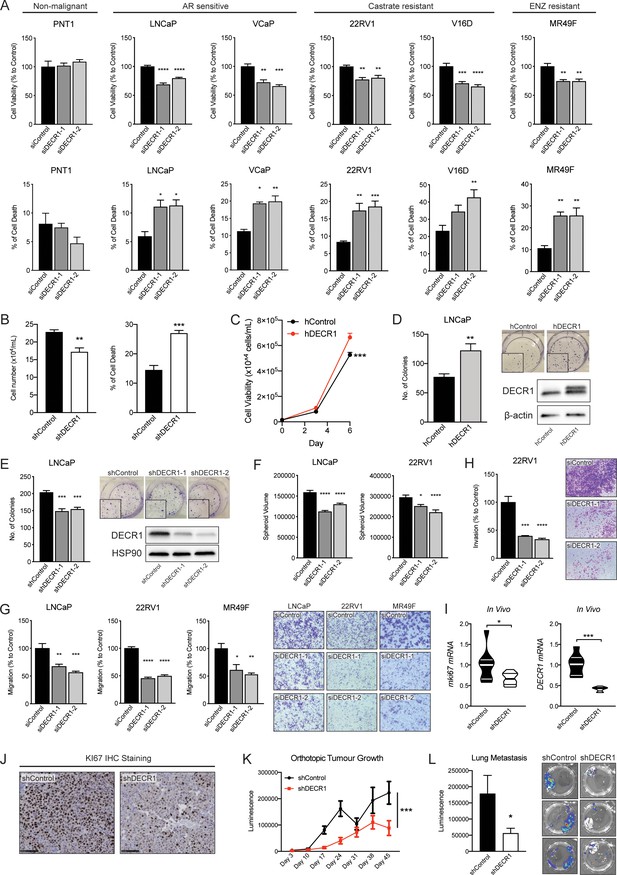
DECR1 knockdown suppresses oncogenic phenotypes of PCa cells.
(A) Cell viability after DECR1 knockdown in non-malignant PNT1 prostate cells; hormone-responsive PCa cell lines (LNCaP and VCaP); castrate-resistant V16D and 22RV1 cell lines and enzalutamide-resistant MR94F cells cultured in full serum media. (B) Cell viability and cell death of stable DECR1 knockdown LNCaP cells cultured in full serum media. (C) Cell viability of stable DECR1-overexpressed LNCaP cells cultured in full serum media. Cell viability and cell death were measured using trypan blue exclusion following 96 hr DECR1 knockdown. Percentages are represented relative to the control siRNA; n = 3 independent experiments per cell line. (D) Clonogenic cell survival of LNCaP cells was assessed using colony formation assay. Stable DECR1-overexpressed cells or (E) stable DECR1 knockdown was achieved using two different short hairpin (sh) vectors and DECR1 expression was confirmed using western blot. Cells were cultured for 2 weeks, washed with PBS, fixed with paraformaldehyde and stained with 1% crystal violet for 30 min. Colonies with more than 50 cells were counted manually; data shown is representative of n = 2 independent experiments. (F) LNCaP and 22RV1 cell growth in 3D spheres. Spheroids were prepared using the hang drop assay following 48 hr DECR1 knockdown. Spheroid volumes were determined after five days of culturing the cells in 20 µl drops; at least 25 spheres per cell line were assessed using the ReViSP software, n = 3 independent experiments per cell line. (G) LNCaP, 22RV1 and MR49F cell migration and (H) 22RV1 cell invasion were assessed using a transwell migration/invasion assay. Cells were transfected with DECR1 siRNA or control siRNA for 48 hr prior to the assay; data shown is representative of n = 3 independent experiments. (I) Violin plots of mKi67 and DECR1 mRNA expression in subcutaneous LNCaP tumors (n = 5 mice, shControl; n = 4 mice, shDECR1). (J) Representative Ki67 IHC staining of subcutaneous LNCaP tumors. Scale bar, 100 μm. Data in bar graphs are represented as the mean ± s.e.m. Statistical analysis was performed using one-way ANOVA, followed by Dunnett's multiple comparisons test: *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001. (K) Tumor growth of intraprostatically injected LNCaP cells (shControl and shDECR1). (J) Lung luminescence readings following DECR1 knockdown in mice. Data are presented as mean ± s.e.m. Statistical analysis was performed using two-way ANOVA or two-tailed student’s t-test: *p<0.05 and ***p<0.001.
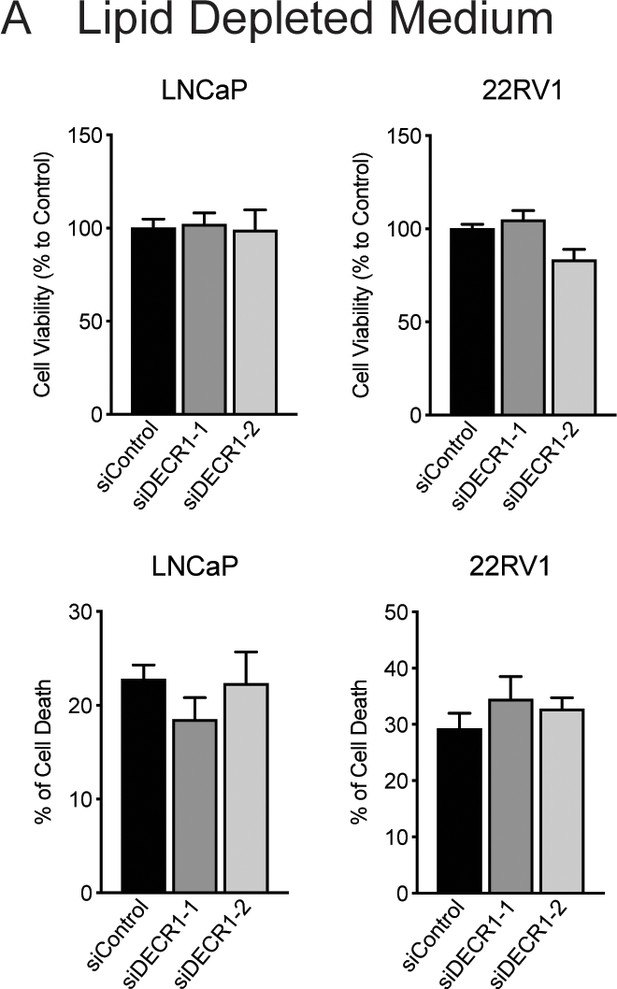
Depletion of extracellular lipids prevents antiproliferative effects of DECR1 in prostate cancer cells.
(A) Cell viability and cell death after DECR1 knockdown in LNCaP and 22RV1 cultured in full serum media.
Cell viability and cell death were measured using trypan blue exclusion following 96 hr DECR1 knockdown. Percentages are represented relative to the control siRNA; n = 3 independent experiments per cell line.
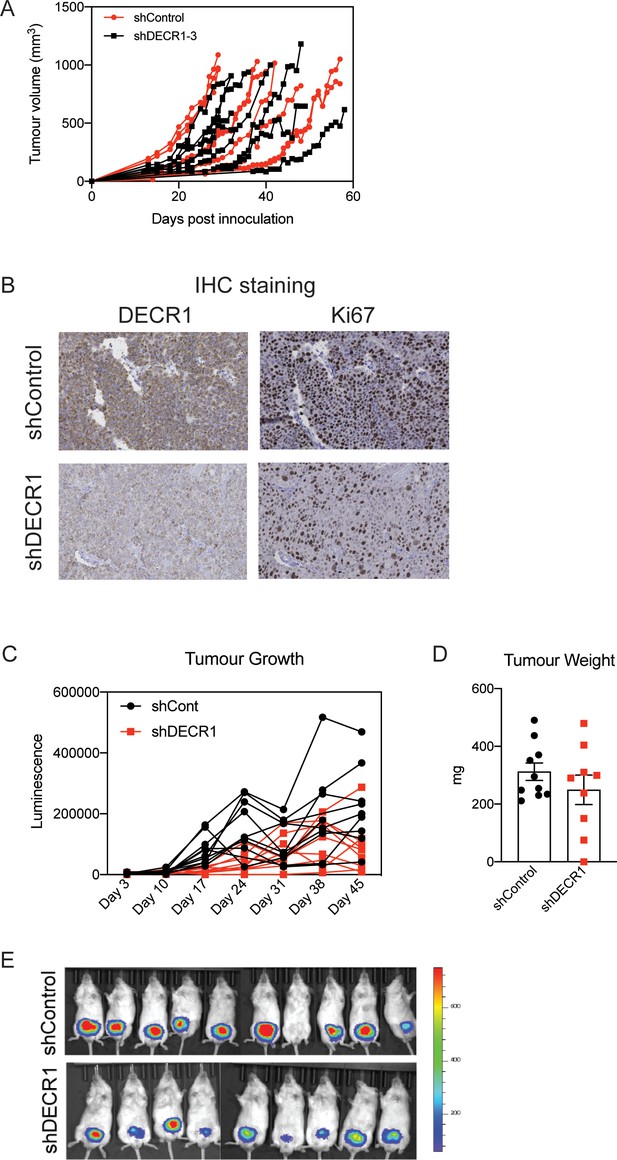
DECR1 suppresses growth of prostate tumor xenografts in mice.
(A) Individual tumor growth of shControl and shDECR1 cells in LNCaP-xenograft tumors. (B) Violin plots of mKi67 and DECR1 mRNA expression in LNCaP tumors (n = 5 mice, shControl; n = 4 mice, shDECR1). (C) Representative DECR1 and KI67 IHC staining of consecutive sections of LNCaP-xenograft tumors. (D) Individual tumor growth of shControl and shDECR1 cells in LNCaP-xenograft tumors from the second cohort of mice. (E) Tumor growth was monitored based on luciferase activity over time as indicated by IVIS imaging (n = 10 mice, shControl; n = 9 mice, shDECR1). Statistical analysis was performed using two-tailed Student’s t-test: *p<0.05 and ***p<0.001.
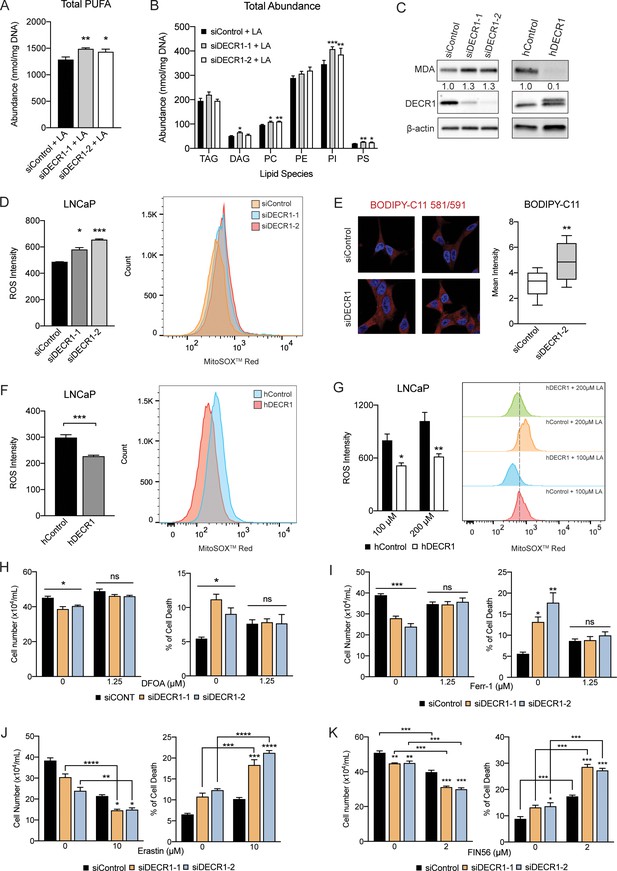
DECR1 knockdown induces PUFA accumulation, lipid peroxidation and ferroptosis.
(A) Abundance of total PUFAs and (B) total abundance of lipid species in phospholipids from control and DECR1 knockdown cells supplemented with linoleic acid (LA). (C) Malondialdehyde (MDA), an oxidative stress marker, was measured by western blot in LNCaP cells transfected with DECR1 siRNAs and in DECR1-overexpressed LNCaP cells. (D) Mitochondrial superoxide levels were quantified following 96 hr DECR1 knockdown using MitoSOX red stain. Fluorescent intensity was quantified using flow cytometry and ROS levels presented as mean fluorescent intensity. (E) Fluorescent images of BODIPY-C11 stained LNCaP cells following DECR1 knockdown (left). Fluorescent intensity was quantified using ImageJ and presented as mean fluorescent intensity (right). (F) Mitochondrial superoxide levels were quantified in stable DECR1-overexpressing LNCaP cells using MitoSOX red stain under basal or (G) linoleic acid (100 µM or 200 µM) conditions. Fluorescence intensity was quantified using flow cytometry and ROS levels were presented as mean fluorescent intensity. Cell viability of LNCaP cells after 48 hr DECR1 knockdown, treated with (H) deferoxamine (DFOA, 1.25 µM), (I) ferrostatin (Ferr-1, 1.25 µM), (J) erastin (10 µM), or (K) FIN56 (2 µM). Data in bar graphs are represented as the mean ± s.e.m. Statistical analysis was performed using two-tailed Student’s t-test (A-G) or one-way ANOVA, followed by Holm-Sidak's multiple comparisons test (H-K): ns, not significant, *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001. TAG: triacylglycerol; DAG: diacylglycerol; PC: phosphotidylcholine; PE: phosphotidylethanolamine; PI: phosphatidylinositol; PS: phosphotidylserine.
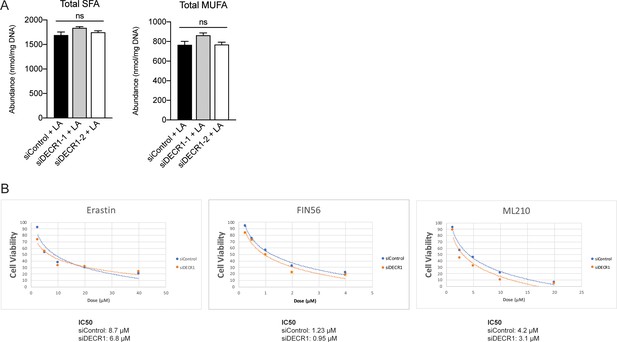
Depletion of DECR1 sensitizes prostate cancer cells to ferroptosis inducing agents.
(A) Abundance of total SFA and (B) MUFA species in phospholipids from control and DECR1 knockdown cells supplemented with linoleic acid (LA). (C) Abundance of total SFAs (left) and MUFAs (right) in Control and DECR1 knockdown cells supplemented with LA. (D) IC50 values of ferroptosis inducers were determined in Control and DECR1 knockdown cells. Statistical analysis was performed using two-tailed Student’s t-test: *p<0.05, **p<0.01 and ***p<0.001.
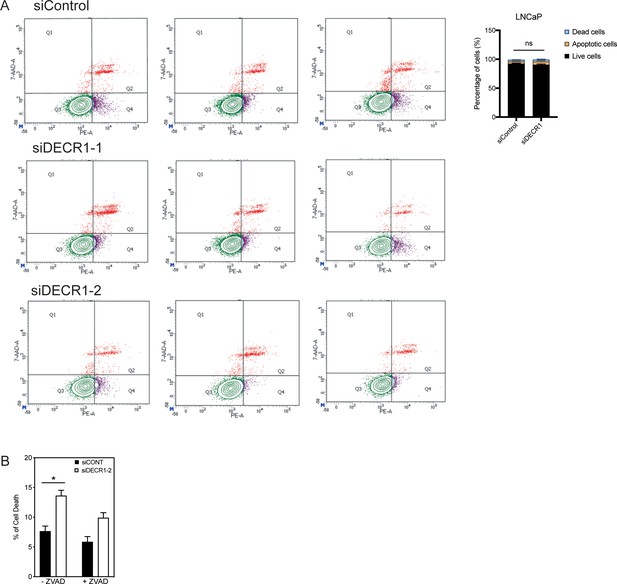
Targeting DECR1 does not induce apoptosis of prostate cancer cells.
(A) LNCaP cells were stained with AnnexinV-Phycoerythrin (PE)/7-Aminoactinomycin D (7-AAD) followed by flow cytometry analysis after 96 hr DECR1 knockdown to detect apoptotic cells. (B) Cell death after DECR1 knockdown in LNCaP cells treated with or without the caspase inhibitor (ZVAD) measured using trypan blue exclusion.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (M. musculus, male) | NOD scid Gamma (M. musculus, male,) Mice | The Jackson Laboratory/Interbred at SAHMRI Bioresources | NOD.Cg-Prkdcscid/J RRID:IMSR_JAX:001303 | |
| Cell line (Homo-sapiens) | LNCaP | ATCC | ATCC CRL-1740 RRID:CVCL_1379 | |
| Cell line (Homo-sapiens) | VCaP | ATCC | ATCC CRL-2876 RRID:CVCL_2235 | |
| Cell line (Homo-sapiens) | 22RV1 | ATCC | ATCC CRL-2505 RRID:CVCL_1045 | |
| Cell line (Homo-sapiens) | PNT1A | The European Collection of Authenticated Cell Cultures (ECACC) | Cat# 95012614 RRID:CVCL_2163 | |
| Cell line (Homo-sapiens) | PNT2 | The European Collection of Authenticated Cell Cultures (ECACC) | Cat# 95012613 RRID:CVCL_2164 | |
| Cell line (Homo-sapiens) | V16D | PMID:27046225 | Kind gift from Prof. Amina Zoubeidi | |
| Cell line (Homo-sapiens) | MR49F | PMID:27046225 | Kind gift from Prof. Amina Zoubeidi | |
| Transfected construct (Homo sapiens) | control siRNA | Dharmacon | D-001810-01-20 ON-TARGET plus Non-targeting siRNA #1 | transfected construct (human) |
| Transfected construct (Homo sapiens) | siDECR1-1 | Dharmacon | J-009642-05-0002 | transfected construct (human) |
| Transfected construct (Homo sapiens) | siDECR1-2 | Dharmacon | J-009642-06-0002 | transfected construct (human) |
| Transfected construct (Homo sapiens) | DECR1 shRNA lentivector | GenTargrt | LVS-1002 | Lentiviral construct to transfect and express the shRNA. |
| Transfected construct (Homo sapiens) | hDECR1 Overexpressing Lentivector | GenTargrt | LVS-2002 | Lentiviral construct to transfect and overexpress DECR1. |
| Transfected construct (Homo sapiens) | Negative control shRNA lentivector | GenTargrt | LVS-1002 | Lentiviral construct to transfect and express the shRNA. |
| Antibody | Anti-human β-Actin (Mouse monoclonal) | Sigma-Aldrich | Cat#: A5441 RRID:AB_476744 | (WB 1:2000) |
| Antibody | Anti-human-HSP90 (Rabbit Polyclonal) | Cell Signalling Technology | Cat#: 48745 RRID:CVCL_E547 | (WB 1:1000) |
| Antibody | Anti-human DECR1 (Rabbit Polyclonal) | Prestige Antibodies (Sigma-Aldrich) | Cat#: HPA023238 RRID:AB_1847587 | (WB 1:1000) (IHC: 1:500) |
| Antibody | Anti-human Malondialdehyde (Rabbit Polyclonal) | Abcam | Cat#: ab6463 RRID:AB_305484 | (WB 1:1000) |
| Antibody | Anti- human Androgen receptor (Rabbit Polyclonal) | Santa Cruz Biotechnology | Cat#: sc-816 RRID:AB_1563391 | (WB 1:1000) |
| Antibody | Anti-human PARP (Rabbit Polyclonal) | Cell Signalling Technology | Cat#: 9542 RRID:AB_592473 | (WB 1:1000) |
| Antibody | Anti-human Cytochrome C (Rabbit Polyclonal) | Abcam | Cat#: ab90529 RRID:AB_10673869 | (WB 1:2000) |
| Other | MitoTracker Red CMXRos | Thermo Fisher Scientific | Cat#: M7512 | ICC 1:1000 |
| Other | MitoSOX Red Mitochondrial Superoxide Indicator | Thermo Fisher Scientific | Cat#: M36008 | Flow Cytometry: 2.5 µM |
| Other | 3,3′-Diaminobenzidine (DAB) Enhanced Liquid Substrate System tetrahydrochloride | Sigma Aldrich | Cat#: D3939 | |
| Other | BODIPY-C11 | Thermo Fisher Scientific | Cat#: D3861 | Imaging: 5 µM |
| Antibody | Anti-human KI67 (Mouse monoclonal) | DAKO | Cat#: M7240 RRID:AB_2142367 | (IHC 1:200) |
| Antibody | Anti-human AR (Rabbit polyclonal) | Santa Cruz | Cat#: sc-816 RRID:AB_1563391 | (WB 1:1000) |
| Sequence-based reagent | DECR1_F | This paper | PCR primers | CTAAATGGCACAGCCTTCGT |
| Sequenced-based reagent | DECR1_R | This paper | PCR primers | AACCTGAACCAGTCTCAGCA |
| Sequence-based reagent | GAPDH_F | This paper | PCR primers | TGCACCACCAACTGCTTAGC |
| Sequenced-based reagent | GAPDH_R | This paper | PCR primers | GGCATGGACTGTGGTCATGAG |
| Sequence-based reagent | PPIA_F | This paper | PCR primers | GCATACGGGTCCTGGCAT |
| Sequence-based reagent | PPIA_R | This paper | PCR primers | ACATGCTTGCCATCCAACC |
| Sequence-based reagent | TUBA1B _F | This paper | PCR primers | CCTTCGCCTCCTAATCCCTA |
| Sequence-based reagent | TUBA1B _R | This paper | PCR primers | CCGTGTTCCAGGCAGTAGA |
| Sequence-based reagent | MKI67_F | This paper | PCR primers | GCCTGCTCGACCCTACAGA |
| Sequence-based reagent | MIK67_R | This paper | PCR primers | GCTTGTCAACTGCGGTTGC |
| Sequence-based reagent | L19_F | This paper | PCR primers | TGCCAGTGGAAAAATCAGCCA |
| Sequence-based reagent | L19_R | This paper | PCR primers | CAAAGCAAATCTCGACACCTTG |
| Sequence-based reagent | GUSB_F | This paper | PCR primers | CGTCCCACCTAGAATCTGCT |
| Sequence-based reagent | GUSB_R | This paper | PCR primers | TTGCTCACAAAGGTCACAGG |
| Sequence-based reagent | DECR1_F | This paper | ChIP-qPCR | TTCTGGAGCGCTAAGAGAGC |
| Sequence-based reagent | DECR1_R | This paper | ChIP-qPCR | AGGGCTTCATCTGACAGTGG |
| Sequence-based reagent | KLK3_F | This paper | ChIP-qPCR | GCCTGGATCTGAGAGAGATATCATC |
| Sequence-based reagent | KLK3_R | This paper | ChIP-qPCR | ACACCTTTTTTTTTCTGGATTGTTG |
| Sequence-based reagent | NC2_F | This paper | ChIP-qPCR | GTGAGTGCCCAGTTAGAGCATCTA |
| Sequence-based reagent | NC2_R | This paper | ChIP-qPCR | GGAACCAGTGGGTCTTGAAGTG |
| Chemical compound, drug | Etomoxir | Sigma Aldrich | Cat#: E1905 | |
| Chemical compound, drug | Dihydrotestosterone | Sigma Aldrich | Cas#: 521-18-6 | |
| Chemical compound, drug | Enzalutamide | Sapphire Bioscience | Cat#: S1250 | |
| Chemical compound, drug | Bovine-serum albumin | Bovostar | Cat#: BSAS-AU | |
| Chemical compound, drug | Linoleic acid | Sigma Aldrich | Cat#: L1376 | |
| Chemical compound, drug | Palmitic acid | Sigma Aldrich | Cat#: P0500 | |
| Chemical compound, drug | D-Luciferin | PerkinElmer | Cat#: 122799 | 3 mg/20 g |
| Chemical compound, drug | Deferoxamine | Sigma Aldrich | Cat#: D9533 | |
| Chemical compound, drug | Ferrostatin | Sigma Aldrich | Cat#: SML0583 | |
| Chemical compound, drug | Erastian | Sigma Aldrich | Cat#: E7781 | |
| Chemical compound, drug | ML210 | Tocris Bioscience | Cat#: 6429 | |
| Chemical compound, drug | FIN56 | Tocris Bioscience | Cat#: 6280 | |
| Chemical compound, drug | cell fractionation kit | Abcam | Cat#: ab109719 | |
| Chemical compound, drug | RNeasy RNA extraction kit | Qiagen | Cat#: 74136 | |
| Chemical compound, drug | iScript cDNA Synthesis kit | Bio-Rad | Cat#: 1708890 | |
| Chemical compound, drug | Seahorse XF Cell Mitochondrial Stress Test kit | Agilent | Cat#: 103015–100 | |
| Software, algorithm | GraphPad Prism | GraphPad Software, Inc | Prism V7 RRID:SCR_002798 | |
| Software, algorithm | R | R Development Core Team, 2019 | R version 3.6.2 RRID:SCR_001905 | |
| Software, algorithm | ReViSP | PMID:25561413 | ReViSP | Volume assessment of cancer spheroids |
| Software, algorithm | IVIS Spectrum In Vivo Imaging System | PerkinElmer | IVIS Spectrum In Vivo Imaging System RRID:SCR_018621 | Tumor volume analysis |
| Other | Lipofectamine RNAiMAX transfection reagent | Thermo Fisher Scientific | 13778075 | |
| Software, algorithm | ImageJ analysis software | NIH | ImageJ RRID:SCR_003070 | |
| Software, algorithm | TraceFinder v5.0 | Thermo Fisher Scientific | OPTON-30688 |