R-spondins engage heparan sulfate proteoglycans to potentiate WNT signaling
Figures
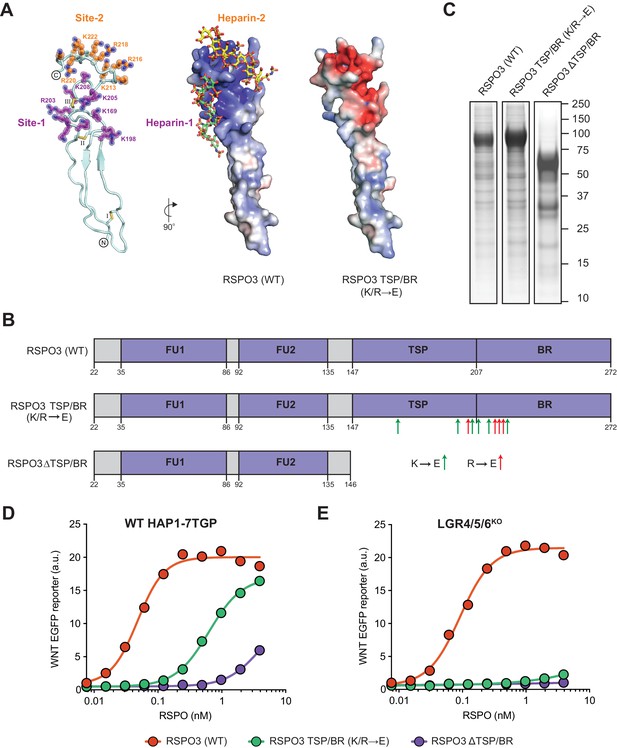
Mutations in the putative HSPG-binding surface of the RSPO3 TSP/BR domain impair its ability to amplify WNT signaling.
(A) Homology model of the isolated TSP/BR domain of human RSPO3 (residues valine 146 - isoleucine 232; UniprotKB Q9BXY4). Left panel: cartoon representation of the protein backbone, with side-chains shown for lysine (K) and arginine (R) residues. N- and C-termini are circled and disulfide bonds are labelled with roman numerals. Two positively charged HS-binding grooves, revealed by the electrostatic potential map (middle panel), are labeled Site-1 and Site-2. K and R residues lining Site-1 and Site-2 are colored purple and orange, respectively, and residues mutated to glutamic acid (E) in the RSPO3 TSP/BR (K/R→E) mutant are labeled. Middle panel: the electrostatic potential map calculated with the Adaptive Poisson-Boltzmann Solver (Jurrus et al., 2018) is displayed from −10 kT/e to +10 kT/e (blue: positively charged; red: negatively charged). Two virtually docked 8-mer heparin chains (estimated free energies are −13.9 and −14.8 kcal/mol for Site-1 and Site-2, respectively) are depicted in stick representation (red: oxygen, blue: nitrogen, gold: sulphur, green: carbon of heparin-1, yellow: carbon of heparin-2). Right panel: electrostatic potential map of the RSPO3 TSP/BR (K/R→E) mutant. The orientation of the middle and right panels is 90° rotated clockwise around the y-axis relative to the left panel. (B) Schematic representation of WT and mutant RSPO3 variants showing the domains and mutations present in these proteins. The N-terminal HA and the C-terminal Fc and 1D4 tags present in these constructs are not shown. Amino acid numbers for human RSPO3 (UniProtKB Q9BXY4) are indicated below. Green and red arrows show K→E and R→E mutations, respectively, introduced into the TSP/BR domain of the RSPO3 TSP/BR (K/R→E) mutant. Polypeptide lengths are drawn to scale. See Supplementary file 1 for the nucleotide sequences of these constructs. (C) Coomassie-stained polyacrylamide gels showing equal volumes of the three purified RSPO3 proteins used in (D) and (E). Molecular weight standards in kilodaltons (kDa) are indicated to the right. (D) and (E) Dose-response curves for the indicated purified RSPO3 variants in WT HAP1-7TGP (D) and LGR4/5/6KO (E) cells, in the presence of 1.43% WNT3A CM. Each circle represents the median WNT reporter fluorescence from 2,500 cells. Dose-response curves were fitted to the data using non-linear regression as described in Materials and methods. Where possible, half-maximal effective concentrations (EC50, all in nM) were derived from curve fits and are as follows. RSPO3 (WT): 0.048 ± 0.005 in (D) and 0.092 ± 0.007 in (E). RSPO3 ΔTSP/BR (K/R→E): 0.63 ± 0.02 in (D). RSPO3 ΔTSP/BR: 3.93 ± 1.0 in (D).
Movie showing a structural homology model of the WT RSPO3 TSP/BR domain and the K/R→E mutant.
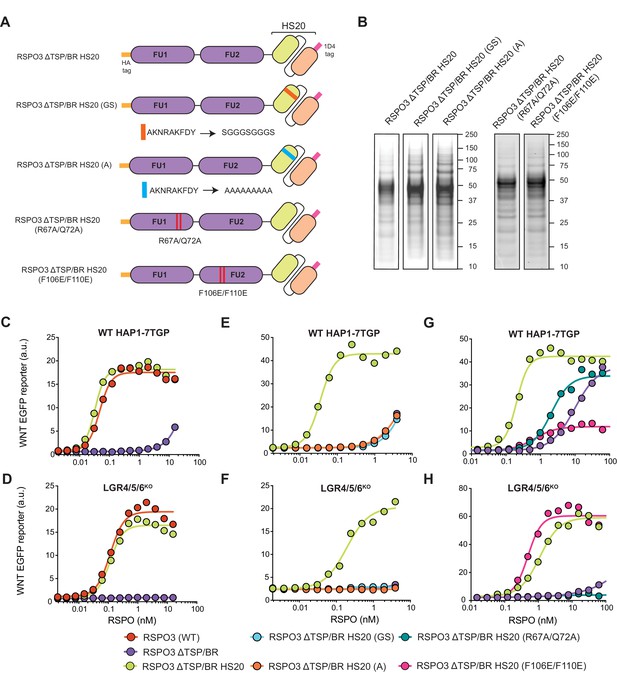
The TSP/BR domain of RSPO3 can be replaced by HS20, an scFv that binds HS chains, without compromising activity.
(A) Cartoons depicting synthetic proteins in which the FU1 and FU2 domains of RSPO3 were fused to HS20, an scFv that binds to HS chains. In two control proteins designed to abolish HS binding, the CDR3 loop of HS20 was replaced with a glycine-serine (GS) or a poly-alanine (A) linker. In two other proteins, we introduced mutations that diminish binding of the FU1 domain to ZNRF3/RNF43 (R67A/Q72A) or of the FU2 domain to LGRs (F106E/F110E). See Supplementary file 1 for the nucleotide sequences of these constructs. (B) Coomassie-stained polyacrylamide gels showing equal volumes of the five purified RSPO3 proteins used in (C–H). Molecular weight standards in kDa are indicated to the right of each gel. (C–H) Dose-response curves for the indicated purified RSPO3 variants in WT HAP1-7TGP (C, E and G) and LGR4/5/6KO (D, F and H) cells, in the presence of 1.43% WNT3A CM. Each circle represents the median WNT reporter fluorescence from 2,500 cells. Dose-response curves were fitted to the data using non-linear regression as described in Materials and methods. Where possible, EC50 values (all in nM) were derived from curve fits and are as follows. RSPO3 (WT): 0.046 ± 0.004 in (C) and 0.11 ± 0.015 in (D). RSPO3 ΔTSP/BR: 9.8 ± 0.92 in (G). RSPO3 ΔTSP/BR HS20: 0.032 ± 0.002 in (C), 0.11 ± 0.013 in (D), 0.035 ± 0.003 in (E), 0.18 ± 0.03 in (F), 0.20 ± 0.017 in (G) and 1.1 ± 0.12 in (H). RSPO3 ΔTSP/BR HS20 (R67A/Q72A): 2.0 ± 0.26 in (G). RSPO3 ΔTSP/BR HS20 (F106E/F110E): 0.68 ± 0.014 in (G) and 0.49 ± 0.07 in (H).
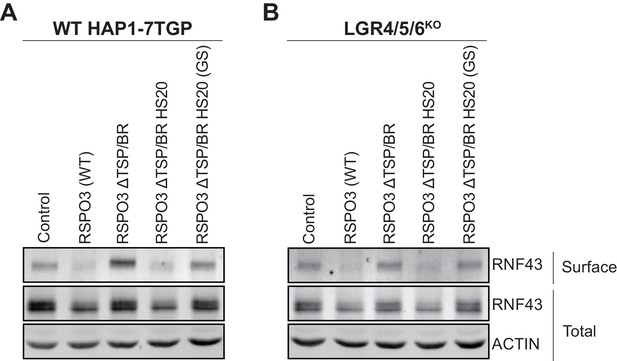
Signaling by WT RSPO3 and RSPO3 ΔTSP/BR HS20 causes internalization and degradation of RNF43 in the presence and absence of LGRs.
Cell-surface RNF43-Flag (captured by cell-surface biotinylation; see Materials and methods), total RNF43-Flag and total actin were visualized by immunoblotting after treatment of WT HAP1-7TGP (A) or LGR4/5/6KO (B) cells with the indicated RSPO3 variants.
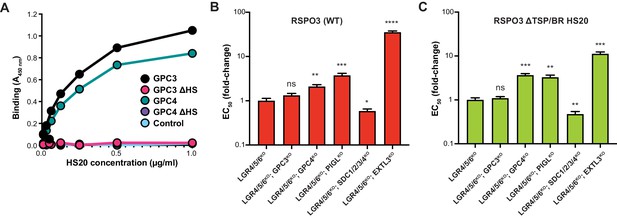
RSPO3 ΔTSP/BR HS20 engages the HS chains of multiple HSPGs.
(A) Binding of HS20 to GPC3 or GPC4. An ELISA plate was coated with GPC3, GPC4, or mutant versions lacking HS chains (GPC3ΔHS or GPC4ΔHS). HS20 scFv was added at increasing concentrations (x-axis), followed by a goat anti-human IgG horseradish peroxidase conjugate. Bound HS20 was quantified with 3,3′,5,5′-tetramethylbenzidine detection reagent by measuring absorbance at 450 nm (y-axis). An irrelevant Fc-fusion protein (CD276-hFc, labeled ‘Control’) was used as an antigen control. (B–C) Fold-change in the EC50 of WT RSPO3 (B) or RSPO3 ΔTSP/BR HS20 (C) in LGR4/5/6KO cells and clonal derivatives in which different HSPGs were depleted through the indicated gene disruptions (see Results and Materials and methods for details). Since deletion of some GPCs in HAP1 cells leads to impaired WNT reception (Lebensohn et al., 2016; Lebensohn and Rohatgi, 2018), we first determined a concentration of WNT3A CM that yielded similar levels of WNT reporter activity in all the cell lines and then titrated RSPO3 variants to determine their EC50 values. Error bars denote the standard error of the curve fits used to calculate the EC50. The statistical significance of differences between the measured EC50 values in LGR4/5/6KO cells and clonal derivatives thereof was determined by a two-tailed, unpaired t-test, and is indicated as **** (p<0.0001), *** (p<0.001), ** (p<0.01), * (p<0.05) or ns (non-significant).
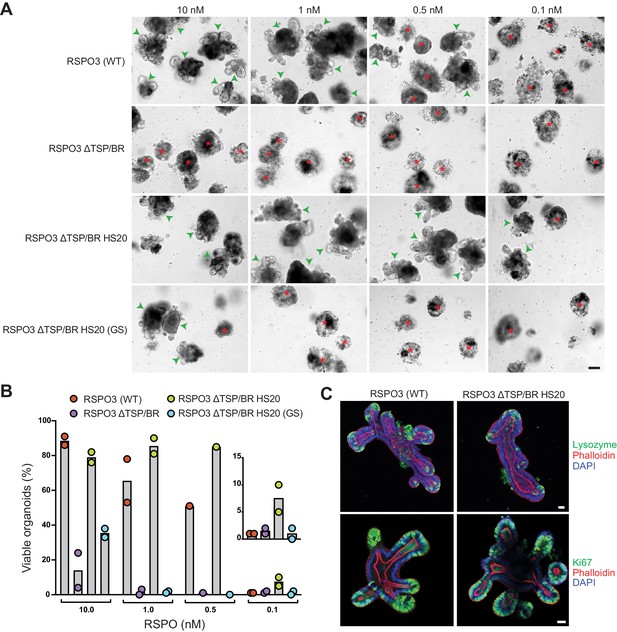
The interaction with HSPGs potentiates the ability of RSPO3 to support the growth of small intestinal organoids.
(A) Bright-field microscopy images of B6 mouse small intestinal organoids grown in EN medium supplemented with various purified RSPO3 proteins at the indicated concentrations. Green arrowheads indicate viable, crypt-containing organoids and red asterisks mark dead organoids. The scale bar in the bottom right image represents 100 μm. (B) The viability of organoids grown at various concentrations of the indicated RSPO3 variants was quantified from images of the type shown in (A). Each circle represents the quantification of an independent experiment. The inset (right) shows a magnified view of organoid viability at a ligand concentration of 0.1 nM. (C) Confocal microscopy images of mouse small intestinal organoids grown in medium supplemented with 1 nM of the indicated purified RSPO3 proteins. DAPI (blue) stains nuclei, phalloidin (red) stains actin filaments at the apical surface of cells, lysozyme (green, top row) stains Paneth cells and Ki67 (green, bottom row) stains proliferating cells. For both images in each row, the scale bars in the right image represent 30 μm.
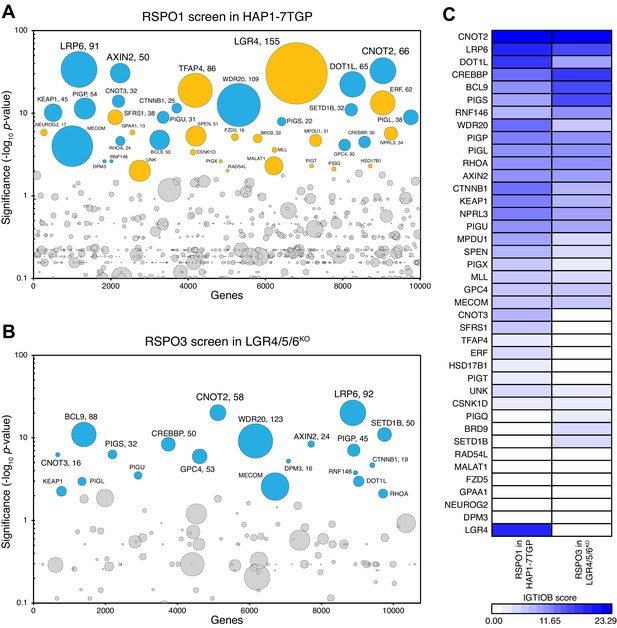
A haploid genetic screen for regulators of RSPO3 signaling in cells lacking LGRs.
(A–B) Circle plots depicting genes enriched for inactivating gene trap (GT) insertions in the screen for regulators of RSPO1-potentiated WNT signaling in HAP1-7TGP cells (A) and in the screen for regulators of RSPO3-potentiated WNT signaling in LGR4/5/6KO cells (B). The y-axis indicates the significance of inactivating GT insertion enrichment in the sorted vs. the control cells (expressed in units of -log10FDR-corrected p-value) and the x-axis indicates genes (in random order) for which inactivating GT insertions were mapped in the sorted cells (see Materials and methods). Genes with FDR-corrected p-value<0.01 are labeled and colored cyan if they reached this level of significance in both screens, or orange if they reached this level of significance in only one of the two screens. The diameter of each circle is proportional to the number of unique inactivating GT insertions mapped in the sorted cells, which is also indicated next to the gene name for the most significant hits with FDR-corrected p-values<10−4. See Supplementary file 4 for ranked lists of hits from both screens. (C) Heat map comparing the two screens. Genes enriched for inactivating GT insertions (FDR-corrected p-value<0.01) in at least one of the two screens (Supplementary file 5) were clustered based on their IGTIOB score in each screen (see Materials and methods).
Additional files
-
Supplementary file 1
Nucleotide sequences of RSPO3 WT, mutant and HS20-fusion constructs used in this study.
The name of the encoded protein and the length (in bp) of the nucleotide sequence is indicated. RSPO3 (WT), RSPO3 TSP/BR (K/R→E) and RSPO3 ΔTSP/BR were cloned into pHLsec-HA-Tev-Fc-Avi-1D4. RSPO3 ΔTSP/BR HS20, RSPO3 ΔTSP/BR HS20 (GS), RSPO3 ΔTSP/BR HS20 (A), RSPO3 ΔTSP/BR HS20 (R67A/Q72A) and RSPO3 ΔTSP/BR HS20 (F106E/F110E) were cloned into pHLsec-HA-Avi-1D4. Bases in lowercase overlap the sequences upstream of the unique AgeI sites and downstream of the unique KpnI sites in the pHLsec-HA-Tev-Fc-Avi-1D4 and pHLsec-HA-Avi-1D4 vectors, respectively. Bases in uppercase encode RSPO3 WT, mutant and HS20-fusion proteins. For mutant constructs, mutated bases are indicated in red and the resulting altered codons are underlined. For HS20-fusion constructs, bases encoding a codon-optimized glycine/serine linker (STGGSGGSGGSG) are indicated in light blue.
- https://cdn.elifesciences.org/articles/54469/elife-54469-supp1-v1.docx
-
Supplementary file 2
List of oligonucleotides and primers used to generate and characterize clonal cell lines engineered using CRISPR/Cas9.
The names and sequences of pairs of oligonucleotides encoding sgRNAs (which were cloned into pX458-mCherry) are shown in the first and second columns, respectively. The names and sequences of pairs of PCR primers used to amplify corresponding genomic regions flanking sgRNA target sites are shown in the third and fourth columns, respectively. The names and sequences of primers used to sequence the amplified target sites are shown in the fifth and sixth columns, respectively.
- https://cdn.elifesciences.org/articles/54469/elife-54469-supp2-v1.xlsx
-
Supplementary file 3
Description of engineered cell lines used in this study.
Clonal cell lines derived from HAP1-7TGP in which multiple genes were targeted using CRISPR/Cas9 (see Materials and methods) are described. The ‘Cell Line Name’ column indicates the generic name used throughout the manuscript to describe the genotype and the ‘Clone #' column identifies the individual clone used. The figures in which each clone was used are also indicated. The ‘CRISPR guide’ column indicates the name of the guide or guides used, which is the same as that of the oligonucleotides encoding sgRNAs (see Materials and methods and Supplementary file 2). The ‘Genomic Sequence’ column shows 80 nucleotides of genomic sequence (5’ relative to the gene is to the left) surrounding the target site; when two adjacent sites within the same gene were targeted, 80 nucleotides of genomic sequence surrounding each target site are shown and the number of intervening bp that are not shown between the two sites is indicated in parenthesis. Each cell line made using a different set of CRISPR guides is separated by a horizontal spacer, under which the reference (WT) genomic sequence (obtained from RefSeq) targeted by each CRISPR guide is indicated. Within this reference genomic sequence, the guide sequence is colored blue and the site of the double strand cut made by Cas9 is between the two underlined bases. Sequencing results for individual mutant clones are indicated below the reference sequence. Mutated, inserted or deleted nucleotides are colored red (dashes represent deleted nucleotides and ellipses are used to indicate that a deletion continues beyond the 80 nucleotides of sequence shown) and the nature of the mutation, the resulting genotype and any pertinent observations are also described. The CRISPR guide or guides used to target different genes, as well as the genomic sequence, mutation, genotype and observations pertaining to each of the targeted genes are designated ‘1’, ‘2’, ‘3’ and ‘4’ in the column headings and are shown under horizontal spacers of different colors.
- https://cdn.elifesciences.org/articles/54469/elife-54469-supp3-v1.xlsx
-
Supplementary file 4
Ranked lists of hits from screens.
Genes containing at least one inactivating GT insertion in the population of sorted cells from each of the two genetic screens described in this work are listed in separate spreadsheets (the screen name is indicated on the tab of each spreadsheet), and are ranked based on the significance of inactivating GT insertion enrichment (p-value) in the sorted vs. the unsorted (control) cells. For the unsorted cells, the number of all GT insertions in genes (regardless of orientation) is indicated for the complete dataset and for each gene (column B). For the sorted cells, the total number of inactivating GT insertions in genes (sense and antisense insertions in exons and sense insertions in introns, column C), as well as the number of sense or antisense GT insertions in exons or in introns (columns D-G), is indicated for the complete dataset and for each gene. Three measures of GT insertion enrichment are shown: the p-value and the FDR-corrected p-value (both derived from columns B and C), the latter of which was used to generate the circle plots in Figure 6A and B, and the Intronic GT Insertion Orientation Bias (IGTIOB) score (derived from columns F and G), used to generate the heat map in Figure 6C. See Materials and methods for details.
- https://cdn.elifesciences.org/articles/54469/elife-54469-supp4-v1.xlsx
-
Supplementary file 5
List of significant hits included in the comparative analysis between screens.
Genes used to generate the heat map in Figure 6C, comparing the RSPO1 screen in WT HAP1-7TGP cells (Figure 6A) and the RSPO3 screen in LGR4/5/6KO cells (Figure 6B), are shown. Genes enriched for inactivating GT insertions (FDR-corrected p-value<0.01) in at least one of the two screens are shown, and the FDR-corrected p-value and IGTIOB score for each gene in each screen is indicated. Genes are shown in the same order as in the heat map in Figure 6C, clustered based on their IGTIOB scores (see Materials and methods for details).
- https://cdn.elifesciences.org/articles/54469/elife-54469-supp5-v1.xlsx
-
Supplementary file 6
Key resources table.
- https://cdn.elifesciences.org/articles/54469/elife-54469-supp6-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54469/elife-54469-transrepform-v1.docx





