Rapamycin rejuvenates oral health in aging mice
Figures
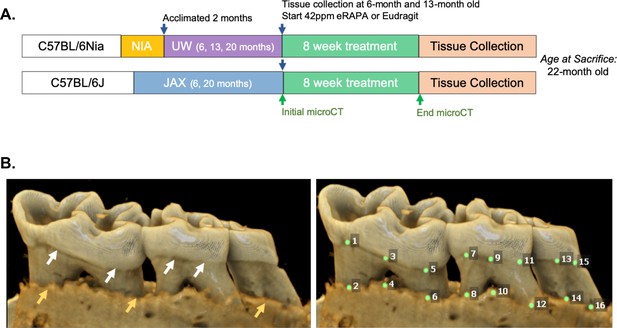
Cross-institution experimental design and assay for measuring periodontal bone loss.
(A) The NIA-UW colonies were received directly from the NIA Aged Rodent Colony at 4, 11, and 18 months, then acclimated for two months within the UW facilities (ARCF) until they reached 6 (Young), 13 (Adult), and 20 months (Old). The Young and Adult cohorts were harvested for oral tissues and microbiome. The Old cohorts were randomized and either given Eudragit or 42ppm eRAPA within the food for 8 weeks. For the JAX colonies, an initial microCT image was taken prior to the 8 week treatment and then a final microCT before harvest. All animals were harvested at the end of 8 weeks, ~22 months old. (B) Representative image of a mandible is shown. Periodontal bone loss was measured as distance from the cementoenamel junction (CEJ, white arrows) to the alveolar bone crest (ABC, orange arrows) on 16 predetermined landmarks on the buccal aspect of maxillary and mandibular periodontium. The CEJ-ABC distances were totaled for each mouse.
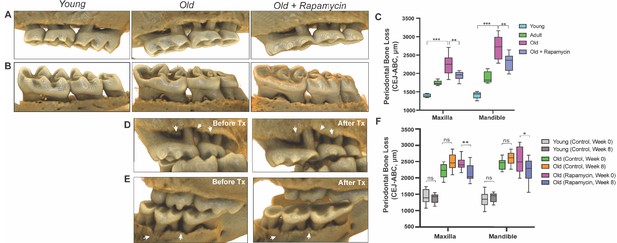
Rapamycin reverses age-associated periodontal bone loss (NIA-UW and JAX).
(A and B) Representative images of NIA-UW (A) maxillary and (B) mandibular teeth of Young, Old, and Old treated with 42ppm eRAPA (rapamycin) revealing age-associated periodontal bone loss. 8 weeks of rapamycin attenuated periodontal bone loss. (C) Box-and-whiskers plots shows median, 25th and 75th percentile with whiskers at the 5th and 95th percentile. Statistical analysis was completed using unpaired t-test, with p-values <0.05 were considered statistically significant. *p<0.05, **p<0.01, ***p<0.005 (D and E) Representative images of the (D) maxillary and (E) mandibular teeth from the same animal in the JAX cohort before treatment (labeled Old) and after 8 weeks of 42ppm eRAPA (labeled Old+Rapamycin). On both the maxilla and mandible, there is periodontal bone loss around and in-between the molars, but after 8 weeks of 42ppm eRAPA the bone loss is reversed. White arrowheads indicate areas of bone loss and bone loss reversal (F) Box-and-whiskers plots shows median, 25th and 75th percentile with whiskers at the 5th and 95th percentile. Longitudinal comparison was completed with the same animal at baseline or after 8 weeks with either eudragit (control) or 42ppm eRAPA (rapamycin). Statistical analysis was completed using paired t-test, with p-values <0.05 were considered statistically significant. *p<0.05, **p<0.01, ***p<0.005.
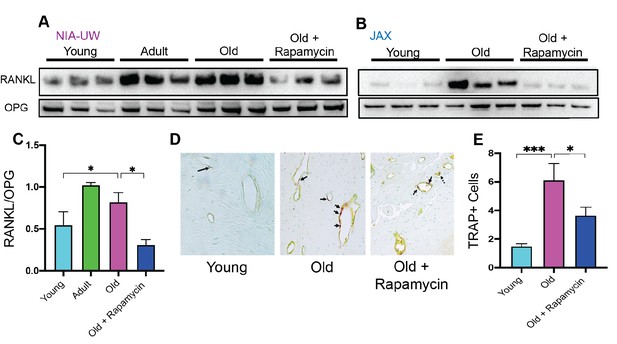
Rapamycin attenuates age-associated increase in RANKL expression and TRAP+ cells in periodontal bone.
(A and B) RANKL and OPG expression was determined by western blot analysis of total lysates from the periodontal bone of aged animals (Young, Adult, and Old) and Old animals treated for 8 weeks with 42ppm rapamycin (eRAPA). The periodontal bone within both the NIA-UW and JAX Colonies showed an increased expression of RANKL while 8 weeks of rapamycin treatment ameliorated the increased RANKL expression. Each lane represents individual periodontal bone samples. (C) Quantification of RANKL/OPG of the NIA-UW western blot analysis. (D) Representative histological sections of the alveolar bone furcation that have undergone TRAP azo-dye staining with FastGreen counterstain. (E) Enumeration of TRAP+ cells within the periodontal bone from two-independent observers reveals an increase number of TRAP+ cells with age and diminishes with rapamycin treatment. Statistical analysis was completed with unpaired t-test, with significance set to p<0.05. *p<0.05, **p<0.01, ***p<0.005.
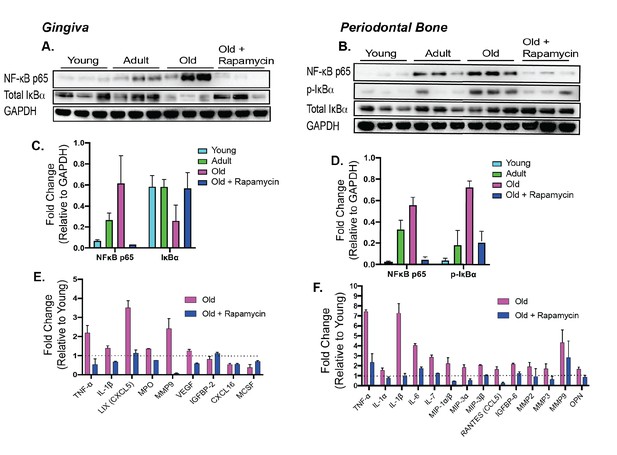
Rapamycin alters increased NF-κB expression and inflammatory cytokine profiles in periodontium.
NF-κB p65 and IκBα expression was determined by western blot analysis of total lysates from the gingiva (A,C) and periodontal bone (B,D) of control animals (Young, Adult, and Old) and Old animals treated for 8 weeks with rapamycin (42ppm eRAPA). GAPDH was used a loading control. Both in the aging gingiva and periodontal bone, there is an overall increased expression of NF-κB p65 with corresponding alteration of IκBα or p-IκBα. 8 weeks of 42ppm eRAPA treatment attenuates the changes seen with age. For the gingiva, each lane represents gingiva from animals co-housed (n = 2), and each lane for the periodontal bone western blot represents individual animals. (E and F) Protein expression levels of mouse cytokines and chemokines was determined by a spotted nitrocellulose membrane assay (Proteome Profiler Mouse, R and D Systems) by loading pooled samples from (E) gingiva and (F) periodontal bone of Young and Old (Control, Eudragit), and Old animals treated for 8 weeks with rapamycin (42ppm eRAPA). Data are shown per manufacture’s protocol, with fold-change relative to Young (Set to 1), expressed as mean ± SEM. All changes shown are statistically significant (p<0.05). CXCL16 and MCSF expression levels in (E) were not statistically significant.
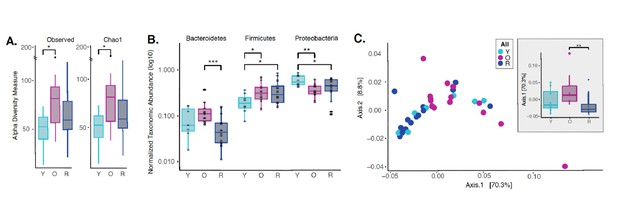
Rapamycin shifts aged oral microbiome towards young oral microbiome.
(A) Alpha diversity for all samples reveal significant differences between young (Y) and old (O) mice without rapamycin treatment (p<0.05). (B) Phylum level abundance using normalized agglomerated data show significant difference for the Bacteroidetes (p<0.001) in old (O) mice and old mice with rapamycin treatment (R) for all samples. Also, significant changes are observed in the Firmicutes (p<0.05) and Proteobacteria phylum (p<0.05, p<0.01) that is age and treatment dependent. (C) Principal coordinate analysis using weighted Unifrac distances reveal beta diversity in the rapamycin-treated old (R) groups clustered with the young (Y). (C, inner panel) A significant separation between old (O) and rapamycin-treated old (R) groups (p<0.01; Axis 1, 70.3%) was observed, but no significant difference between young (Y) and rapamycin-treated old (R) groups was observed. *p<0.05, **p<0.01,***p<0.001.
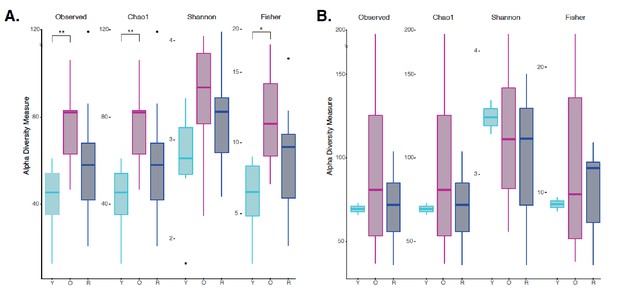
Independent Alpha Diversity Analysis for JAX and UW-NIA animals.
(A) Alpha Diversity measures for JAX Lab animals, including 6 month old (Y), 22 month old (O), and 22-month-old mice treated with rapamycin (R). (B) Alpha Diversity Measures for UW-NIA animals, including Y, O, and R mice. Significant differences between the different alpha diversity measures between Y and O mice, including Observed, Chao1, and Fisher, was observed among the JAX samples. No significant difference between taxonomic richness among Y and R mice was observed in Jax samples. This result was also observed independently in the UW-NIA animals. *p<0.05, **p<0.01.
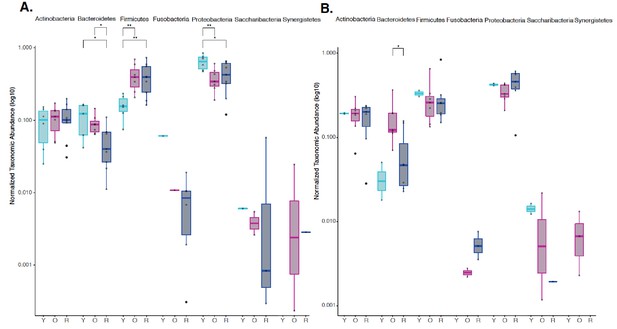
Independent phylum level abundance analysis for JAX and UW-NIA animals.
Normalized agglomerated data were used to evaluate phylum level abundances using an amplicon sequence variant (ASV) approach for both JAX and UW-NIA animals independently. (A) Significant differences in Bacteroidetes, Firmicutes, and Proteobacteria was observed in JAX animals, including 6- month-old (Y), 22-month-old (O), and 22-month-old mice treated with rapamycin (R). (B) Significant differences in Bacteroidetes were observed in UW-NIA animals. *p<0.05, **p<0.01.
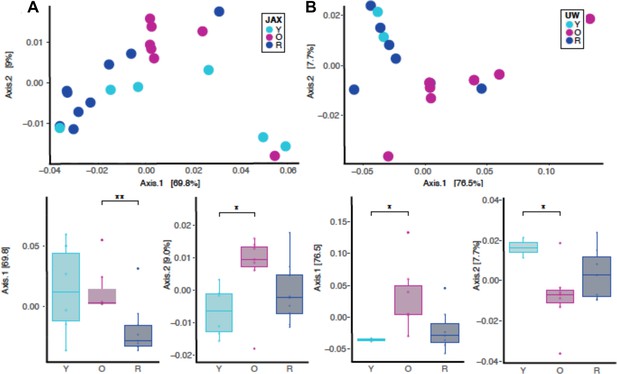
Independent beta diversity for JAX and UW-NIA animals by principal coordinate analysis using weighted unifrac distances.
(A) Beta diversity for only JAX samples there was significant separation between O and R (p<0.01; Axis 1, 69.8%) as well as between Y and O groups (p<0.05; Axis 2, 9.0%). (B) Beta diversity for only the UW samples also reveal significant difference between Y and O groups (p<0.05; Axis 1, 76.5%; p<0.05; Axis 2, 7.7%). *p<0.05, **p<0.01.
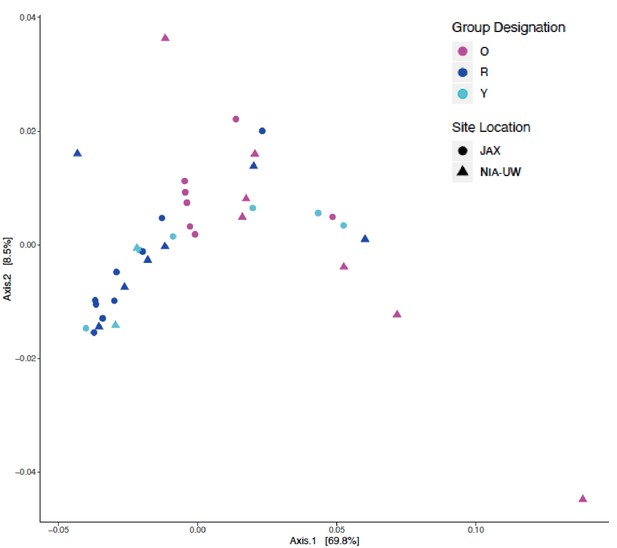
Combined beta diversity for JAX and NIA-UW animals by principal coordinate analysis using weighted unifrac distances by site location and group designation.
No distinct differences in clustering by site location was observed. A PERMANOVA (nperm = 999) analysis was performed using the Adonis package (v 2.5–6) as part of the R suite comparing site location (JAX, NIA-UW) and group designation (Y = Young, O = Old, R = Old + Rapa) between JAX and NIA-UW mice used in this study via Bray Curtis Distances (p=0.341).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background(M. musculus) | C57BL/6NIA | NIA Aged Rodent Colony | PMID:27549339 | |
| Strain, strain background(M. musculus) | C57BL/6J | Jackson Laboratory | RRID:IMSR_JAX:000664 | |
| Chemical compound, drug | Rapamycin | Rapamycin Holdings | Amount based upon active rapamycin content to provide 42 parts per million concentration in chow. | |
| Chemical compound, drug | Eudragit | Rapamycin Holdings | ||
| Chemical compound, drug | RIPA Lysis and Extraction Buffer | ThermoFisher Scientific | Cat#: 89901 | |
| Chemical compound, drug | HALT Protease Inhibitor Cocktail | ThermoFisher Scientific | Cat#: 78438 | |
| Chemical compound, drug | HALT Phosphatase Inhibitor Cocktail | ThermoFisher Scientific | Cat#: 78420 | |
| Chemical compound, drug | Restore Plus Stripping Buffer | Thermfisher Scientific | Cat#: 46430 | |
| Antibody | anti -NFkBp65(Rabbit Monoclonal) | Cell Signaling | Cat#: 8242 | WB (1:1000) |
| Antibody | anti-phospho-IκBα (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc8404 | WB (1:1000) |
| Antibody | Anti- IκBα (Rabbit Monoclonal) | Abcam | Cat#: 32518 | WB (1:1000) |
| Antibody | anti-GAPDH (Rabbit monoclonal) | Cell Signaling | Cat#: 5174 | WB (1:1000) |
| Antibody | anti-RANKL (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc377079 | WB (1:1000) |
| Antibody | anti-OPG (Goat polyclonal) | R and D Systems | AF459 | WB (1:1000) |
| Antibody | anti-IgGκ (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc516102 | WB (1:10000) |
| Antibody | anti-rabbit IgG (Donkey polyclonal) | Thermo Fisher Scientific | Cat#: 31458 | WB (1:10000) |
| Antibody | anti-goat IgG (Donkey polyclonal) | Abcam | Cat#: ab97110 | WB (1:10000) |
| Commercial assay, kit | Proteome Profiler Mouse XL Cytokine Array | R and D Systems | Cat#: ARY028 | |
| Commercial assay, kit | Acid Phosphatase, Leukocyte (TRAP) Kit | Millipore Sigma | Cat#: 387A-1KT | |
| Commercial assay, kit | QIAamp DNA Microbiome Kit | Qiagen | Cat#: 51704 | |
| Commercial assay, kit | DNA Clean and Concentrator Kit | Zymo Research | Cat#: D4014 | |
| Commercial assay, kit | KAPA HiFi HotStart ReadyMix | KAPA Biosystems | Cat#: KK2601 | |
| Commercial assay, kit | Nextera XT Index Kit V2 | Illumina | Set A: FC-131–2001 Set D: FC-131–2004 | |
| Commercial assay, kit | AMPure XP Beads | Agencourt | A63881 | |
| Commercial assay, kit | SequalPrep Normalization Kit | Invitrogen | A1051001 | |
| Commercial assay, kit | TapeStation 4200 High Sensitivity D1000 assay | Agilent Technologies | G2991AA | |
| Commercial assay, kit | Tapestation Reagents | Agilent Technologies | 5067–5585 | |
| Commercial assay, kit | High Sensitivity D1000 ScreenTape | Agilent Technologies | 5067–5584 | |
| Commercial assay, kit | Qubit High Sensitivity dsDNA assay | ThermoFisher Scientific | Q32854 | |
| Commercial assay, kit | MiSeq Reagent Kit v3 (600 cycle) | Illumina | Cat#: MS-102–3003 | |
| Commercial assay, kit | PhiX Control Kit v3 | Illumina | Cat#: FC-110–3001 | |
| Software, Algorithm | Qiime2 | https://qiime2.org/ | V.2019.1 | |
| Software, Algorithm | DADA2 Package | PMID:27214047 | ||
| Software, Algorithm | Human Oral Microbiome Database (HOMD) | Homd.org PMID:30534599 | v. 15.1 | |
| Software, Algorithm | R-studio | https://rstudio.com/ | RRID:SCR_000432 | Version 3.5.3 |
| Software, Algorithm | Phyloseq | PMID:23630581 | RRID:SCR_013080 | |
| Software, Algorithm | Clustvis | PMID:25969447 | RRID:SCR_017133 | |
| Software, Algorithm | Ggplot2 | https://www.springer.com/gp/book/9780387981413 | RRID:SCR_014601 | https://www.springer.com/gp/book/9780387981413 |
| Software, Algorithm | Ampvis2 | http://dx.doi.org/10.1101/299537v1 | ||
| Software, Algorithm | vegan | https://cran.r-project.org, https://github.com/vegandevs/vegan | RRID:SCR_011950 | |
| Software, Algorithm | Ade4 | https://www.jstatsoft.org/article/view/v086i01 | ||
| Software, Algorithm | Bioinformatic scripts and microbiome data used in analysis | This paper | https://github.com/kkerns85/Rapamycin_rejuvenates_oral_health_in_aging_mice.git. | |
| Software, Algorithm | R Markdown | This paper | https://rpubs.com/kkerns85/Rapamycin_Rmrkdown | |
| Software, Algorithm | Graphpad Prism | Graphpad (graphpad.com) | RRID:SCR_002798 | Version 8.4 |





