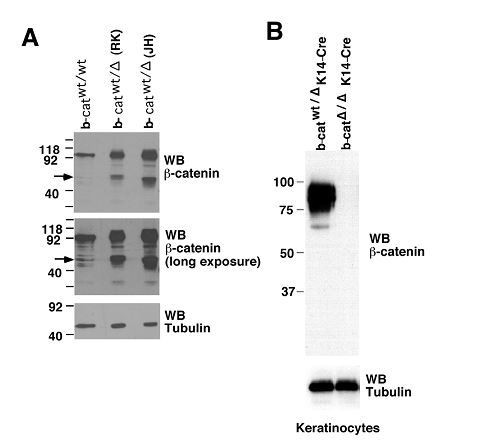
β-catenin and γ-catenin are dispensable for T lymphocytes and AML leukemic stem cells
Figures
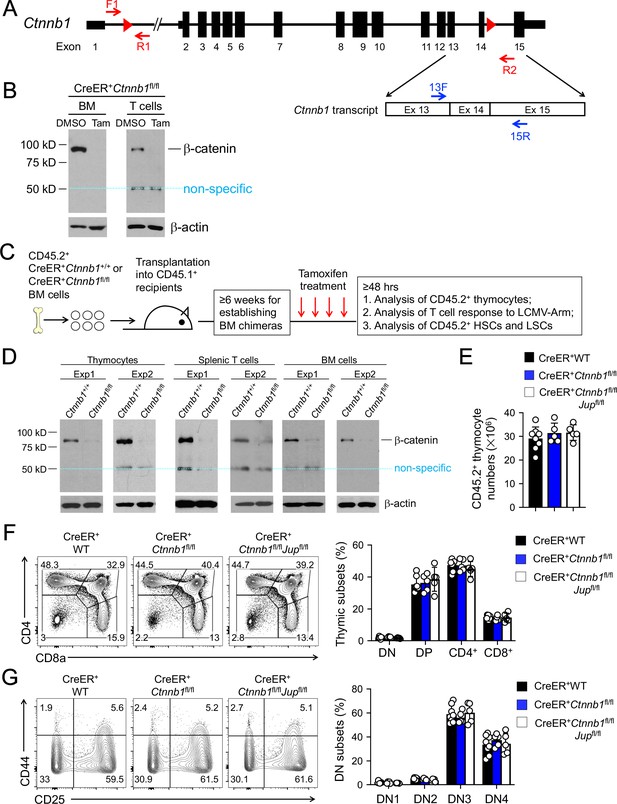
β-catenin null mutation alone or combined deletion with γ-catenin did not detectably affect thymocyte development.
(A) Targeting strategy to generate β-catenin null mutant mouse strain. All Ctnnb1 exons are shown, with red triangles denoting LoxP sites and red arrows denoting genotyping primers. Shown in the lower panel is partial Ctnnb1 transcript with blue arrows marking RT-PCR primers. (B) Ex vivo β-catenin ablation. Lineage-negative BM cells and splenic CD3+ T cells were isolated from CreER+Ctnnb1fl/fl mice and cultured with DMSO or tamoxifen followed by immunoblotting for β-catenin and β-actin with the latter as equal loading control. (C) Experimental design for generation of BM chimeras and analysis. (D) In vivo β-catenin ablation. BM chimeras reconstituted with WT or CreER+Ctnnb1fl/fl BM cells were treated with tamoxifen as in C). CD45.2+ total thymocytes, TCRβ+ splenocytes, or total BM cells were sorted and immunoblotted for β-catenin and β-actin. Data from two independent experiments are shown. In (B and D), the 50 kDa band that appeared in some blots are considered non-specific reactivity to the anti-β-catenin antibody. Refer to Figure 1—figure supplement 1 for size comparison with a truncated β-catenin protein produced from Ctnnb1 exons 2–6-targeted allele. (E) Thymic cellularity. WT, CreER+Ctnnb1fl/fl, or CreER+Ctnnb1fl/flJupfl/fl BM chimeras were treated with tamoxifen as in C), and CD45.2+ thymocytes were enumerated. (F) Detection of thymic maturation stages. CD45.2+ thymocytes were surface-stained with biotinylated lineage markers (minus CD3ε) to exclude non-T cells, and with CD4 and CD8 to identify DN, DP, CD4+ and CD8+ subsets. (G) Detection of DN subsets. CD45.2+ DN thymocytes were surface-stained with CD44 and CD25 to identify DN1 to DN4 subsets. In panels (E–G), values in representative contour plots denote percentages, and bar graphs are cumulative data of means ± s.d. from ≥3 experiments. None of the parameters was statistically significant among the groups as determined by one-way ANOVA, and thus unmarked for clarity.
-
Figure 1—source data 1
Source files, containing original data for Figure 1E,F and G, to document thymic cellularity (E), frequency of thymocyte subsets at different developmental stages (F, G).
- https://cdn.elifesciences.org/articles/55360/elife-55360-fig1-data1-v2.xlsx
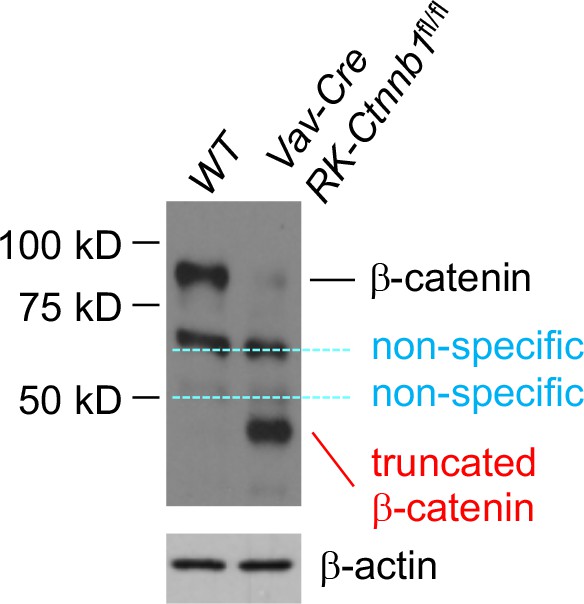
Deletion of Ctnnb1 exons 2–6 gives rise to a truncated β-catenin protein in bone marrow cells.
Ctnnb1 exons 2–6 was previously targeted by R Kelmer and colleagues (Brault VR et al, Development 128, 1253, 2001, Stock No. 004152, the Jackson Laboratory), and referred to as RK-Ctnnb1-floxed mice. Bone marrow cells from Vav-Cre+ RK-Ctnnb1fl/fl and WT mice were immunoblotted with anti-β-catenin (Clone 14, BD Transduction Laboratories). A truncated β-catenin protein of approximately 40 kDa was detected in Vav-Cre+ RK-Ctnnb1fl/fl but not WT cells. Other two bands present in both WT and Vav-Cre+ RK-Ctnnb1fl/fl cells are considered non-specific.
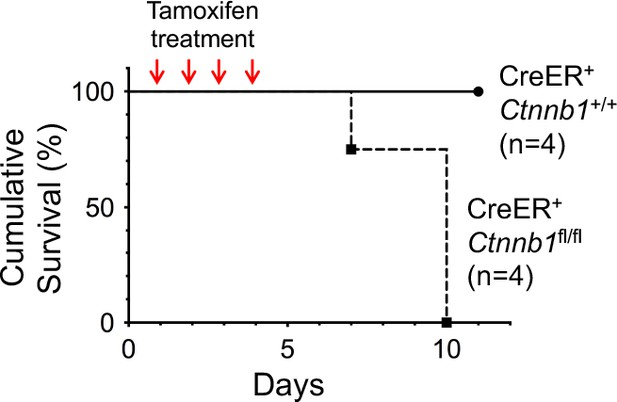
Induced deletion of β-catenin in whole body results in mouse lethality.
CreER+Ctnnb1fl/fl and CreER+Ctnnb1+/+ mice were treated with Tamoxifen on four consecutive days, and the survival curve is shown.
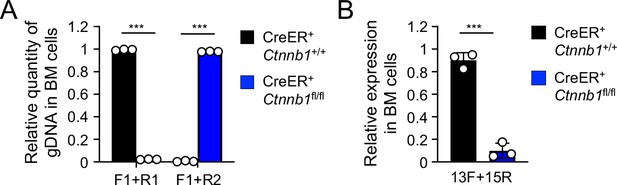
Validation of β-catenin null mutation on genomic DNA (A) and transcript (B) levels upon treatment of the BM chimera with Tamoxifen.
(A) Detection of deletion efficiency of Ctnnb1 locus on genomic DNA level. BM chimeras reconstituted with WT or CreER+Ctnnb1fl/fl BM cells were treated with tamoxifen as in Figure 1C. Genomic DNA (gDNA) was isolated from sorted CD45.2+ BM cells, and DNA segments in the Ctnnb1 locus were detected with indicated primer sets (as in Figure 1A) by quantitative PCR. Lef1 gene locus was detected to normalize quantity of genomic DNA input. (B) Detection of deletion efficiency of Ctnnb1 transcripts. Total RNA was isolated from sorted CD45.2+ BM cells, and Ctnnb1 transcript was detected with the indicated primers (as in Figure 1A) by quantitative RT-PCR. Hprt transcript was detected to normalize quantity of total RNA input. Data are means ± s.d. (n = 3 from two experiments). Similar results were obtained from sorted CD45.2+ total thymocytes and splenic T cells (not shown). ***, p<0.001 by Student’s t-test.
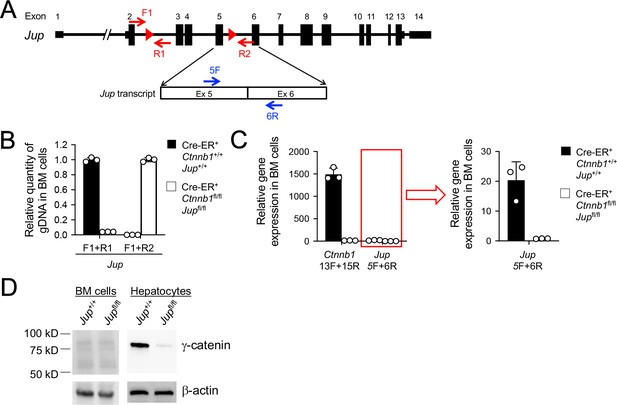
Validation of γ-catenin ablation induced by tamoxifen treatment.
(A) Diagram showing the conditionally targeted Jup allele. All Jup exons are shown, with red triangles denoting LoxP sites and red arrows denoting genotyping primers. Shown in the lower panel is partial Jup transcript with blue arrows marking RT-PCR primers. (B) Detection of deletion efficiency of the Jup locus on genomic DNA level. BM chimeras reconstituted with WT or CreER+Ctnnb1fl/flJupfl/fl BM cells were treated with tamoxifen as in Figure 1C. Genomic DNA (gDNA) was isolated from sorted CD45.2+ BM cells, and DNA segments in the Jup locus were detected with indicated primer sets in A) by quantitative PCR. Lef1 gene locus was detected to normalize quantity of genomic DNA input. (C) Detection of deletion efficiency of Ctnnb1 and Jup transcripts. Total RNA was isolated from CD45.2+ BM cells sorted as in B), and Ctnnb1 and Jup transcripts were detected with the indicated primers (Figure 1A and panel A) in this supplement) by quantitative RT-PCR. Hprt transcript was detected to normalize quantity of total RNA input. Jup detection, marked with a red box, was displayed on a smaller y-axis scale to demonstrate deletion efficiency upon tamoxifen induction. Data are means ±s.d. (n = 3 from two experiments). Similar results were obtained from sorted CD45.2+ total thymocytes (not shown). (D) In vivo γ-catenin ablation. In the left panel, CD45.2+ BM cells were sorted as in B) and immunoblotted for γ-catenin, where no γ-catenin-specific band was detected. In the right panel, WT or CreER+Ctnnb1fl/flJupfl/fl mice were treated directly with tamoxifen for four consecutive days, and on day 5, hepatocytes were immunoblotted for γ-catenin.
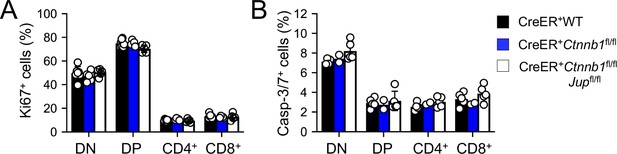
Induced deletion of β-catenin or both β- and γ-catenin did not detectably affect thymocytes proliferation (A) or apoptosis (B).
WT, CreER+Ctnnb1fl/fl, or CreER+Ctnnb1fl/flJupfl/fl BM chimeras were treated with tamoxifen as in Figure 1C. CD45.2+ thymic subsets were intracellularly stained for Ki67 to detect thymocyte proliferation (A), or stained for active Caspase-3/7 to detect thymocyte apoptosis (B). Data are means ±s.d. from ≥3 experiments. None of the parameters was statistically significant among the groups as determined by one-way ANOVA, and thus unmarked for clarity.
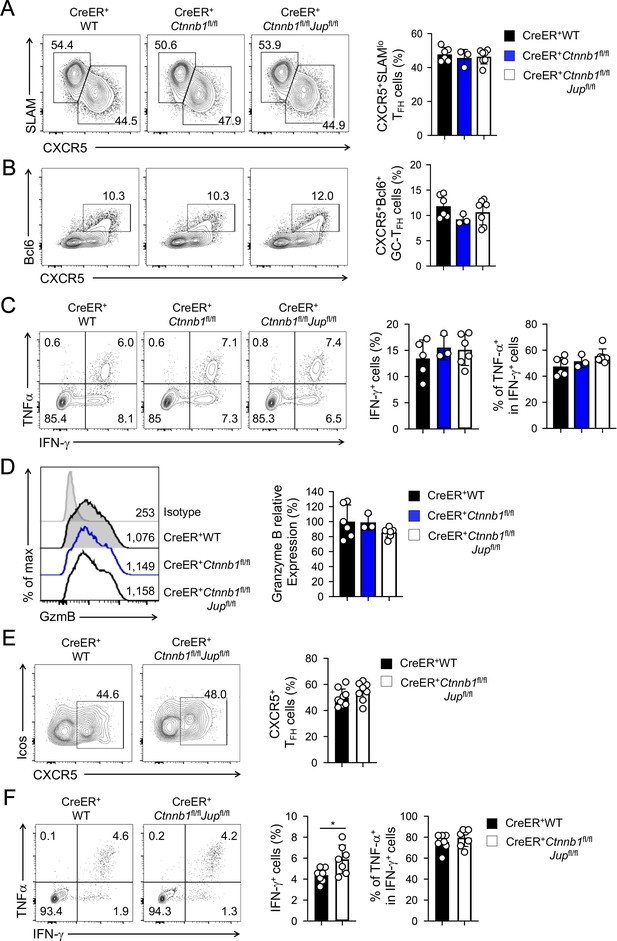
β-catenin and γ-catenin are not required for T cell responses to acute viral infection.
BM chimeras were established and treated with tamoxifen as in Figure 1C, and infected with LCMV. The infected mice were analyzed on eight dpi for effector (A–D) and ≥40 dpi for memory phase responses (E, F). (A) Detection of CXCR5+SLAMlo TFH and CXCR5–SLAMhi TH1 cells in CD45.2+ CD44hiCD62L– activated CD4+ splenocytes on eight dpi by cell surface staining. (B) Detection of CXCR5+Bcl6+ GC-TFH cells in CD45.2+ CD44hiCD62L– activated CD4+ splenocytes on eight dpi by intranuclear staining. (C) Detection of IFN-γ and/or TNF-α-producing cells in CD45.2+CD8+ splenocytes on eight dpi by intracellular staining after 5 hr incubation with GP33 peptides. (D) Detection of granzyme B expression in CD45.2+ CD11ahi activated CD8+ splenocytes on eight dpi by intracellular staining. Values in half-stacked histograms denote geometric mean fluorescence intensity (gMFI). (E) Detection of CXCR5+ memory TFH cells in CD45.2+CD44hi antigen-experienced CD4+ splenocytes on ≥40 dpi by cell surface staining. (F) Detection of IFN-γ and/or TNF-α-producing memory CD8+ T cells in CD45.2+CD8+ splenocytes on ≥40 dpi by intracellular staining after 5 hr incubation with GP33 peptides. In all panels, values in representative contour plots denote percentages, and cumulative data are means ± s.d. from two experiments. *, p<0.05 by Student’s t-test; all other unmarked parameters were not statistically significant among the groups as determined by one-way ANOVA (A–D) or Student’s t-test (E, F).
-
Figure 2—source data 1
Source files, containing original data for Figure 2A–F, to document frequency of antigen-specific Tfh cells (A, B) and CD8 T cells (C), expression of Granzyme B in CD8 T cells (D) at the effector response phase, and the frequency of antigen-specific Tfh cells (E) and CD8 T cells (F) at the memory phase.
- https://cdn.elifesciences.org/articles/55360/elife-55360-fig2-data1-v2.xlsx
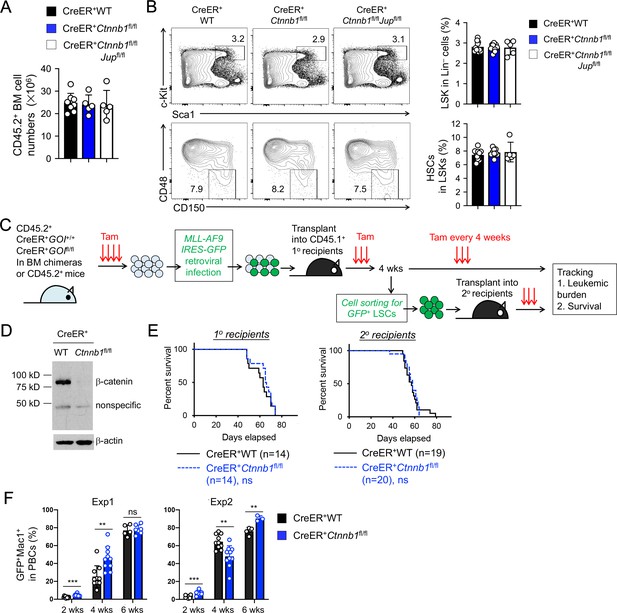
β-catenin is not essential for HSC homeostasis and AML LSC self-renewal.
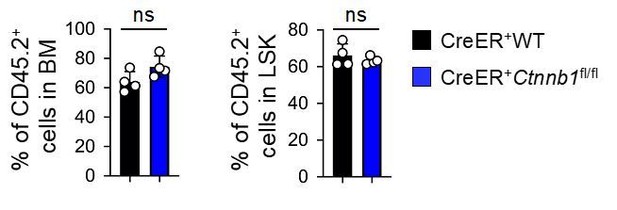
(A–B) BM chimeras were established and treated with tamoxifen as in Figure 1C, and analyzed for CD45.2+ BM cellularity (A), LSKs and HSCs by cell surface staining (B). Values in representative contour plots (B) denote percentages, and cumulative data on frequency of LSKs or HSCs are means ± s.d. from two experiments. None of the parameters was statistically significant among the groups as determined by one-way ANOVA. (C) Experimental design for modeling AML initiation and propagation in mice. After initial tamoxifen (Tam) treatments for four consecutive days, the 1o recipients and 2o recipients were subjected to recurring Tam treatment for three consecutive days at 4 week intervals to ensure long-term elimination of targeted proteins from the floxed alleles. For 1o recipients, MLL-AF9 retrovirus-infected Lin–BM cells containing 104 GFP+Lin–cKit+ cells were transplanted. For 2o recipients, 103 LSCs were sorted from 1o recipients at 4 weeks after initial transplantation and then transplanted. GOI, gene of interest. (D) Complete deletion of β-catenin in AML cells as determined by immunoblotting of sorted CD45.2+GFP+Mac1+ BM cells from 1o recipients on day 28 after BM transplantation, where WT and Ctnnb1-floxed cells were subjected to two rounds of tamoxifen treatment. (E) Kaplan-Meier survival curves of 1o and 2o recipients of WT or β-catenin-deficient LSCs. Data are pooled from two independent experiments. ns, not statistically significant as determined by log-rank test. (F) Longitudinal tracking of CD45.2+GFP+Mac1+ AML leukemic cells in PBCs of 2o recipients. For week 8, the surviving recipients were analyzed. Data from two independent experiments were displayed separately because modest differences were observed in kinetics of leukemic cell expansion at week 4. These differences did not affect recipient survival (see E). **, p<0.01; ***, p<0.001 by Student’s t-test.
-
Figure 3—source data 1
Source files, containing original data for Figure 3A,B and F, to document BM cellularity (A), and frequency of LSKs and Slam HSCs in BM cells (B), and leukemia burden in peripheral blood of AML receipt mice (F).
- https://cdn.elifesciences.org/articles/55360/elife-55360-fig3-data1-v2.xlsx
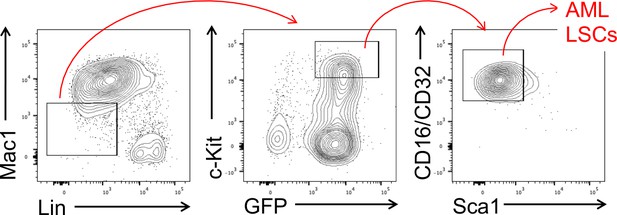
Gating strategy for sorting of AML LSCs, as defined as CD45.2+GFP+Lin–Mac1loc-Kithi Sca1loCD16/CD32hi BM cells in the 1o recipients on day 28 after BM transplantation.
Mac1 and Gr.1 were excluded from lineage markers so that Mac1 expression can be independently assessed. It is of note that Mac1hi cells have the LSC capacity to propagate AMLs upon transplantation, but with modestly prolonged latency (not shown).
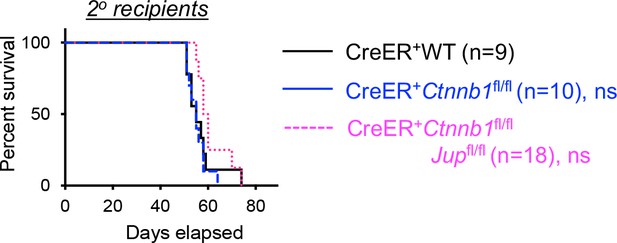
Kaplan-Meier survival curve of 2o recipients of WT, β-catenin or β/γ-catenin-deficient AML LSCs. ns, not statistically significant as determined by log-rank test.
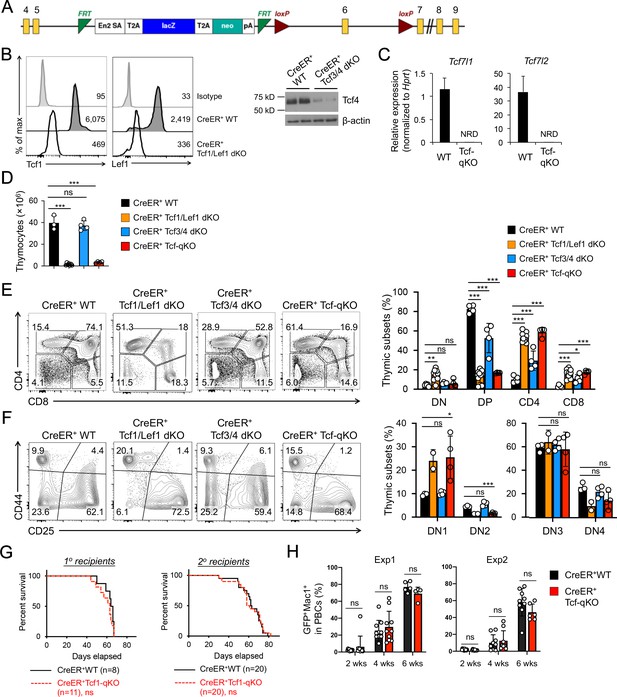
Tcf/Lef TFs are critical for T cell development but not essential for AML LSC self-renewal.
(A) Targeting strategy for Tcf7l1 gene locus. Yellow boxes denote exons, with exon numbers marked on top. Exon six is flanked with two LoxP sites (marked with red triangles). The LacZ/Neo cassette flanked by Frt Sites (cyan wedges) was removed with Flippase in germline-transmitted mice. (B) In vivo ablation of Tcf/Lef proteins. CreER+ WT, Tcf1/Lef1 dKO, and Tcf3/4 dKO mice were treated with tamoxifen for four consecutive days. Three days later, total thymocytes were intracellularly stained for Tcf1 and Lef1 in Tcf1/Lef1 dKO and control mice, and values denote geometric mean fluorescent intensity in representative half-stacked histograms (left panels). Total thymocytes were immunoblotted for Tcf4 protein in Tcf3/4 and control mice (right panel). (C) Validation of efficient deletion of targeted Tcf7l1 and Tcf7l2 exons in hematopoietic stem/progenitor cells. CreER+Tcf-qKO and WT mice were treated with tamoxifen for four consecutive days, and two days later, Flt3–LSK cells, which were enriched in both long-term and short-term HSCs, were sort-purified and analyzed by quantitative RT-PCR. Relative expression of Tcf7l1 and Tcf7l2 was determined by normalizing to Hprt, and shown as means ± s.d. (n = 5). NRD, not reliably detected. (D) Thymic cellularity. CreER+ WT, Tcf1/Lef1 dKO, Tcf3/4 dKO, and Tcf-qKO mice were treated with tamoxifen as in B), and thymocytes were enumerated. (E) Detection of thymic maturation stages. Thymocytes were surface-stained with biotinylated lineage markers (minus CD3ε) to exclude non-T cells, and with CD4 and CD8 to identify DN, DP, CD4+ and CD8+ subsets. (F) Detection of DN subsets. DN thymocytes were surface-stained with CD44 and CD25 to identify DN1 to DN4 subsets. Note that deletion of Tcf1 and Lef1, as in Tcf1/Lef1 dKO and Tcf-qKO mice, caused premature, modest upregulation of CD25 in a portion of DN1 cells, and the gate was adjusted accordingly to demarcate DN1 and DN2 subsets. Data in D–F are means ±s.d. from ≥2 experiments. Statistical significance for multiple groups was first assessed by one-way ANOVA, and that for indicated pair comparison was determined with Tukey’s correction. *, p<0.05; **, p<0.01; ***, p<0.001; ns, not statistically significant. In F), n = 2 for Tcf1/Lef1 dKO, and thus no p values are shown. (G) Kaplan-Meier survival curves of 1o and 2o recipients of WT or Tcf-qKO LSCs. Data are pooled from two independent experiments. ns, not statistically significant as determined by log-rank test. (H) Longitudinal tracking of CD45.2+GFP+Mac1+ AML leukemic cells in PBCs of 2o recipients. For week 8, the surviving recipients were analyzed. Data from two independent experiments were displayed separately because modest differences were observed in kinetics of leukemic cell expansion at week 4. These differences did not affect recipient survival (see G). ns, not statistically significant as determined by Student’s t-test.
-
Figure 4—source data 1
Source files, containing original data for Figure 4D–H, to document thymic cellularity (D), frequency of thymocyte subsets at different developmental stages (E, F), and leukemia burden in peripheral blood of AML receipt mice (G).
- https://cdn.elifesciences.org/articles/55360/elife-55360-fig4-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus. musculus) | C57BL/6J | Jackson Laboratory | RRID:IMSR_ JAX:000664 | |
| Genetic reagent (Mus. musculus) | Ctnnb1fl/fl | This paper | Ctnnb1 exons 2–14 floxed. Send reagent request to haihui.xue@hmh-cdi.org | |
| Genetic reagent (Mus. musculus) | Jup1fl/fl | PMID:22036570 | ||
| Genetic reagent (Mus. musculus) | Tcf7fl/fl | PMID:24836425 | ||
| Genetic reagent (Mus. musculus) | Lef1fl/fl | PMID:23103132 | ||
| Genetic reagent (Mus. musculus) | Tcf7l1fl/fl | This paper | Tcf7l1 Exonsix floxed. Send reagent request to haihui.xue@hmh-cdi.org | |
| Genetic reagent (Mus. musculus) | Tcf7l2fl/fl | PMID:21383188 | ||
| Strain, strain background (virus) | Lymphocytic choriomeningitis virus Armstrong strain E-350 | ATCC | VR-1271 | |
| Antibody | anti-β-catenin (Mouse monoclonal) | BD Biosciences | Cat. No. 610154 Clone 14 | IB (1:2000) |
| Antibody | anti-γ-catenin (Mouse monoclonal) | BD Biosciences | Cat. No. 610253 Clone 15 | IB (1:2000) |
| Antibody | Anti-Tcf4 (Rabbit monoclonal) | Cell Signaling Technology | Cat. No. 2565 Clone C9B9 | IB (1:1000) |
| Recombinant DNA reagent | MLL-AF9-GFP plasmid | PMID:17463288 | ||
| Chemical compound, drug | 4-hydroxy-tamoxifen | Millipore-Sigma | Cat. No. T176 | |
| Chemical compound, drug | Tamoxifen | Millipore-Sigma | Cat. No. T5648 | |
| Commercial assay or kit | Vybrant FAM caspase-3/7 assay kit | Invitrogen | Cat. No. V35118 | |
| Commercial assay or kit | QuantiTect Reverse Transcription Kit | Qiagen | Cat. No. 205313 | |
| Software, algorithm | FlowJo | https://www.flowjo.com | RRID:SCR_008520 | |
| Software, algorithm | GraphPad Prism | http://www.graphpad.com/ | SCR_015807 |