ATR expands embryonic stem cell fate potential in response to replication stress
Figures
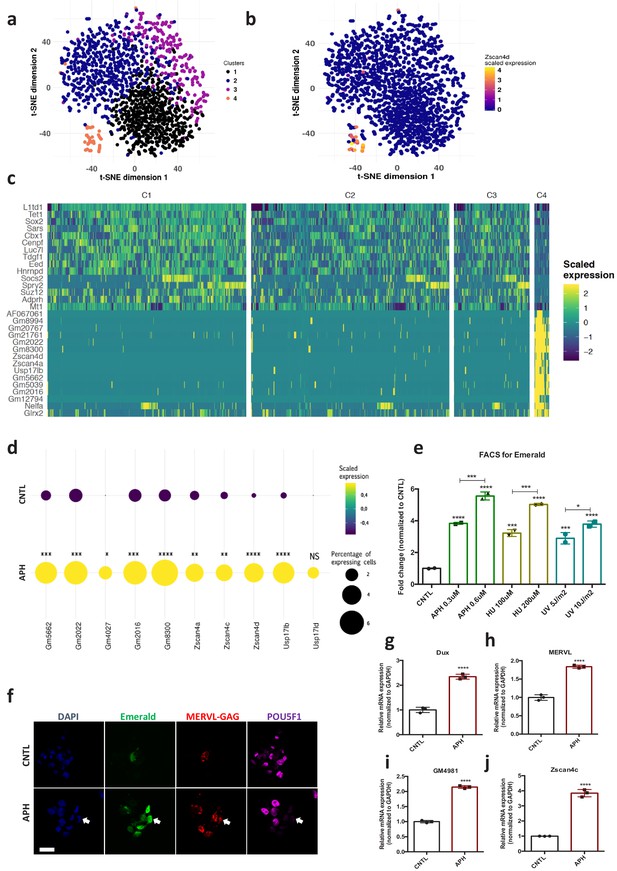
Induction of RS increases the number of 2C-like cells in ESCs culture and activates the expression of 2C-specific genes in mouse embryos.
(a) Clustering of 1399 Drop-seq single-cell expression profiles into four cell populations. The plot shows a two-dimensional representation (t-SNE) of global gene expression relationship; cells are colored according to their cluster. Clusters were identified by shared nearest neighbor algorithm (see methods). (b) t-SNE plot showing Zscan4d expression level across all CNTL and APH-treated cells. (c) Heatmap showing the list of top 30 genes that are differentially expressed between cluster 4 cells (orange cluster in a) and the rest of the population (cluster 1,2, and 3). (d) Plot showing the scaled expression of 2C-specific markers and the percentage of cells expressing 2C-related genes in CNTL and APH-treated condition. Fisher's exact test was used to determine p-values. (e) FACS analysis on pZscan4-Emerald ESCs upon treatment with various RS-inducing agents. (f) Immunostaining of ESCs for ZSCAN4-Emerald, MERVL-GAG and canonical pluripotency marker POU5F1 upon treatment with APH (bar = 25 µm). (g–j) RT-qPCR analysis on blastocyst-stage embryos treated with APH for key 2C-like markers. Statistical significance compared to CNTL unless otherwise indicated. All bar-plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
-
Figure 1—source data 1
FACS, qPCR and Western quantification.
- https://cdn.elifesciences.org/articles/54756/elife-54756-fig1-data1-v1.xlsx
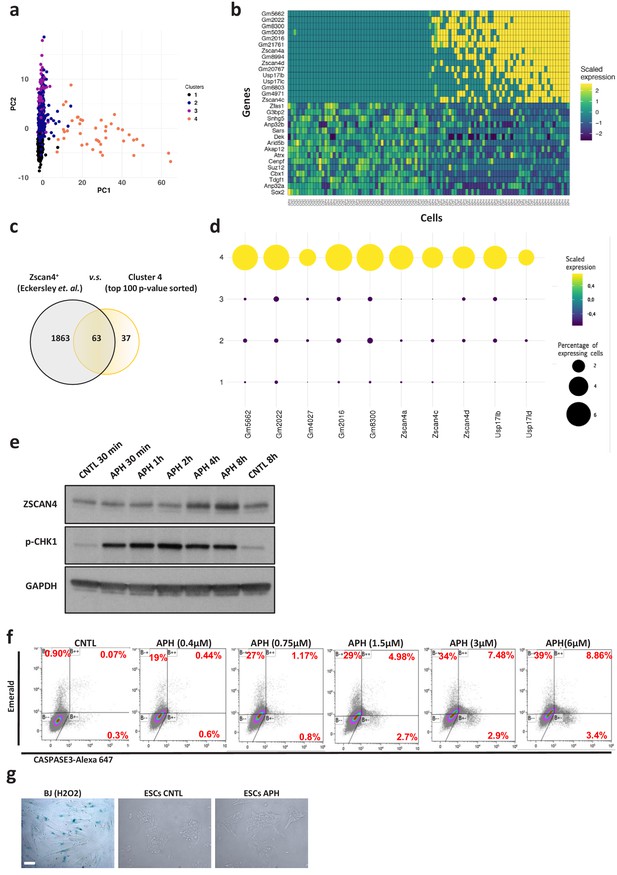
Induction of RS increases the number of 2C-like cells in ESCs culture.
(a) PCA plot on 1399 cells treated with or without APH. Cells are colored based on clusters. (b) Heatmap showing scaled expression values of top 30 genes and 100 ranked cells according to PC1. (c) Venn diagram showing the overlap of the top 100 DEGs of cluster four with those expressed in 2C-like cells (according to Eckersley-Maslin et al., 2016). (d) Plot showing the scaled expression of 2C-specific markers and the percentage of cells expressing 2C-related genes in the four clusters. (e) Immunoblot showing the expression of ZSCAN4 and p-CHK1 proteins upon treatment with APH at different time points. (f) FACS analysis for Emerald and CASPASE 3-Alexa 647 on pZscan4-Emerald ESCs upon treatment with increasing concentrations of APH. (g) β-galactosidase staining of H2O2-treated BJ fibroblast cells, untreated and APH-treated ESCs. For western blots quantification refer to Figure 1—source data 1.
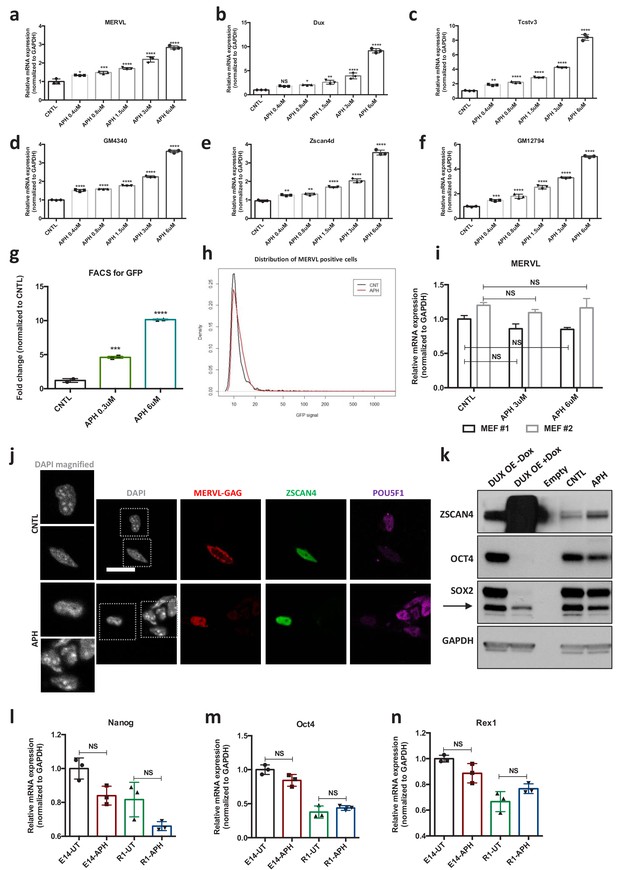
Characterization of APH-induced 2C-like cells.
(a–f) RT-qPCR analysis for 2C-like genes upon treatment with increasing concentrations of APH (g) FACS analysis on pMERVL-GFP ESCs upon treatment with APH. (h) FACS analysis showing the mean GFP signal intensity within pMERVL-GFP positive population in CNTL and APH-treated pMERVL-GFP ESCs. (i) RT-qPCR analysis for MERVL upon treatment with APH in two different MEF lines. (j) Immunostaining for 2C markers, MERVL-GAG and ZSCAN4 and ESC marker, POU5F1 in untreated and APH-treated ESCs, DAPI staining shows the lack of chromocenters in 2C-like cells (bar = 20 µm), (k–n) Immunoblot and RT-qPCR analysis of ESCs for ZSCAN4 and canonical pluripotency markers upon treatment with APH. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 1—source data 1.
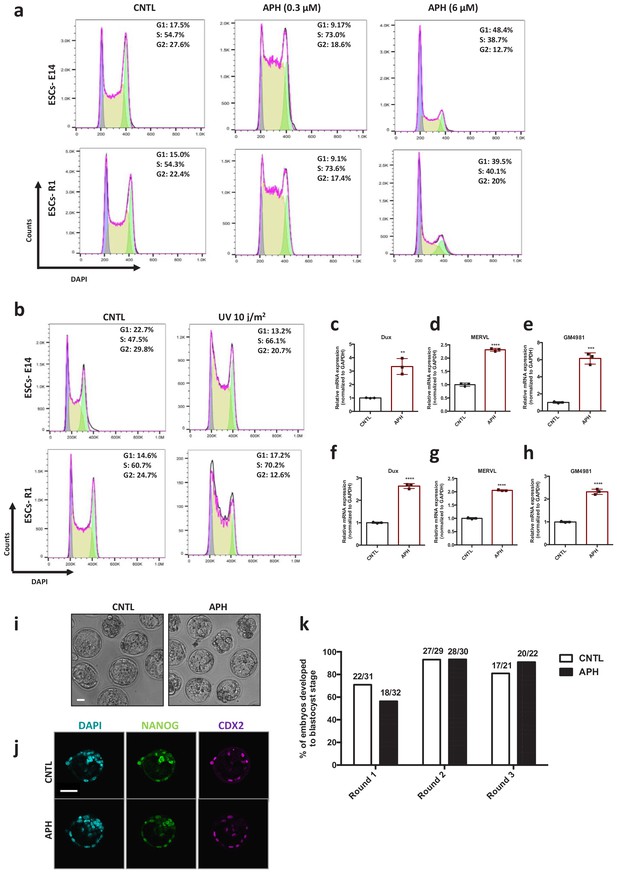
Characterization of APH-induced 2C-like cells.
(a) Cell cycle analysis of E14 and R1 ESCs upon treatment with low (0.3 μM) and high (6 μM) concentrations of APH. (b) Cell cycle analysis of E14 and R1 ESCs upon UV treatment. (c-e), (f-h) Two replicates of RT-qPCR analysis on blastocyst-stage embryos for key 2C-specific genes Dux, Gm4981 and MERVL. (i) Wide field microscope images of mouse embryos at blastocyst stage after APH treatment (bar = 25 µm) (j) Immunostaining of blastocyst-stage embryos for key ESCs marker, NANOG and the key TSC marker, CDX2 after APH treatment (bar = 50 µm). (k) Percentage of embryos developed to the blastocyst stage after APH treatment in three replicates. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
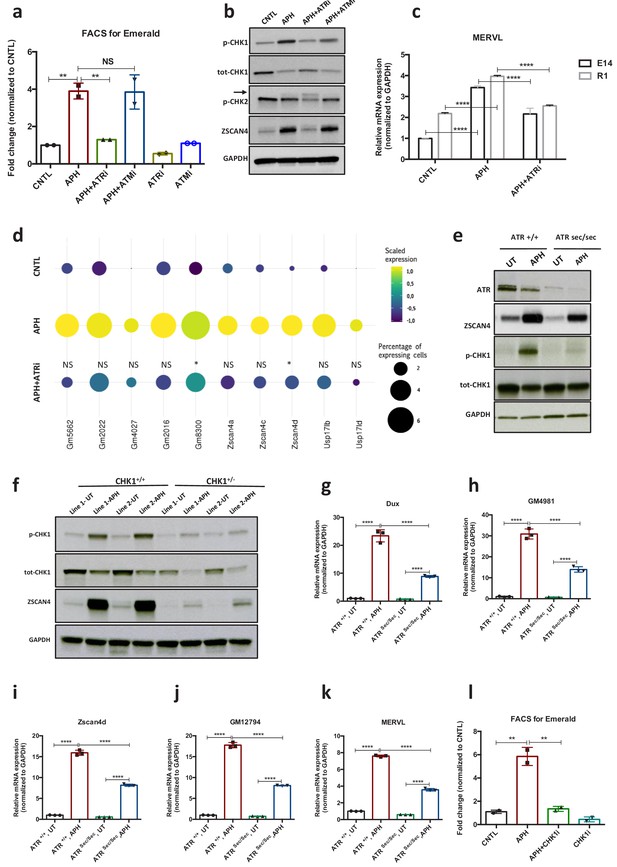
ATR and CHK1-mediated RSR triggers activation of key 2C-specific genes in ESCs.
(a) FACS analysis on pZscan4-Emerald ESCs showing the number of Em+ cells upon treatment with APH and specific ATR and ATM inhibitors. (b) Immunoblot for the phosphorylation status of key DDR kinases (CHK1 and CHK2) and the ZSCAN4 protein level upon treatment with APH and ATM/ATR inhibitor in ESCs. (c) RT-qPCR analysis of two ESCs lines for the expression of MERVL upon treatment with APH and ATRi. (d) Plot showing the scaled expression of 2C-specific markers and the percentage of cells expressing 2C-related genes in CNTL, APH-treated and APH+ATRi conditions. Fisher's exact test was used to determine p-values. (e) Immunoblot for ZSCAN4, ATR and the phosphorylation status of CHK1 upon APH treatment of AtrSec/Sec and Atr+/+ ESCs. (f) Immunoblot showing the expression of ZSCAN4 and p-CHK1 in Chk1+/- and Chk1+/+ ESCs upon treatment with APH. (g–k) RT-qPCR for 2C-specific genes in AtrSec/Sec and Atr+/+ ESCs treated with APH. (l) FACS analysis of pZscan4-Emerald ESCs showing the number of Em+ cells upon treatment with APH and a specific CHK1 inhibitor. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 2—source data 1.
-
Figure 2—source data 1
FACS, qPCR and Western quantification.
- https://cdn.elifesciences.org/articles/54756/elife-54756-fig2-data1-v1.xlsx
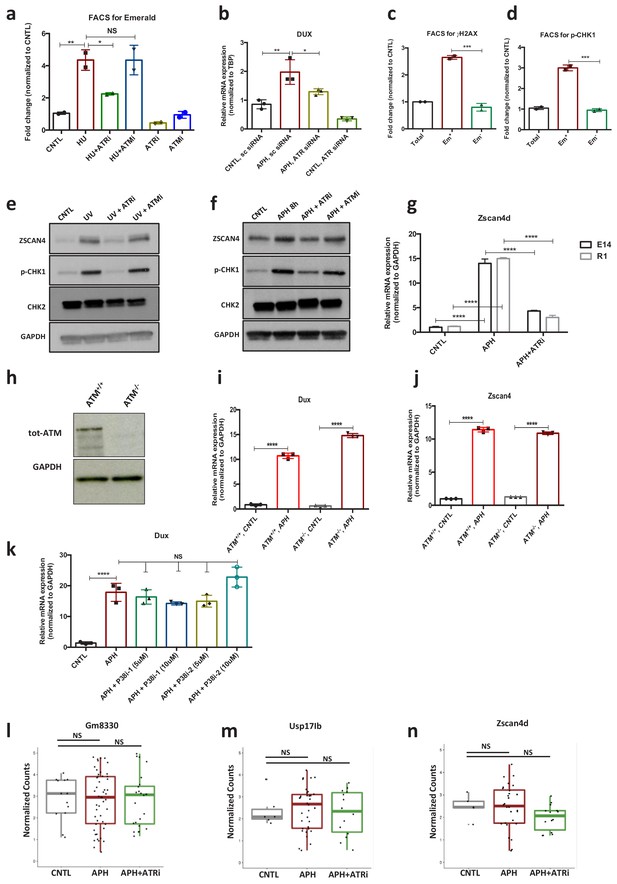
ATR-mediated RSR triggers activation of key 2C-specific genes in ESCs.
(a) FACS analysis of pZscan4-Emerald ESCs upon treatment with ATM and ATR inhibitors after induction of RS upon HU treatment. (b) RT-qPCR analysis of Dux mRNA upon APH treatment and ATR KD (c,d) FACS analysis of pZscan4-Emerald ESCs for DNA damage markers γH2AX and p-CHK1 (Em+ and Em- correspond to Emerald-GFP positive and negative populations, respectively). (e, f) Immunoblot showing the expression of ZSCAN4, p-CHK1 and p-CHK2 proteins in ESCs upon treatment with UV or APH for 8 hr. (g) RT-qPCR analysis for Zscan4d gene upon treatment with a specific ATRi in two distinct ESC lines (E14 and R1). (h) Immunoblot showing the absence of ATM kinase in ATM KO cells. (i,j) RT-qPCR analysis for Dux and Zscan4 mRNA upon APH treatment in ATM WT and KO ESCs. (k) RT-qPCR analysis for Dux mRNA upon APH and two different concentrations of p38 inhibitors (1 and 2). (l-n) Box plot showing the expression level of 2C-related genes (within the subpopulation of cells expressing the specific marker) at the single-cell level in CNTL, APH and APH+ATRi condition. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 2—source data 1.
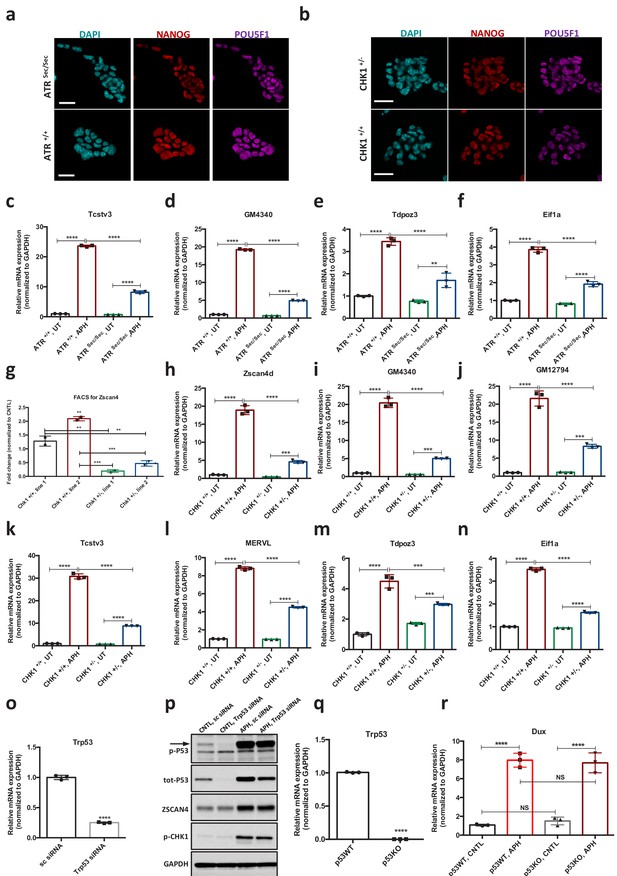
ATR and CHK1-mediated RSR triggers activation of key 2C-specific genes in ESCs.
(a, b) Immunostaining of AtrSec/Sec, Atr+/+ ESCs and Chk1-/+, Chk1+/+ ESCs for the canonical pluripotency markers POU5F1 and NANOG (bar = 25 µm). (c–f) RT-qPCR analysis for 2C-related genes in Atrsec /sec and Atr+/+ ESCs upon treatment with APH. (g) FACS analysis of Chk1+/- and Chk1+/+ ESCs for the basal expression level of ZSCAN4 protein (h–n) RT-qPCR analysis for 2C-related genes in Chk1+/- and Chk1+/+ ESCs upon treatment with APH. (o) RT-qPCR analysis for Trp53 expression upon siRNA-mediated KD. CNTL sample was transfected with sc siRNA. (p) Immunoblot showing ZSCAN4 expression and phosphorylation status of P53 and CHK1 upon APH treatment in CNTL and Trp53 KD ESCs. (q,r) RT-qPCR analysis for Trp53 and Dux expression in Trp53 WT and KO ESCs. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 2—source data 1.
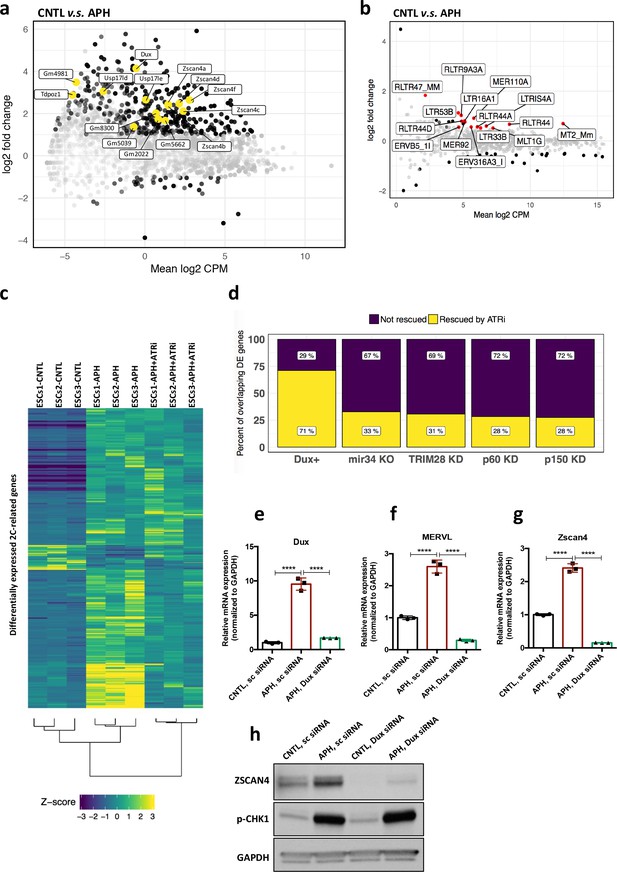
ATR induces transcriptional signature of 2C-like cells in ESCs.
(a) MA plot showing gene expression in ESCs treated with APH in comparison to control. Key 2C-specific genes are highlighted (b) MA plot showing retrotransposons expression upon APH treatment. (c) Heatmap showing the robust z-scores for 2C-specific genes in the indicated samples. 2C-related genes were identified by performing a differential expression analysis on ZSCAN4+/MERVL+v.s. ZSCAN4-/ MERVL- ESCs from Eckersley-Maslin et al. (2016). (d) Bar plot displaying the percentage of ATR-dependent differentially expressed genes among the ones shared between APH-treated ESCs and each dataset. (e–g) RT-qPCR analysis of Dux KD ESCs for Dux and Zscan4d genes, and MERVL upon treatment with APH. (h) Immunoblot for p-CHK1 and ZSCAN4 proteins upon treatment with APH in Dux KD ESCs in comparison with control ESCs. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 3—source data 1.
-
Figure 3—source data 1
qPCR and Western quantification.
- https://cdn.elifesciences.org/articles/54756/elife-54756-fig3-data1-v1.xlsx
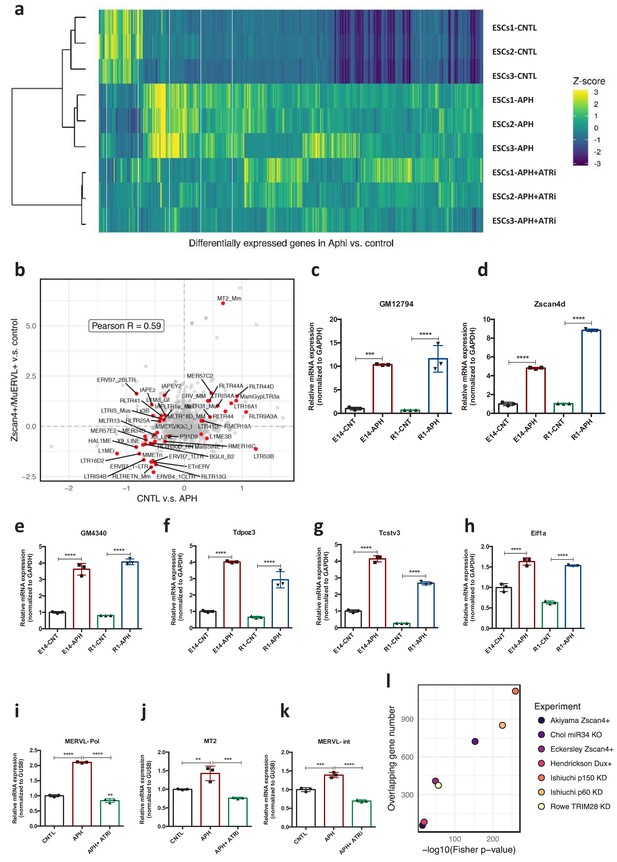
ATR induces transcriptional signature of 2C-like cells in ESCs.
(a) Heatmap showing the robust z-scores for all the DEGs in three ESCs lines upon treatment with APH or APH+ATRi. (b) Plot showing the repeat subfamilies that are significantly differentially expressed both in our data (CNTL v.s. APH) and the ZSCAN4+/MERVL+ v.s. ZSCAN4-/MERVL- comparison (Eckersley-Maslin et al., 2016). The Pearson correlation (0.59) between the log2 fold changes from the two comparisons is also shown on the plot. (c–h) Validation of RNA-Seq results by RT-qPCR analysis. (i–k) RT-qPCR analysis of ESCs for expression of MERVL elements upon treatment with APH and ATRi (l) Plot shows the number of genes expressed in ESCs upon treatment with APH overlapping with previously published datasets. The -log10 (Fisher p-value) shows the significance of overlaps obtained from Fisher test. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
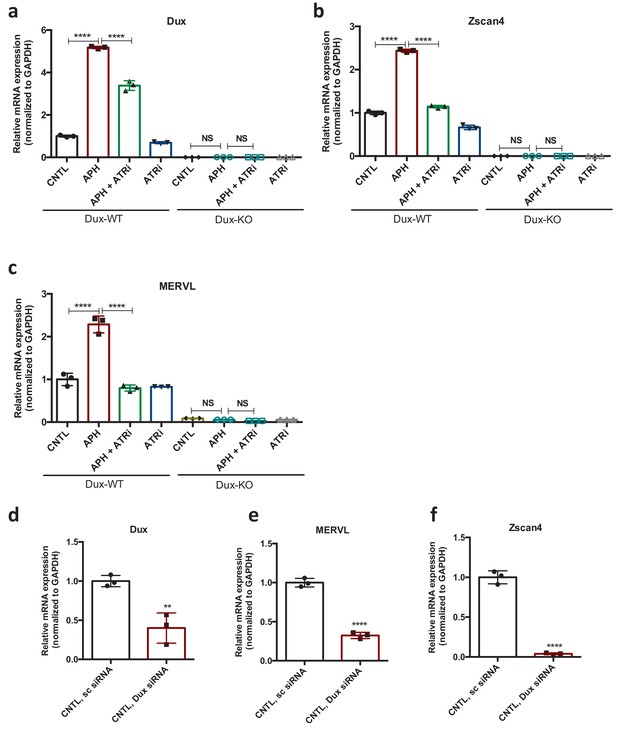
ATR induces transcriptional signature of 2C-like cells in ESCs.
(a–c) RT-qPCR analysis for Dux, Zscan4, and MERVL expression upon treatment with APH and ATRi in Dux KO and WT ESCs. (d-f) RT-qPCR analysis of ESCs for expression of 2C-like genes in siRNA-mediated Dux KD ESCs compared to CNTL ESCs treated with sc siRNA. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
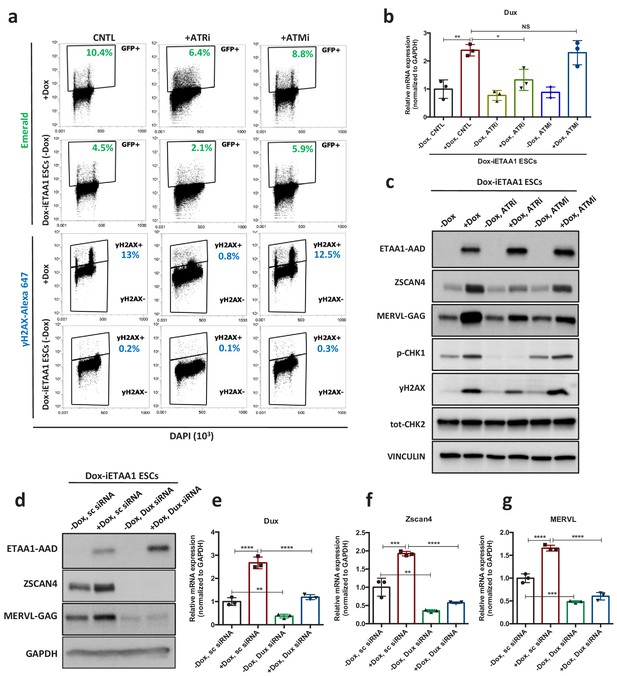
ETAA1-mediated activation of ATR induces 2C-like cells in a RS-free context.
(a) FACS analysis for γH2AX and Emerald-GFP in Dox-iETAA1 ESCs in the presence or absence of Dox and upon treatment with ATRi or ATMi. (b) RT-qPCR results for Dux expression in Dox-iETAA1 ESCs upon Dox induction in the presence or absence of ATRi or ATMi. (c) Immunoblot showing the expression of ETAA1-AAD, ZSCAN4, MERVL-GAG and the phosphorylation status of CHK1, CHK2 and H2AX in Dox-iETAA1 ESCs upon treatment with Dox, ATRi or ATMi. (d) Immunoblot showing the expression of ETAA1-AAD, ZSCAN4, MERVL-GAG in Dox-iETAA1 ESCs upon treatment with Dox and Dux knock down. (e–g) RT-qPCR analysis of Dox-iETAA1 ESCs for expression of 2C-related genes (Dux, MERVL and Zscan4) upon treatment with Dox and Dux knock down. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 4—source data 1.
-
Figure 4—source data 1
qPCR and Western quantification.
- https://cdn.elifesciences.org/articles/54756/elife-54756-fig4-data1-v1.xlsx
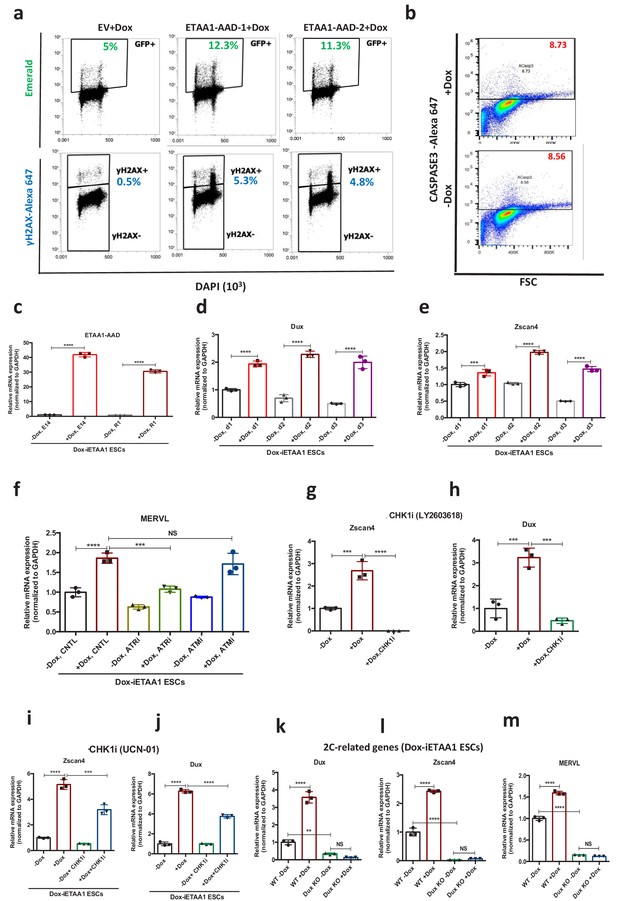
Characterization of ETAA1-AAD inducible ESCs.
(a) FACS analysis for γH2AX and Emerald upon overexpression of ETAA1-AAD by Dox in two different lentivirus clones with respect to empty vector (EV)-infected ESCs. (b) FACS analysis of CASPASE-3 in Dox-iETAA1 ESCs. (c) RT-qPCR analysis for the expression of ETAA1-AAD in Dox-iETAA1 ESCs (E14 and R1) upon Dox treatment. (d, e) RT-qPCR results for Dux and Zscan4 expression in Dox-iETAA1 ESCs after 24, 48 and 72 hr of Dox administration (f) RT-qPCR results for MERVL element in Dox-iETAA1 ESCs upon Dox administration in the presence or absence of ATRi or ATMi. (g–h) RT-qPCR results for 2C-related genes, Zscan4 and Dux in Dox-iETAA1 ESCs upon Dox and CHK1i treatment. (i–j) RT-qPCR analysis for Zscan4 and Dux expression in Dox-iETAA1 ESCs in the absence or presence of Dox and CHK1i (LY2603618). (k–m) RT-qPCR analysis for the expression of 2C-related genes in Dox-inducible Dux KO ESCs in comparison with WT ESCs upon treatment with Dox. Statistical significance compared to CNTL unless otherwise indicated. (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
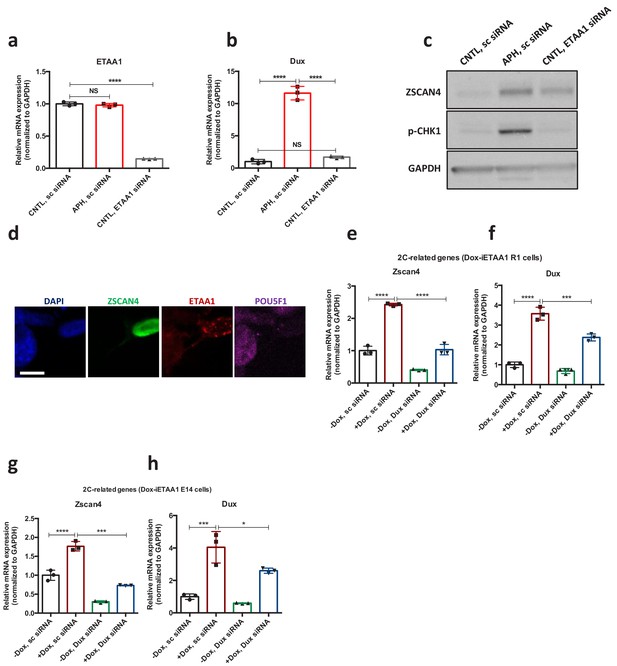
ETAA1-mediated activation of ATR induces 2C-like cells in a RS-free context.
(a,b) RT-qPCR analysis for ETAA1 and Dux genes expression in siRNA-mediated Etaa1 KD ESCs upon treatment with APH. CNTL ESCs were treated with sc siRNA. (c) Immunoblot for expression of ETAA1, ZSCAN4 and p-CHK1 in siRNA-mediated ETAA1 KD ESCs upon treatment with APH. (d) Immunostaining showing the ETAA1 expression in ZSCAN4 positive cells. (e-h) RT-qPCR analysis of 2C-related genes expression in Dox-iETAA1 ESCs (R1 and E14) upon Dox treatment and/or siRNA mediated Dux KD. For western blots quantification refer to Figure 4—source data 1.
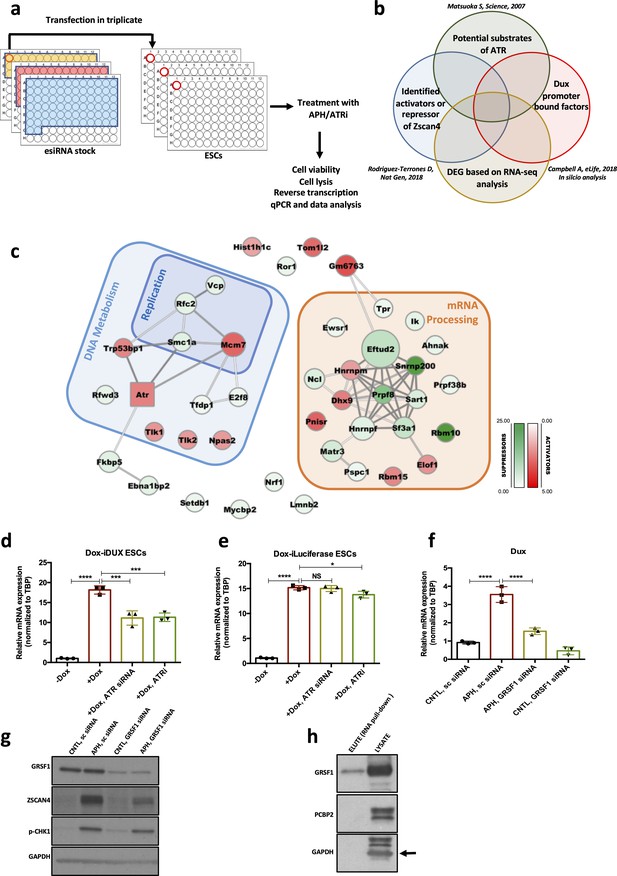
Identification of RSR downstream molecular players regulating the 2C-like state.
(a.b) Schematic design of the esiRNA-based knock-down screening and esiRNA library selection. (c) Protein interaction network for the hits identified through the esiRNA screening. Activators and suppressors are highlighted in red and green, respectively. The interactions are based on the STRING database. (d) RT-qPCR analysis of iDox-Dux ESCs for exogenous Dux mRNA upon treatment with Dox. (e) RT-qPCR analysis of iDox-Luciferase ESCs for Luciferase mRNA upon treatment with Dox. (f) RT-qPCR analysis of Dux mRNA upon APH treatment and Grsf1 KD. (g) Immunoblot showing the expression of GRSF1, ZSCAN4 and pCHK1 upon Gsrf1 KD. (h) Immunoblot showing the binding of GRSF1 protein to the Dux mRNA. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 5—source data 1.
-
Figure 5—source data 1
qPCR and Western quantification.
- https://cdn.elifesciences.org/articles/54756/elife-54756-fig5-data1-v1.xlsx
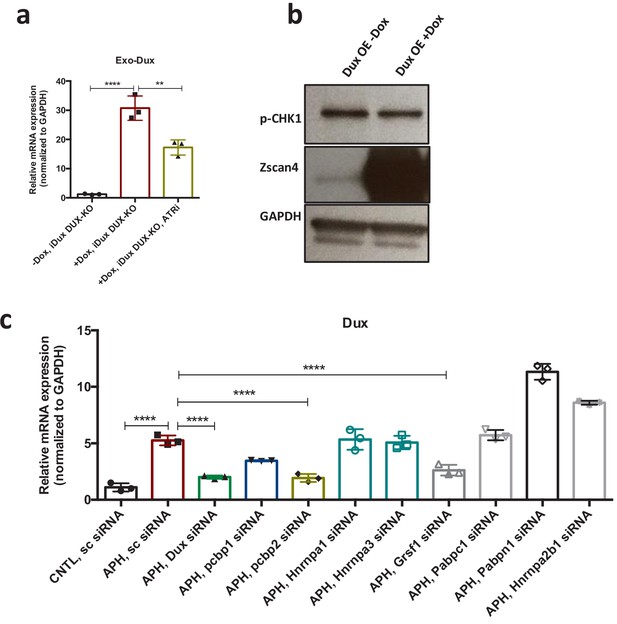
mRNA processing factors regulate the level of Dux transcript.
(a) RT-qPCR analysis for exogenous Dux mRNA in iDux-Dux KO cells upon treatment with Dox, and ATR inhibitor. (b) Immunoblot showing the level of p-CHK1 upon DUX OE. (c) RT-qPCR analysis for Dux mRNA level upon siRNA-based KD of Dux 3’UTR binding factors. Statistical significance compared to CNTL unless otherwise indicated. (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 5—source data 1.
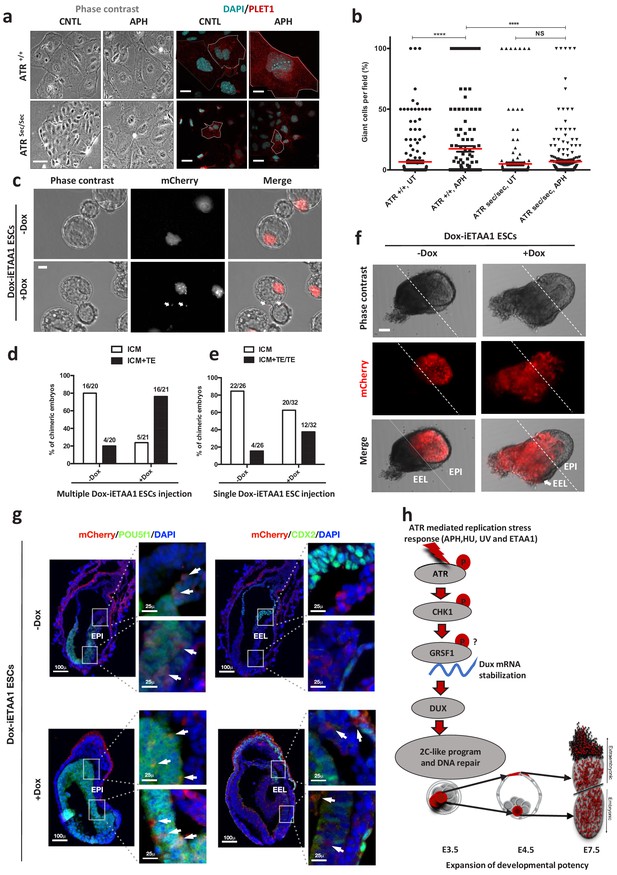
ATR-activated 2C-like cells gain expanded developmental potential in vitro and in vivo.
(a) Phase contrast and immunofluorescence images of TGCs formed by in vitro differentiation of Atr+/+ or Atrsec/sec ESCs treated with or without APH (bar = 25 µm). (b) Quantification of the number of TGCs detected in the conditions represented in a. (c) Images of blastocysts displaying the contribution of mCherry-labeled Dox-iETAA1 ESCs to the ICM and TE layers in the presence and absence of Dox (bar = 20 µm). (d) Bar plot showing the percentage of chimeric embryos in which injected mCherry-labeled Dox-iETAA1 ESCs could contribute to either ICM or ICM+TE with or without Dox. The ratios on top of each bar show the actual number of embryos that were analyzed. (e) Bar plot showing the percentage of chimeric embryos in which single-cell injection of mCherry-labeled Dox-iETAA1 ESCs could contribute to either ICM or ICM+TE/TE with or without Dox. The ratios on top of each bar show the actual number of embryos that were analyzed. (f) Images showing the contribution of injected mCherry-labeled Dox-iETAA1 ESCs to the epiblast (EPI) or extra-embryonic layers (EEL) of mouse embryos at E7.5 with or without Dox treatment (bar = 50 µm). (g) Immunostaining of mouse embryos at E7.5. Arrows in the left panel indicate the contribution of mCherry-labeled Dox-iETAA1 ESCs to the EPI (marked by POU5F1) in Dox-treated and untreated conditions. Arrows in the right panel indicate the contribution of injected mCherry-labeled Dox-iETAA1 ESCs to the EEL (marked by CDX2) only in Dox-treated condition. (lower magnification, bar = 100 µm, higher magnification, bar = 25 µm). (h) Schematic model defining a novel ATR-dependent transcriptional response to maintain the genomic integrity of developing embryos in response to RS. 1) ATR and CHK1-mediated RSR triggers the Dux mRNA accumulation through GRSF1 that in turn increases bipotent 2C-like cells by global transcriptional activation of 2C-specific genes including Zscan4. 2) ATR-induced bipotent ESCs extend their contribution to placental compartment. In the dot plot the mean is represented by a red line. Statistical significance compared to CNTL unless otherwise indicated. (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
-
Figure 6—source data 1
qPCR and TGCs quantification, and TE contribution counting.
- https://cdn.elifesciences.org/articles/54756/elife-54756-fig6-data1-v1.xlsx
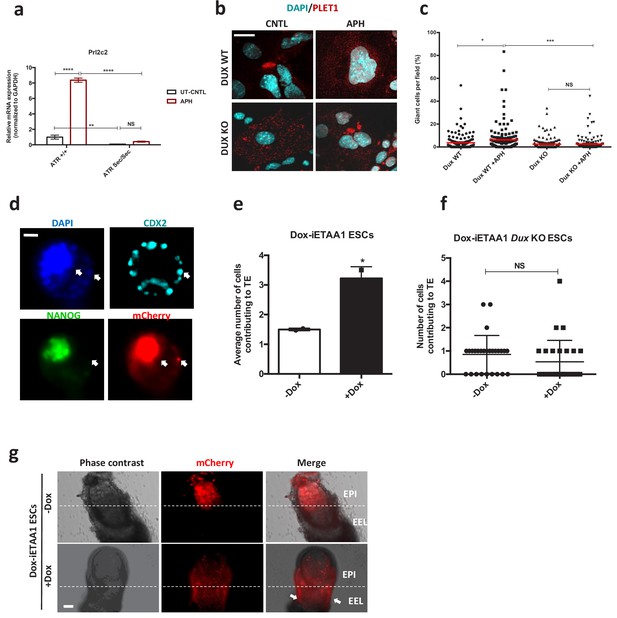
ATR-activated 2C-like cells gain expanded developmental potential.
(a) RT-qPCR assay for the TGCs specific marker Prl2c2 in Atr+/+ and AtrSec/Sec ESC-derived TGCs upon APH treatment. (b) Immunostaining of Dux KO and WT-derived TGCs upon APH treatment (bar = 25 µm). (c) Plot showing the number of TGCs generated from WT and Dux KO ESCs upon APH treatment. (d) Immunostaining of blastocysts displaying the contribution of mCherry-labeled Dox-iETAA1 ESCs to the ICM and TE layers in the presence and absence of Dox (bar = 20 µm). (e) Bar plot showing the average number of injected mCherry-labeled Dox-iETAA1 ESCs that could contribute to TE with or without Dox. (f) Beeswarm plot showing the average number of injected mCherry-labeled Dox-iETAA1 Dux KO ESCs that could contribute to TE with or without Dox. (g) Images showing the contribution of injected mCherry-labeled Dox-iETAA1 ESCs to the epiblast (EPI) or extra-embryonic layers (EEL) of mouse embryos at E7.5 with or without Dox treatment (bar = 50 µm). Statistical significance compared to CNTL unless otherwise indicated. (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA).
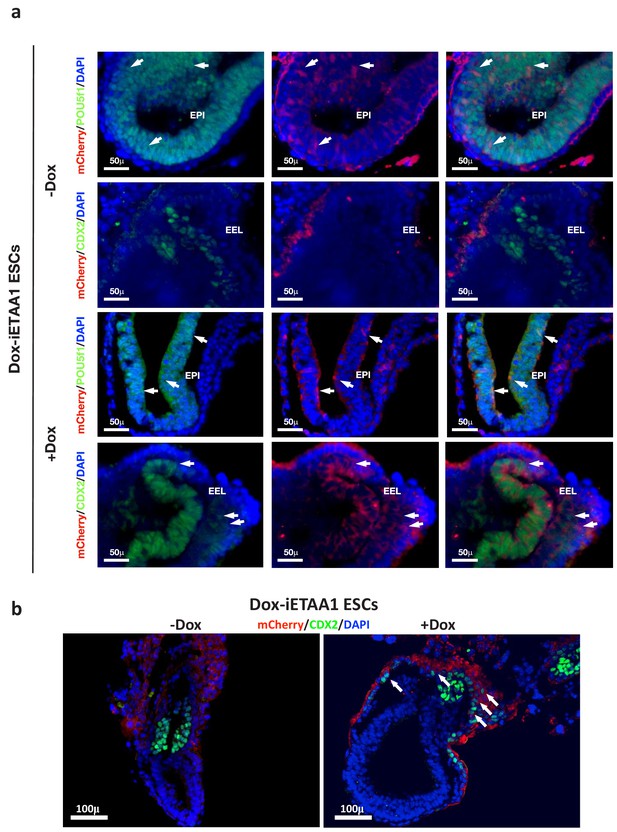
ATR-activated 2C-like cells gain expanded developmental potential.
(a, b) Immunostaining of mouse embryos at E7.5. Arrows indicate the contribution of mCherry-labeled Dox-iETAA1 ESCs to the EPI (marked by POU5F1) or EEL (marked by CDX2) in Dox-treated and untreated conditions.
Additional files
-
Supplementary file 1
The list of genes that are differentially expressed between each cluster cells and the rest of the population.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp1-v1.xlsx
-
Supplementary file 2
Single cells sequencing clusters GO analysis.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp2-v1.xlsx
-
Supplementary file 3
List of DEGs in APH induced ESCs.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp3-v1.xlsx
-
Supplementary file 4
Comparison of DEGs expressed in APH induced cells with published datasets.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp4-v1.xlsx
-
Supplementary file 5
Comparison of DE retroelements in APH induced cells.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp5-v1.xlsx
-
Supplementary file 6
List of Dux activators and supressors based on the screening experiment.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp6-v1.xlsx
-
Supplementary file 7
List of Dux RNA-bound factors identified through mass-spectrometry.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp7-v1.xlsx
-
Supplementary file 8
List of primers used in this study.
- https://cdn.elifesciences.org/articles/54756/elife-54756-supp8-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54756/elife-54756-transrepform-v1.docx





