Hedgehog signaling is required for endomesodermal patterning and germ cell development in the sea anemone Nematostella vectensis
Figures
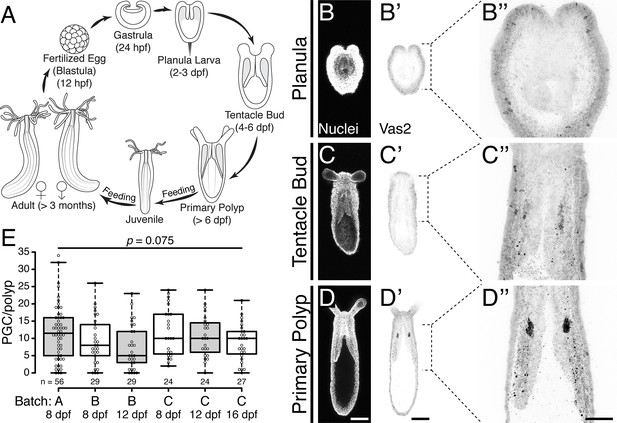
Putative Nematostella PGCs clusters are specified during metamorphosis.
(A) Diagram depicting the Nematostella life cycle. (B–D’’) During the transition from tentacle bud stage larvae to metamorphic primary polyps, Vas2 expression is gradually enriched in the primary mesenteries in two endomesodermal cell clusters in proximity to the pharynx. (E) Quantification of PGC numbers on 8, 12 or 16 dpf from three spawning batches. ANOVA analysis did not show significant differences among mean PGC number in primary polyps on different days post-fertilization. Center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; data points are plotted as open circles. Scale bar = 100 µm in D and D’; 50 µm in D’’. B–D’ are at the same scale; B’’–D’’ are at the same scale.
-
Figure 1—source data 1
PGC numbers on 8, 12 or 16 dpf from three spawning batches.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig1-data1-v1.csv
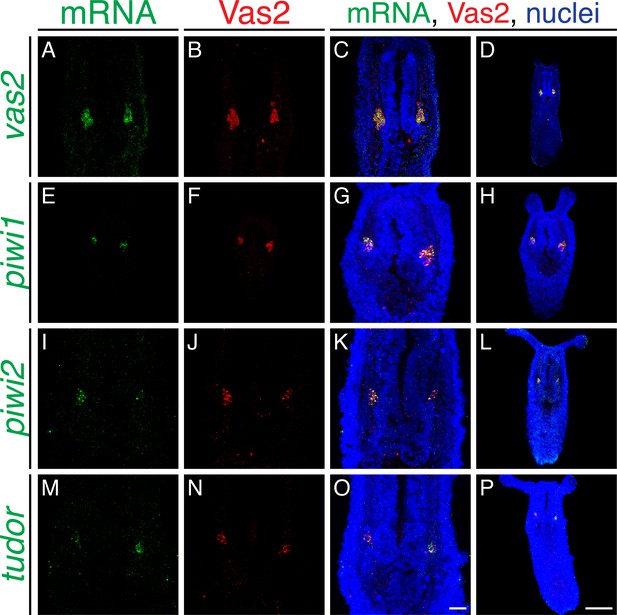
Conserved germline-related genes are expressed in putative PGC clusters.
Fluorescent in situ hybridization (FISH, green) of vas2 (A), piwi1 (E), piwi2 (I) and tudor (M) show enhanced expression within PGC clusters (Vas2-immunofluorescence, red). Scale bar = 20 µm in O; 100 µm in P. A-C, E–G, I–K, and M–O are at the same scale; D, H, L, and P are at the same scale.
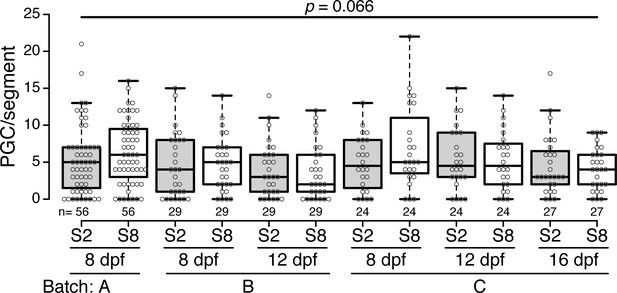
PGC numbers in the segment 2- and segment 8-associated primary mesenteries at three different timepoints post fertilization.
Results are shown for three spawning batches. These data compose Figure 1E.
-
Figure 1—figure supplement 2—source data 1
PGC numbers in the segment 2- and segment 8-associated primary mesenteries at three different timepoints post fertilization.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig1-figsupp2-data1-v1.csv
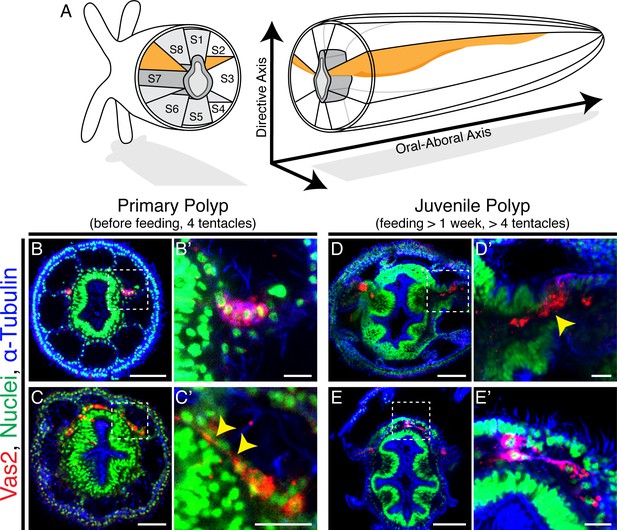
Putative Nematostella PGCs delaminate through epithelial-mesenchymal transition (EMT) and appear to migrate to non-primary mesenteries.
(A) Schematic diagram of Nematostella polyp anatomy depicts the pharynx and mesentery arrangements at the pharyngeal level. The eight mesenteries (two primary mesenteries in orange and six non-primary mesenteries in light gray) harbor gonad epithelium, muscle and digestive tissue. The internal structures of Nematostella are arrayed around the pharynx (dark gray). The Directive and Oral-Aboral axes are indicated. Segment nomenclatures follow He et al., 2018. (B–B’) Paired clusters of putative PGCs labeled by Vas2 immunofluorescence (red) initially exhibit epithelial charateristics. (C–C’) Putative PGCs from >10 dpf primary polyps appear to stretch their cell bodies basally (yellow arrowheads). (D–D’) Following nutrient intake, putative PGCs delaminate into the mesoglea through an apparent EMT (yellow arrowhead). (E–E’) In the mesoglea, these Vas2+ cells exhibit fibroblast-like morphology and are detected between mesenteries at the level of the aboral pharynx. Scale bar = 50 µm in B, D, and E; 20 µm in C; 10 µm in B’, C’, D’, and E’.
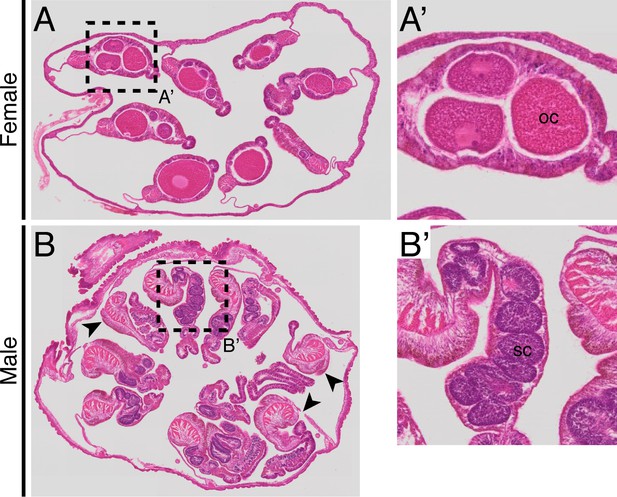
Development of oocytes or sperm in the mesenteries of female (A–A’) and male (B–B’) adult polyps.
Oocytes (oc) or sperm cysts (sc) are observed in adult cross-sections. Note that in (B), sperm cysts of some mesenteries (arrowheads) localize out of the cross-sectional plane.
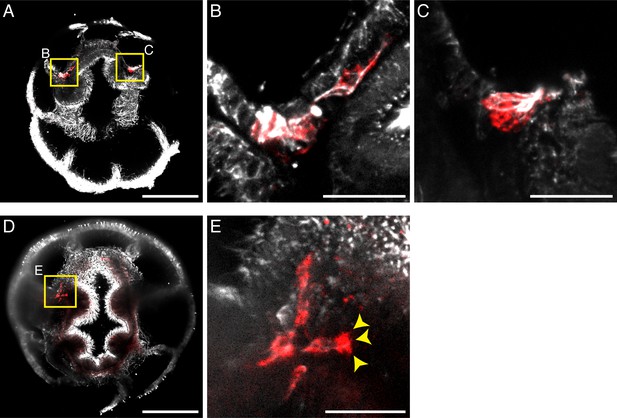
Migratory PGCs show fibroblast-like morphology and pseudopodia.
(A–E) Representative images show the microtubule cytoskeleton (α-Tubulin, grey) of PGCs (red) and their pseudopodia-like protrusions (yellow arrowheads, E). Scale bar = 100 µm in A and D; 20 µm in B–C and E.
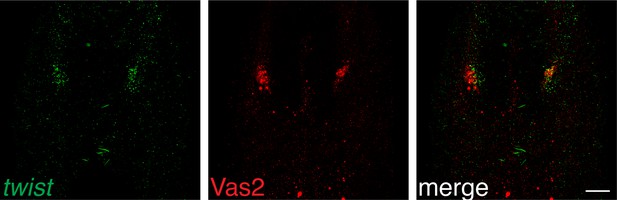
twist expression in putative PGCs suggests their migratory potential.
Scale bar = 20 µm.
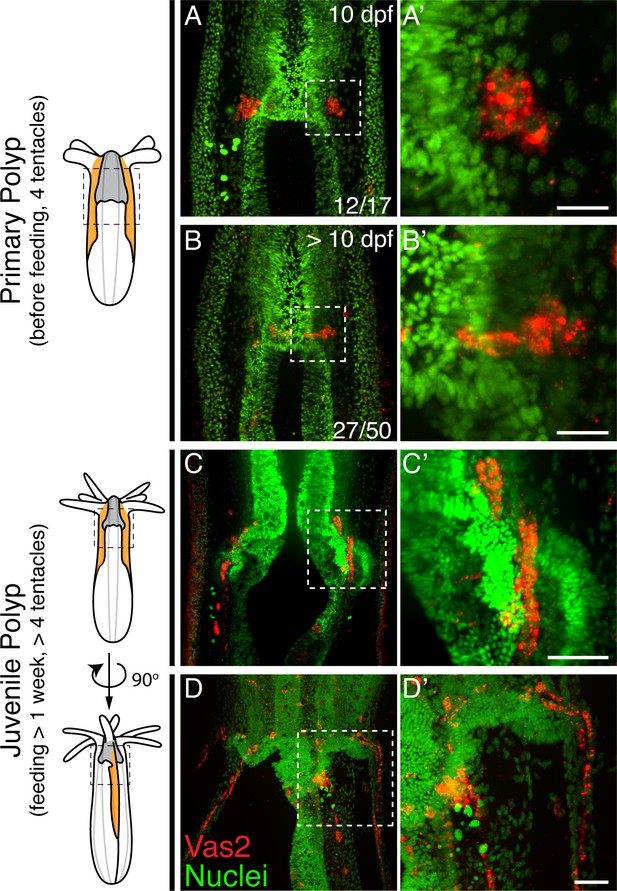
Nematostella PGCs migrate aborally to the gonad rudiments during the juvenile stage.
(A–A’) The majority of young primary polyps (≤10 dpf) exhibit two PGC clusters (Vas2+, red) in close proximity to the pharynx. (B–B’) In more mature primary polyps (>10 dpf), some PGCs elongate and localize between the mesenteries. (C–C’) Following feeding, putative PGCs spread aborally into the gonad rudiments. (D–D’) A juvenile polyp viewed 90 degrees from the orientation of C, showing aborally migrating PGCs in non-primary mesenteries. Scale bar = 10 µm in A’ and B’; 20 µm in C’ and D’.
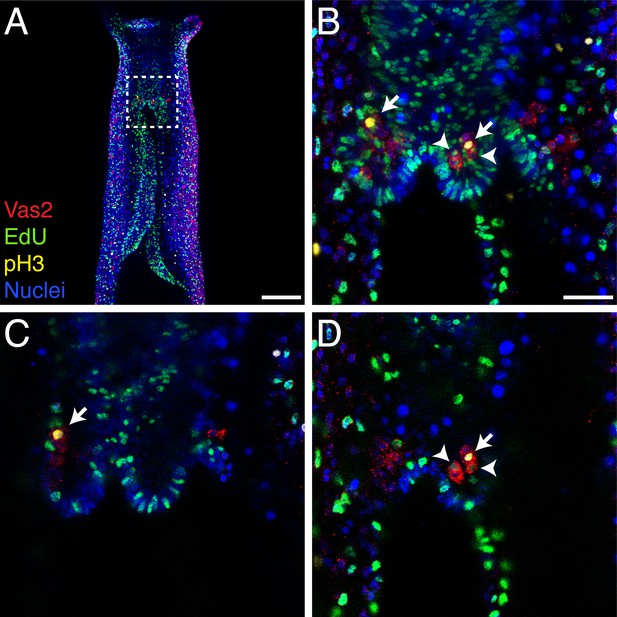
PGCs are proliferative in juvenile polyps.
(A) A representative six tentacle-stage juvenile stained with anti-Vas2 (red), EdU (green), anti-Phospho-Histone H3 (pH3, yellow) and nuclei (blue). (B–D) Enlarged view of the boxed area in A. Proliferative PGCs accumulate EdU (arrowheads) or pH3 (arrow) in the nuclei. (C and D) Selected single focal planes from B. Scale bar = 100 µm in A; 20 µm in B. B–D are at the same scale.
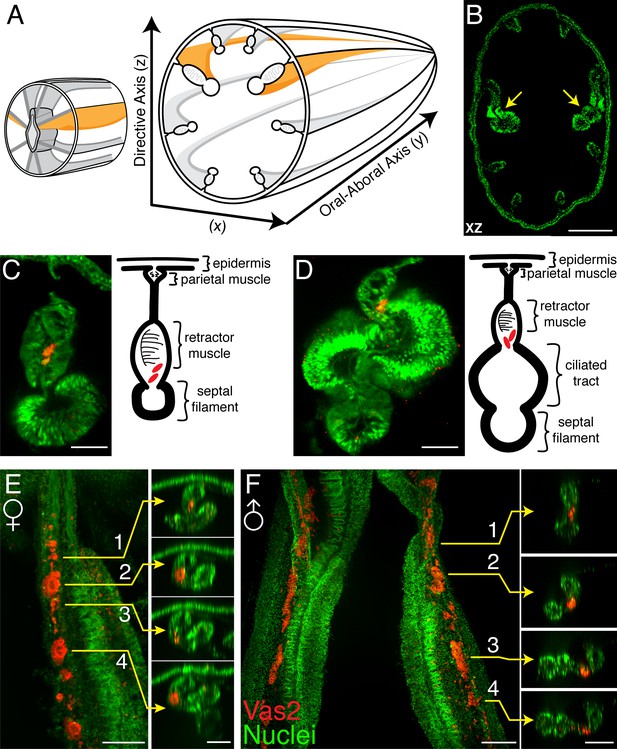
Vas2+ germ cells in juvenile gonad rudiments.
(A) Schematic diagram of Nematostella polyp anatomy depicts the gametogenic mesenteries at the mid-body level. (B) A mid-body level cross section through a juvenile polyp, note the enlarged primary mesenteries (yellow arrows). Nuclei are counter stained by DAPI (green). (C and D) Representative images of maturing mesenteries with corresponding schematic diagrams. Putative PGCs are labeled by Vas2 immunofluorescence in red. (E) Whole-mount juvenile female mesentery shows Vas2-labeled putative PGCs and germ cells in close proximity (red), suggesting maturing oocytes originate from the continuous PGC lineage. (F) Whole-mount juvenile male mesentery shows Vas2-labeled putative PGC and germ cell populations, including the rudimentary sperm cysts. Insets 1–4 of E and F are xz plane views at the indicated levels. Scale bar = 100 µm in B; 20 µm in C–F.
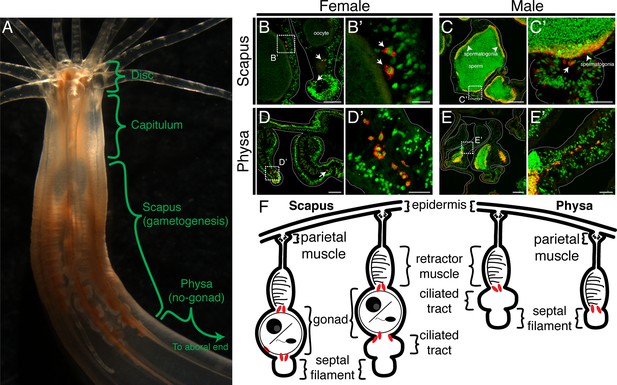
Adult PGC-like lineages localize adjacent to the mature gonad and migrate aborally.
(A) Along the oral-aboral axis, adult Nematostella can be regionally subdivided into the disc (mouth and tentacle base), capitulum (pharynx), scapus (gametogenic region) and physa (non-gametogenic region; Williams, 1975). (B–E’) Female and male cross sections were immunolabeled with Vas2 (red) and counter-stained for nuclei (green). PGC-like cells (arrows in B–B’ and C’) localize next to the maturing oocytes and sperm stem cells (spermatogonia, arrowheads in C–C’), which divide and give rise to sperm in the sperm cyst lumen. Note there are Vas2 puncta surrounding the nuclei of spermatogonia. (D–E’) PGC-like cells also localize aborally in the physa, the site of occasional asexual fission (Hand and Uhlinger, 1995; Burton and Finnerty, 2009). Aboral migration may thus ensure fertility in asexually-produced progeny. (F) Schematic diagrams depict the cross-sectional localization of PGC-like cells (red) in adult mesenteries. The boundaries of mesentery are delineated by gray lines. Scale bar = 50 µm in B; 100 µm in C, D, and E; 20 µm in B’, C’, D’, and E’.
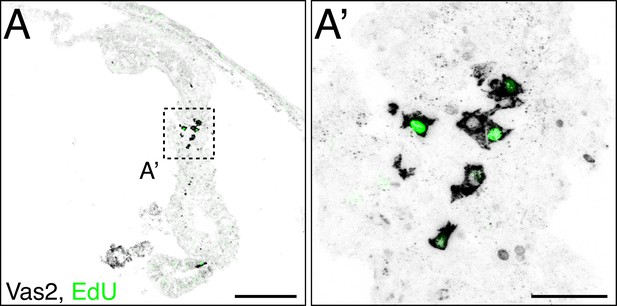
PGC-like cells are proliferative.
In the aboral region of mesenteries, some PGC-like cells (gray) incorporate EdU (green) in their nuclei, consistent with proliferative activity. Scale bar = 50 µm in A; 20 µm in A’.
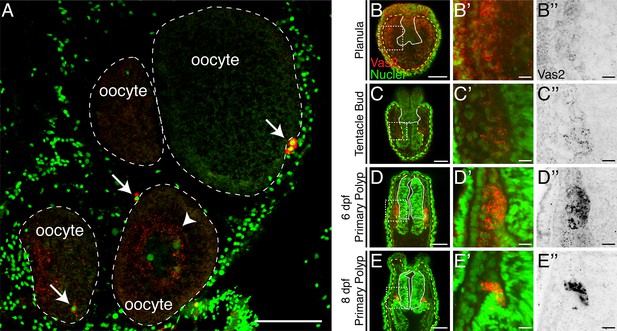
Maternally inherited perinuclear Vas2 granules diminish during PGC specification.
(A) Cross-section of female gonad. Vas2 protein (red) forms puncta that surround the nuclei of maturing oocytes (arrowhead). In addition, PGC-like cells (arrows) are found next to oocytes. Oocyte boundaries are delineated by dotted lines (white). Nuclei and nucleolus are counter stained by Hoechst (green). (B–E’’) Perinuclear Vas2 granules in endomesodermal cells gradually diminish as the PGC clusters are specified next to the pharynx during the tentacle bud through primary polyp stages. The pharynx is delineated by white lines and the boundary between ectoderm and endoderm is delineated by yellow-dotted lines. (B’–E’’) Enlarged views of boxed areas in B-E. B-E’’ are single focal planes under the same imaging conditions. Scale bar = 50 µm in A–E; 10 µm in B’–E’'.
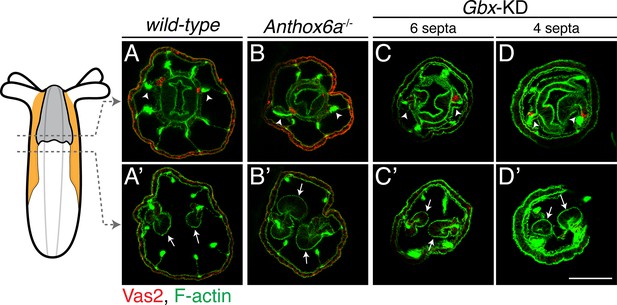
PGC clusters are specified on mesenteries with primary septal filaments of wild-type, Anthox6a mutant and Gbx shRNA knockdown primary polyps.
Although primary mesenteries are missing in Anthox6a mutants or Gbx shRNA knockdown primary polyps, PGCs (labeled by Vas2 immunofluorescence in red) are specified on the mesenteries (arrowheads) where primary septal filaments attach (arrows). Scale bar = 50 µm in D’. All images are at the same scale.
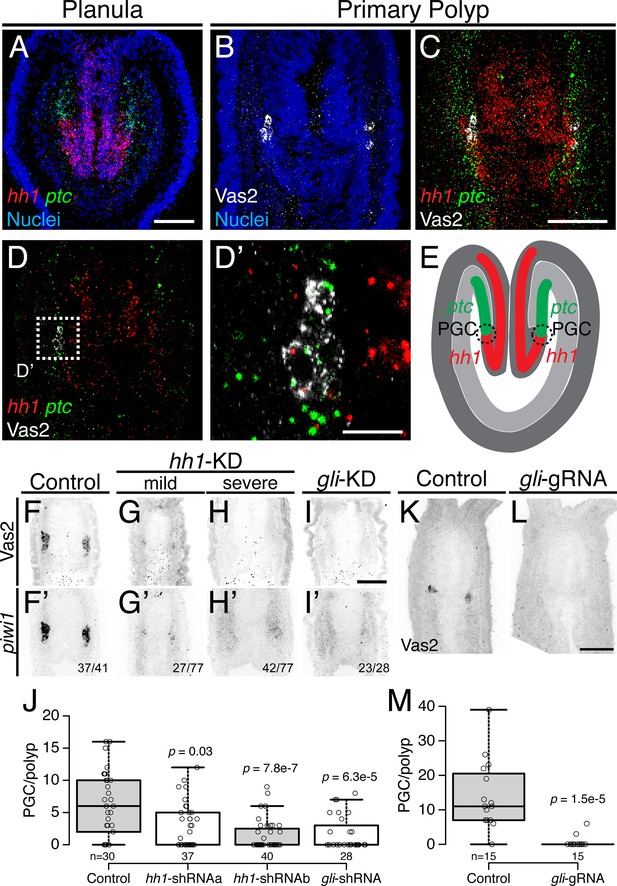
Hh signaling is required for Nematostella PGC formation.
(A) Prior to PGC specification in planula larvae, hh1 (red) and ptc (green) are expressed in pharyngeal ectoderm and endomesoderm, respectively. (B–D’) In primary polyps, PGC clusters (white) are specified within the ptc expression domain, neighboring the hh1 domain. (D) Selected single focal plane of C. (D’) Enlarged view of boxed area in D, showing PGCs expressing ptc (green) and low hh1 signal (red). (E) Diagram depicting the relative expression domains of hh1 and ptc and PGC specification during metamorphosis. (F–I’) PGC formation on 8 dpf—indicated by Vas2 immunostaining and piwi1 fluorescent in situ hybridization—is impaired by hh1 or gli shRNA knockdown. (J) PGC numbers were significantly reduced following hh1 or gli knockdown. p values of ANOVA tests were compared with control GFP-shRNA injection. (K–M) gli gRNAa, gRNAb and Cas9 protein injected embryos showed reduced PGC numbers in six dpf primary polyps. p value of ANOVA test was compared with control GFP-gRNA injection. Scale bar = 50 µm in A, C, I and L; 10 µm in D’. B-D’ are at the same scale; F–I’ are at the same scale; K and L are at the same scale.
-
Figure 7—source data 1
PGC numbers in hh1- or gli-shRNA knockdown primary polyps.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig7-data1-v1.csv
-
Figure 7—source data 2
PGC numbers in gli-gRNA injected primary polyps.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig7-data2-v1.csv
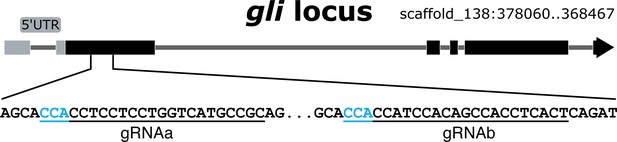
The Nematostella gli locus and gRNA targeting sequences used in this work.
Two gli gRNAs are underlined and the PAM sequences are in cyan.
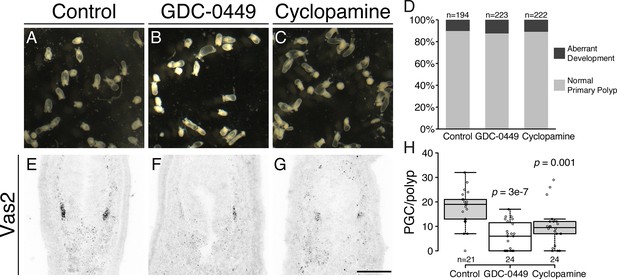
Inhibiting Hh signaling by GDC-0449 or Cyclopamine impairs PGC formation.
(A–D) The majority of primary polyps did not show visible developmental defects after treatment with GDC-0449 or Cyclopamine from the gastrula stage onward. (E–H) Primary polyps treated with GDC-0449 or Cyclopamine from 1 to 8 dpf formed significantly fewer PGCs than controls. p values of ANOVA tests were compared with control treatment. Scale bar = 50 µm in G. A-C are at the same scale; E–G are at the same scale.
-
Figure 8—source data 1
Quantification of normal primary polyps and aberrant development after GDC-0449 or Cyclopamine treatment.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig8-data1-v1.csv
-
Figure 8—source data 2
PGC numbers in GDC-0449 or Cyclopamine treated primary polyps.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig8-data2-v1.csv
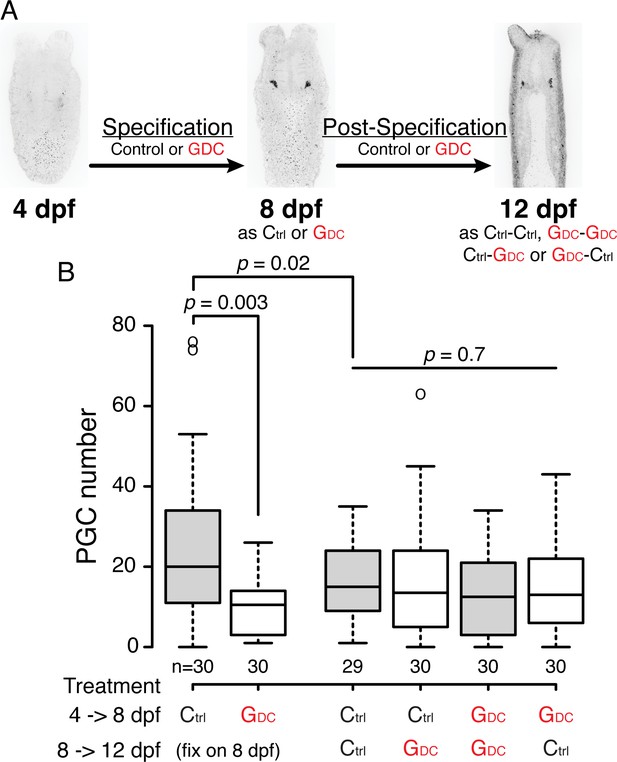
The Hh signaling pathway does not affect PGC behaviors after specification.
(A) Design of Hh signaling inhibitor GDC-0449 treatments: 4 to 8 dpf larvae were tested for PGC specification, and 8 to 12 dpf polyps were tested for PGC behaviors post-specification. Ctrl: DMSO treatment. GDC: 25 µM GDC-0449 treatment. (B) Quantification of PGC numbers after treatments. During PGC specification (4–8 dpf), GDC-0449 resulted in significantly less PGC formation than control (analyzed by one-way ANOVA). After PGCs are specified (after 8 dpf), PGC number does not show significant difference among treatments. However, long-term treatment of DMSO resulted in significant side effects on the PGC number (compare Ctrl and Ctrl-Ctrl, p=0.02).
-
Figure 8—figure supplement 1—source data 1
PGC numbers in GDC-0449 treated 8 and 12 dpf primary polyps.
- https://cdn.elifesciences.org/articles/54573/elife-54573-fig8-figsupp1-data1-v1.csv
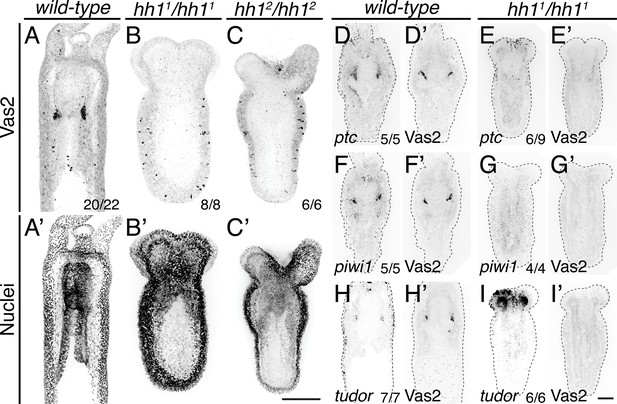
PGC clusters do not form in hh1 mutants.
(A–C’) hh1 homozygous mutant polyps exhibit a shorter body column and pronounced tentacle defects compared to wild-type siblings on 8 dpf. Additionally, no Vas2+ PGC clusters were detected in hh1 homozygous knockout polyps. (D–I’) FISH of ptc, piwi1 or tudor were followed by Vas2 immunohistochemistry on 12 dpf wild-type and hh11/hh11 mutants. (D–E’) hh1 mutant polyps show reduced ptc expression in the endomesoderm. (F–I’) hh1 mutant polyps lose PGC markers, including piwi1, tudor and Vas2. (A–C’) are multiple-focal planes projections. (D–I’) are from single-focal plane at the primary mesentery level. Scale bar = 50 µm in C’ and I’; A–C’ are at the same scale; D–I’ are at the same scale.
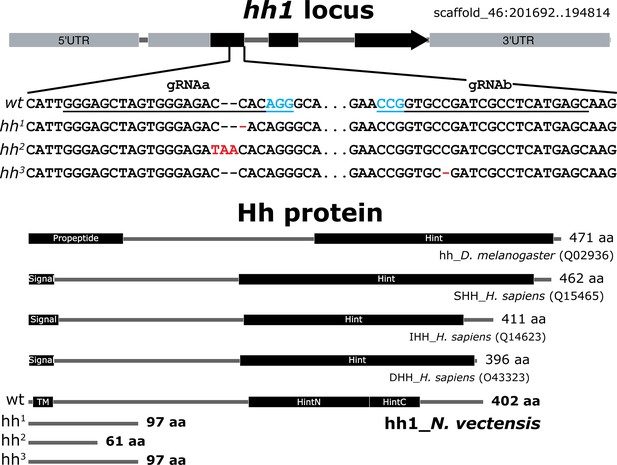
The Nematostella hh1 locus, mutant alleles and the deduced protein structure compared with Hh proteins of other species.
Injection of hh1-gRNAa (underlined; PAM sequence in cyan) resulted in a −1 nucleotide frame shift in the hh1 allele and a +2 nucleotide frame shift in the hh2 allele. Injection of hh1-gRNAb resulted in a −1 nucleotide frame shift in the hh3 allele. Protein domain structures of D. melanogaster and human (H. sapiens) Hh protein are from UniProt (www.uniprot.org) with accession numbers in parenthesis. Nematostella Hh1 protein domains were predicted by SMART analysis (http://smart.embl-heidelberg.de/). TM: transmembrane domain.
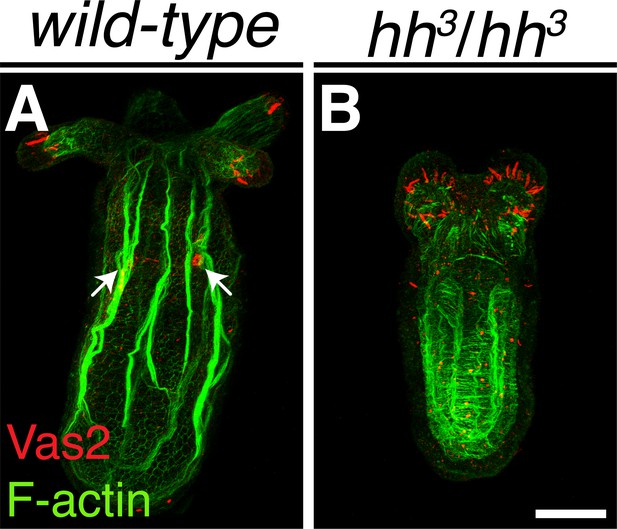
The hh3 mutant recapitulates the phenotype of hh1 and hh2 homozygotes.
Wild-type and homozygous mutant were stained with anti-Vas2 antibody (red) and phalloidin (green) on 12 dpf. The PGC clusters can be detected in wild-type polyps (white arrows) but not in the hh3 mutant. Note that Vas2 signal in the tentacle tips results from non-specific binding of the antibody.
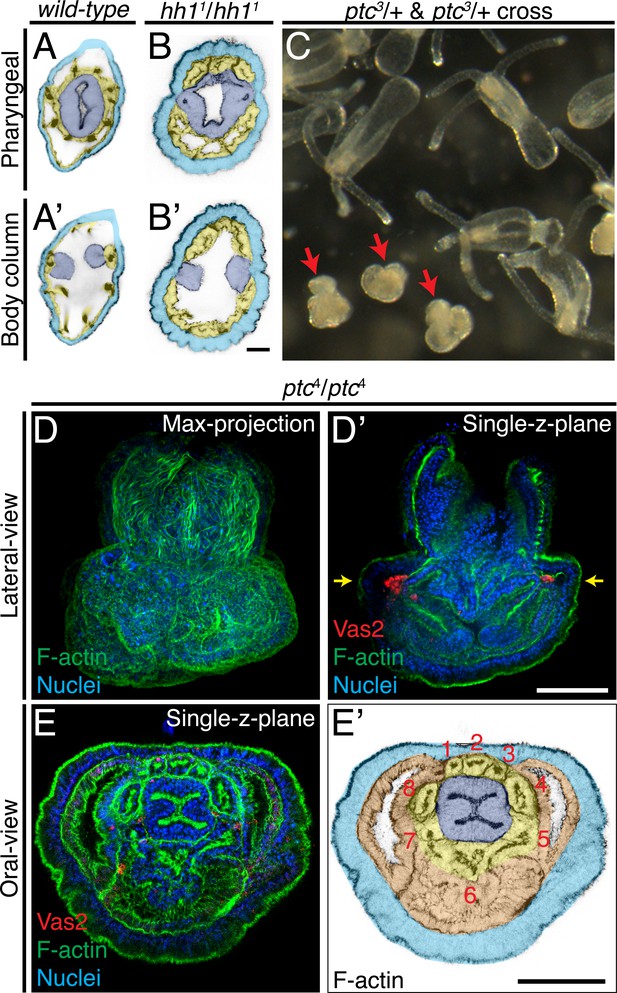
Patterning defects in hh1 and ptc mutants.
(A–B’) In addition to loss of putative PGCs, hh1 mutant polyps show endomesodermal patterning defects. Parts of the pharyngeal ectoderm and septal filaments (navy blue) abnormally contact the outer epidermis (azure blue), without endomesoderm tissue in between (yellow). These contacts segregated the normally contiguous eight endomesodermal segments into blocks of three and five segments along the directive axis. (C) Eighteen dpf F2 progeny from a cross between ptc3/+ heterozygous siblings. The abnormal mushroom-shaped polyps are indicated by red arrows. (D–D’) At the primary polyp stage (12 dpf), homozygous ptc mutants lack the four primary tentacles and do not develop the normal polyp body plan. (E–E’) A single focal plane taken at the level indicated by yellow arrows in D’. Depite significant morphological defects, ptc mutant animals develop a pharynx (navy blue), eight endomesodermal segments (yellow), body wall endomesoderm (orange) and putative PGC clusters (labeled by Vas2 immunofluorescence in red in D’). Scale bar = 20 µm in B’; 50 µm in D’ and E’. A-B’ are at the same scale; D–D’ are at the same scale; E–E’ are at the same scale.
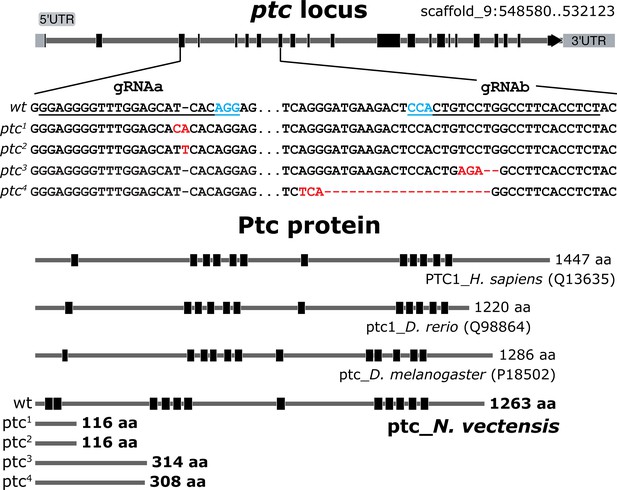
The Nematostella ptc locus, mutant alleles and the deduced protein structure compared to other species.
Injection of ptc-gRNAa (underlined; PAM sequence in cyan) produced a +1 nucleotide frame shift in the ptc1 allele and a distinct +1 nucleotide frame shift in the ptc2 allele. Injection of ptc-gRNAb produced a −2 nucleotide frame shift in the ptc3 allele and a −20 nucleotide frame shift in the ptc4 allele. Information on the Human (H. sapiens), zebrafish (D. rerio) and D. melanogaster Ptc proteins and their corresponding transmembrane domains (black box) are from UniProt with accession number in parenthesis. The transmembrane domains of Nematostella Ptc were predicted by SOSUI analysis (http://harrier.nagahama-i-bio.ac.jp/sosui/).
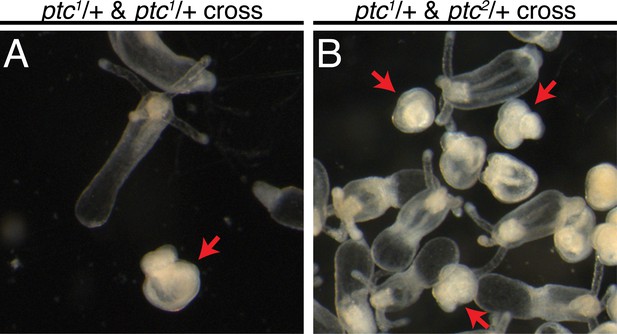
Crosses between ptc1/+ and ptc1/+ or ptc1/+ and ptc2/+ heterozygous siblings.
The observed progeny phenocopy ptc3 and ptc4 homozygotes at 8 dpf (red arrows).
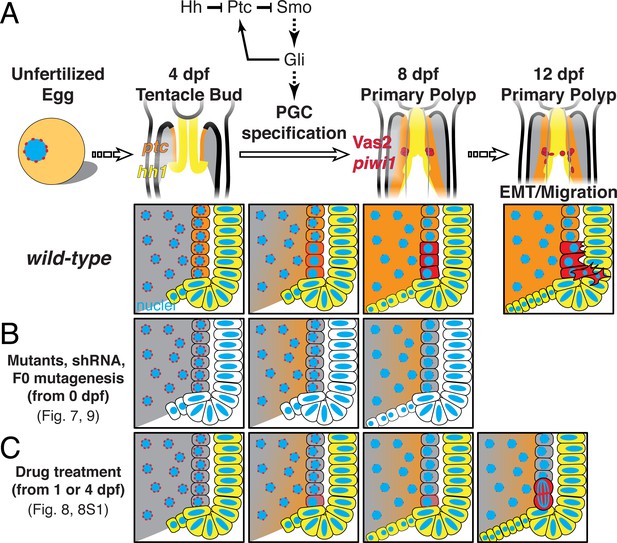
A model of Nematostella PGC specification and migration.
(A) In wild-type 4 dpf tentacle bud larvae, hh1 (yellow) and ptc (orange) are expressed in the pharyngeal ectoderm and endomesoderm, respectively. Between 4 to 8 dpf when larvae metamorphose to primary polyps, Hh1 signals to neighboring endomesodermal cells and specifies Vas2/piwi1-positive PGC clusters (red) in the primary mesenteries. Meanwhile, perinuclear Vas2 granules (red dots) within endodermal cells (gray) gradually diminish. After initial specification, the PGC clusters undergo EMT and migrate to gonad rudiments during the juvenile stage. (B) hh1 KD, gli KD, hh1 mutants and gli gRNA-Cas9 injected embryos develop reduced or absent PGCs clusters, indicating a requirement for Hh signaling in this process, whether direct or indirect. (C) Drug treatments inhibiting Smo activity between 4 to 8 dpf impair PGC specification. However, some polyps still form reduced numbers of PGCs at later time points, possibly due to compensatory PGC proliferation. Note that the reduced ptc expression depicted in B is supported by FISH data in hh1 mutants but is presumptive in C.
Videos
3D reconstruction of pharyngeal structures.
PGCs (magenta) are specified on the epithelium of the two primary mesenteries, close to the pharynx (cyan cells in the center). The endomesodermal nuclei are pseudocolored in blue, and the ectodermal nuclei are in cyan.
Tables
Primer sets for cloning probe templates into pPR-T4P (gene-specific regions are underlined).
| Target | Sequence | Probe size (bp) | |
|---|---|---|---|
| piwi1 | Forward | CATTACCATCCCGCGAGCCTACAACCAGGAGAG | 1327 |
| Reverse | CCAATTCTACCCGCGTTGTGTTGATGCCCATAG | ||
| piwi2 | Forward | CATTACCATCCCGTGGGCGGTACTTCTACAACC | 1367 |
| Reverse | CCAATTCTACCCGTGCCCTTGATAAGGAGCATC | ||
| vas2 | Forward | CATTACCATCCCGTGAAGGGTCTCCAATTCCTG | 1530 |
| Reverse | CCAATTCTACCCGTGTGCAGATTACAGCCAAGG | ||
| tudor | Forward | CATTACCATCCCGGAACCTACTTGCTTCCGCAG | 1450 |
| Reverse | CCAATTCTACCCGACGACTCGGTGTTCCCATAG | ||
| twist | Forward | CATTACCATCCCGAAATCTCGGTGTCGGTCTTG | 1020 |
| Reverse | CCAATTCTACCCGTATCGCAGCTTTGCTTTCAG | ||
| hh1 | Forward | CATTACCATCCCGTTTCATTGGGAGCTAGTGGG | 1327 |
| Reverse | CCAATTCTACCCGAAAGCGTGAATTGGGTCTTG | ||
| ptc | Forward | CATTACCATCCCGGATGTGCGTGTGTGGGATAG | 1455 |
| Reverse | CCAATTCTACCCGACCGCGAGGTAATTGAACAC | ||
shRNA sequences.
| Target | shRNA name | Sequence |
|---|---|---|
| hh1 | hh1-shRNAa | GGCTTGCTATAACACTGAT |
| hh1 | hh1-shRNAb | GGCAGAGCTGTTGATATAA |
| gli | gli-shRNA | GAGAAGAGGGATTTCACAT |
| Gbx | Gbx-shRNAa | GCCAAGGTTAATAGATCCT |
| Gbx | Gbx-shRNAb | GGAAACGTGTACGATCACT |
| eGFP | eGFP-shRNA | GACGTAAACGGCCACAAGTT |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Nematostella vectensis) | vas1 | GenBank | AY730696 | |
| Gene (Nematostella vectensis) | vas2 | GenBank | AY730697 | |
| Gene (Nematostella vectensis) | piwi1 | GenBank | MF683122 | |
| Gene (Nematostella vectensis) | piwi2 | GenBank | MF683123 | |
| Gene (Nematostella vectensis) | tudor | GenBank | XM_032376221 | |
| Gene (Nematostella vectensis) | twist | GenBank | AY286509 | |
| Gene (Nematostella vectensis) | hh1 | GenBank | EU162651 | |
| Gene (Nematostella vectensis) | ptc | GenBank | EU162650 | |
| Gene (Nematostella vectensis) | gli | GenBank | EU162649 | |
| Gene (Nematostella vectensis) | Anthox6a | GenBank | GQ240845 | |
| Gene (Nematostella vectensis) | gbx | GenBank | DQ500757 | |
| Genetic reagent (Nematostella vectensis) | hh11 | This paper | See Figure 9—figure supplement 1 | |
| Genetic reagent (Nematostella vectensis) | hh12 | This paper | See Figure 9—figure supplement 1 | |
| Genetic reagent (Nematostella vectensis) | hh13 | This paper | See Figure 9—figure supplement 1 | |
| Genetic reagent (Nematostella vectensis) | ptc1 | This paper | See Figure 10—figure supplement 1 | |
| Genetic reagent (Nematostella vectensis) | ptc2 | This paper | See Figure 10—figure supplement 1 | |
| Genetic reagent (Nematostella vectensis) | ptc3 | This paper | See Figure 10—figure supplement 1 | |
| Genetic reagent (Nematostella vectensis) | ptc4 | This paper | See Figure 10—figure supplement 1 | |
| Peptide, recombinant protein | vas2 | This paper | MCFKCQQTGHFARECPNESAAGENGDRPKPVTYVPPTPTEDEEEMFRSTIQQGINFEKYDQIEVLVSGNNPVRHINSFEEANLYEAFLNNVRKAQYKKPTPHHHHHH | |
| Antibody | anti-vas2 (Rabbit polyclonal) | This paper | IF (1:1000), stock: 0.4 mg/mL | |
| Antibody | anti-α-Tubulin (mouse monoclonal) | Sigma-Aldrich | T9026 | IF (1:1000) |
| Antibody | anti-Phospho-Histone H3 (mouse monoclonal) | Sigma-Aldrich | 05–806 | IF (1:1000) |
| Antibody | anti-DIG-POD Fab fragments (Sheep polyclonal) | Sigma-Aldrich | 11207733910 | FISH (1:1000) |
| Antibody | anti-Fluorescein-POD Fab fragments (Sheep polyclonal) | Sigma-Aldrich | 11426346910 | FISH (1:1000) |
| Chemical compound, drug | GDC-0449 | Cayman Chemical | 13613 | 25 µM |
| Chemical compound, drug | Cyclopamine | Cayman Chemical | 11321 | 5 µM |
| Chemical compound, drug | Benzyl alcohol | Sigma-Aldrich | 305197 | |
| Chemical compound, drug | Benzyl benzoate | Sigma-Aldrich | B6630 | |
| Commercial assay or kit | Click-iT EdU Kit | Thermo Fisher Scientific | C10339 | |
| Commercial assay or kit | TSA Plus Cyanine 3 System | PerkinElmer | NEL744001KT | |
| Commercial assay or kit | ImProm-II Reverse Transcription System | Promega | A3800 | |
| Commercial assay or kit | T7 RNA polymerase Kit | Promega | P2077 | |
| Commercial assay or kit | AmpliScribe T7-Flash Transcription Kit | Lucigen | ASF3507 | |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | Imaris | Bitplane | RRID:SCR_007370 | 8.3 |
| Software, algorithm | BoxPlotR | BoxPlotR.shiny (https://github.com/VizWizard/BoxPlotR.shiny/blob/master/README.md) | RRID:SCR_015629 | |
| Other | Hoechst-34580 stain | Sigma-Aldrich | 63493 | 1 µg/mL |
| Other | SYBR Green I stain | Thermo Fisher Scientific | S7567 | 1:5000 |
| Other | Phalloidin stain | Thermo Fisher Scientific | A12379 | 1:200 |
| Other | DIG RNA labeling mix | Sigma-Aldrich | 11277073910 | |
| Other | Fluorescein RNA labeling mix | Sigma-Aldrich | 11685619910 | |
| Other | Cas9 protein with NLS | PNA Bio | CP02 | 500 ng/µL |
gRNA sequence for CRISPR/Cas9 mutagenesis (PAM sequences are underlined).
| Target | gRNA name | Sequence |
|---|---|---|
| hh1 | hh1-gRNAa | GGGAGCTAGTGGGAGACCACAGG |
| hh1 | hh1-gRNAb | GCTCATGAGGCGATCGGCACCGG |
| ptc | ptc-gRNAa | GGAGGGGTTTGGAGCATCACAGG |
| ptc | ptc-gRNAb | AGAGGTGAAGGCCAGGACAGTGG |
| gli | gli-gRNAa | GCGGCATGACCAGGAGGAGGTGG |
| gli | gli-gRNAb | AGTGAGGTGGCTGTGGATGGTGG |
Summary of major observations during PGC development.
| PGC event | Developmental stage | Data |
|---|---|---|
| Specification on primary mesenteries | 4–8 dpf | Figure 1B–D |
| EMT and radial migration | juveniles with feeding | Figure 2C–E', Figure 3B–B' |
| Aboral migration to gonad rudiments | juveniles with feeding | Figure 3C–D', Figure 4 |
Summary of experimental designs.
| Experiment | Treatment duration | Data |
|---|---|---|
| Verify PGC specification in hh1 and gli shRNA knockdown | 0–8 dpf | Figure 7F–J |
| Verify PGC specification in gli mutagenesis | 0–6 dpf | Figure 7K–M |
| Verify PGC specification in Smo inhibition by GDC-0449 and Cyclopamine | 1–8 dpf | Figure 8 |
| Test Hh pathway on specification and post-specification by GDC-0449 temporal treatments | 4–8, 4–12 and 8–12 dpf | Figure 8—figure supplement 1 |
| Verify PGC specification in hh11 and hh12 mutants | 0–8 dpf | Figure 9A–C' |
| Verify ptc and PGC marker expression in hh11 mutant | 0–12 dpf | Figure 9D–I' |
| Verify PGC specification in hh13 mutant | 0–12 dpf | Figure 9—figure supplement 2 |
| Demonstrate patterning defects in hh11 mutant | 0–12 dpf | Figure 10A–B' |
| Demonstrate the mutant morphology of ptc3 mutant | 0–18 dpf | Figure 10C |
| Verify PGC specification and demonstrate the mutant morphology of ptc4 mutant | 0–12 dpf | Figure 10D–E' |
| Demonstrate the mutant morphology of ptc1 and ptc2 mutants | 0–8 dpf | Figure 10—figure supplement 2 |